Abstract
Lymphoid organs are characterized by a complex network of phenotypically distinct dendritic cells (DC) with potentially unique roles in pathogen recognition and immunostimulation. Classical DC (cDC) include two major subsets distinguished in the mouse by the expression of CD8α. Here we describe a subset of CD8α+ DC in lymphoid organs of naïve mice characterized by expression of the CX3CR1 chemokine receptor. CX3CR1+ CD8α+ DC lack hallmarks of classical CD8α+ DC, including IL-12 secretion, the capacity to cross-present antigen, and their developmental dependence on the transcriptional factor BatF3. Gene-expression profiling showed that CX3CR1+ CD8α+ DC resemble CD8α− cDC. The microarray analysis further revealed a unique plasmacytoid DC (PDC) gene signature of CX3CR1+ CD8α+ DC. A PDC relationship of the cells is supported further by the fact that they harbor characteristic D–J Ig gene rearrangements and that development of CX3CR1+ CD8α+ DC requires E2-2, the critical transcriptional regulator of PDC. Thus, CX3CR1+ CD8α+ DC represent a unique DC subset, related to but distinct from PDC. Collectively, the expression-profiling data of this study refine the resolution of previous DC definitions, sharpen the border of classical CD8α+ and CD8α− DC, and should assist the identification of human counterparts of murine DC subsets.
Classic splenic dendritic cells (cDC), also termed “conventional DC,” are a subpopulation of mononuclear phagocytes defined in the mouse by high expression of the β integrin CD11c, migratory capacity, and an unrivalled ability to stimulate naïve T cells (1, 2). Beyond phenotypic and functional definitions, recent studies indicate that the short-lived CD11chi cDC are derived from dedicated nonmonocytic bone marrow-derived precursors termed “precDC” (3). Splenic cDC display considerable phenotypic heterogeneity, and their subsets are believed to have distinct functions in pathogen recognition and immunostimulation (4). Splenic CD11b+ cDC, which can be subdivided further into CD4+ and CD8/CD4 double-negative (DN) cDC, efficiently form peptide–MHC class II complexes (5). CD11b+ cDC secrete IL-10 and have been shown to induce T-helper cell type 2 CD4 responses preferentially (6). Development and/or maintenance of CD11b+ cDC require the transcription factors RelB (7), interferon regulatory factor 4 (IRF4) (8, 9), and RBP-J (10).
The second main murine cDC subset is characterized by the expression of CD8α homodimers and the C-type lectin CD205 (4). In vivo, CD8α+ cDC preferentially endocytose dying cells (11) and are considered specialized to cross-present engulfed cellular antigens in the context of MHC class I to CD8+ T cells (12). CD8α+ cDC have a predetermined capacity to secrete IL-12 (p70) and consequently stimulate T-helper type 1 CD4+ T-cell responses (13, 14). CD8α+ cDC also were reported to promote the development of T regulatory cells via production of TGF-β (15). Generation of CD8α+ cDC requires the transcription factors IRF8/ICSBP (16, 17) and Id2 (18, 19) and is specifically controlled by the transcription factor BatF3 (20).
In addition to cDC, lymphoid organs also harbor plasmacytoid DC (PDC) which are specialized in type I IFN secretion in response to viral challenge (21). PDC share a common developmental origin with cDC, although they branch off before the precDC (3, 22, 23), develop locally in the bone marrow, and are relatively long lived in the periphery (24, 25). PDC display a number of lymphocytic features, including the presence of Ig D–J rearrangements as the result of RAG protein expression during their development (26). The generation of PDC is controlled specifically by the transcriptional regulator E2-2 (27).
Here we report the characterization of a murine CD8α+ DC subset that is marked by high-level expression of the chemokine receptor CX3CR1 and low expression of the costimulator CD86. CX3CR1+ CD8α+ DC lacked hallmark features of classical CD8α+ DC, including the ability to produce IL-12, to cross-present antigen, and BatF3 dependence. Instead, their gene-expression profile, the presence of IgH gene rearrangements, and dependence on E2-2 define CX3CR1+ CD8α+ DC as a steady-state DC population related to but distinct from plasmacytoid DC.
Results
Identification of the CX3CR1+ CD8α+ DC Subset.
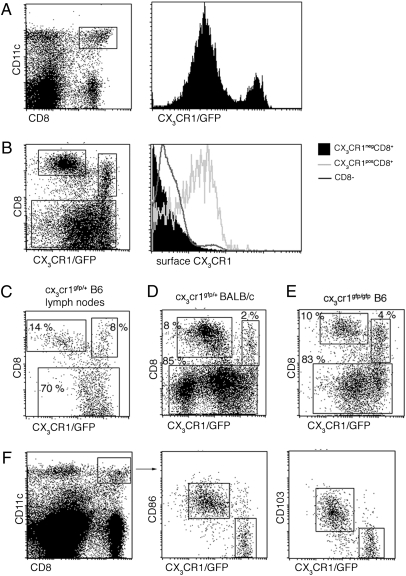
Flow cytometric analysis of spleen cells of Cx3cr gfp mice—a transgenic mouse strain in which the gene encoding the CX3CR1 chemokine receptor was replaced by an EGFP reporter gene (28)—allows the subdivision of splenic CD8α+ DC into CX3CR1/GFP− and CX3CR1/GFP+ cells, respectively (Fig. 1A). Staining with a CX3CL1-Fc fusion protein confirmed the expression of CX3CR1 on the GFP+ DC but not on the GFP− CD8α+ DC in Cx3crgfp/+ mice (Fig. 1B). CX3CR1+ CD8α+ DC generally comprised 15–30% of splenic CD8α+ DC in naïve adult C57BL/6 mice but reached 50% depending on the genetic background and housing facility. CX3CR1− and CX3CR1+ CD8α+ DC populations also could be detected in lymph nodes of Cx3cr gfp C57BL/6 mice (Fig. 1C) and Cx3cr1gfp BALB/c mice (Fig. 1D). Moreover CX3CR1+ CD8α+ DC were present in CX3CR1-deficient Cx3cr1 gfp/gfp mice, indicating that the CX3CR1 chemokine receptor is dispensable for their generation (Fig. 1E). In WT mice that lack the CX3CR1/GFP label, the CX3CR1+ subset of CD8α+ DC can be identified by low side scatter, low levels of the costimulatory molecule CD86, and the lack of the integrin CD103 (Fig. 1F).
Fig. 1.
Identification of the CX3CR1+ CD8α+ DC subset. (A) Flow cytometric analysis of splenocytes from Cx3cr1gfp/+ mice. cDC were identified as CD11chi cells. Histogram shows GFP expression in CD11chi CD8α+ DC. (B) Staining of splenic cDC subpopulations for surface expression of CX3CR1 using a CX3CL1-Fc fusion protein. (C) Flow cytometric analysis of lymph node cells from Cx3cr1gfp/+ mouse. (D) Flow cytometric analysis of splenocytes from Cx3cr1gfp/+ BALB/c mouse. (E) Flow cytometric analysis of splenocytes from CX3CR1-deficient Cx3cr1gfp/gfp mice. (F) Flow cytometric analysis of splenocytes from Cx3cr1gfp/+ mouse. Note characterization of CX3CR1/GFP+ CD8α+ DC as a CD86lo CD103neg population.
CX3CR1+ CD8α+ DC Lack Hallmark Features of Classical CD8α+ DC.
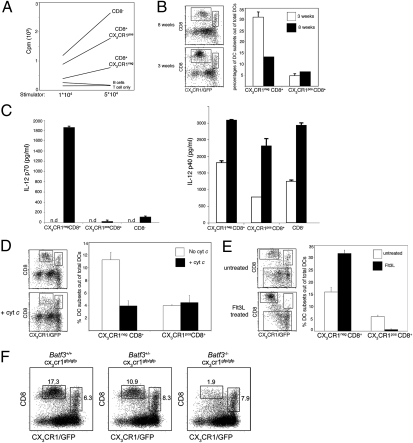
We first investigated whether CX3CR1+ and CX3CR1− CD8α+ DC are distinct entities. CX3CR1+ CD8α+ expressed high levels of MHC class II and could stimulate naïve alloreactive CD4+ T cells efficiently, establishing that they are bona fide DC (2) (Fig. 2A). Classical CD8α+ DC have been reported to be overrepresented in spleens of young mice (29). Interestingly, this age-related skewing of the cDC subset balance was restricted to the CX3CR1− CD8α+ cDC; CX3CR1+ CD8α+ DC were found at equal frequencies in young and old mice (Fig. 2B).
Fig. 2.
CX3CR1+ CD8α+ DC lack hallmarks of classical CD8α+ DC. (A) Mixed leukocyte reaction with indicated numbers of sorted splenic DC subsets isolated from Cx3cr1gfp/+ C57BL/6 mice and BALB/c CD4+ T cells (105). Data are representative of two experiments. (B) Flow cytometric analysis of splenic DC subsets in 3- and 8-wk-old Cx3cr1gfp/+ C57BL/6 mice. Bars represent the percentages of CX3CR1− CD8α+ and CX3CR1+ CD8α+ subsets out of total cDC (CD11chi cells). Data are representative of two experiments. (C) Analysis of IL-12p70 (Left) and IL-12p40 (Right) secretion by sorted splenic DC subsets in response to in vitro exposure to CpG. Data are representative of two experiments.(D) Selective deletion of CX3CR1− CD8α+ splenic DC by CytC injection. Bars represent the percentages of CX3CR1− CD8α+ and CX3CR1+ CD8α+ subsets out of total DC (CD11chi cells). Data are representative of two experiments. (E) Flow cytometric analysis of splenic DC subsets of Cx3cr1gfp/+ C57BL/6 mice bearing a WT tumor or a tumor secreting FLt3L. Bars represent the percentages of CX3CR1− CD8α+ and CX3CR1+ CD8α+ subsets out of total cDC (CD11chi cells) (n = 3). Data are representative of two experiments. (F) Flow cytometric analysis of splenic DC from Batf3+/+Cx3cr1gfp/gfp, Batf3+/−Cx3cr1gfp/gfp, and Batf3−/−Cx3cr1gfp/gfp mice. cDC were gated as CD11chi B220− cells.
A prominent functional feature of classical CD8α+ DC is IL-12 production in response to microbial challenge (13, 14). We previously reported that after mice were injected with Toxoplasma gondii tachyzoite extract only CX3CR1− CD8α+ DC, but not CX3CR1+ CD8α+ DC, produce IL-12 (28). This difference could result from a general inability of the latter cells to produce IL-12 or their lack of the specific Toxoplasma sensor TLR11 (30). To address this issue, we sorted CX3CR1+ CD8α+, CX3CR1− CD8α+, and CD8α− cDC and exposed them in vitro to a Toll-like receptor 9 (TLR9) agonist. As seen in Fig. 2C, C–phosphate–G (CpG) stimulation in the presence of accessory cytokines induced IL-12 production as measured by secretion of the p70 subunit specifically by classical CD8α+ but not CX3CR1+ CD8α+ or CD8α− cDC. Interestingly, however, the TLR9 stimulus boosted production of the p40 subunit (indicative of IL-23) by all the populations tested, confirming the TLR9 responsiveness of the cells (Fig. 2C). This result establishes that, like cDC, CX3CR1+ CD8α+ DC respond to TLR9 engagement but, unlike classical CD8α+ DC, do not produce IL-12.
The main functional hallmark of classical CD8α+ DC is their unique capability to channel exogenous antigens into the MHC class I presentation pathway for cross-presentation (12). Lew and colleagues recently established an elegant method that allows the identification of cross-presenting cells in the in vivo context based on their unique sensitivity to extracellular cytochrome c (CytC) (31). However, in this study only a fraction of splenic CD8α+ DC was depleted; the remainder was unable to cross-present and therefore was deemed functionally impaired, as indicated by impaired IL-12 production (31). To explore the possibility that the CytC-resistant CD8α+ DC were, in fact, CX3CR1+ CD8α+ DC, we injected Cx3cr gfp/+ mice with CytC. Flow cytometric analysis of the CytC-challenged mice revealed that only CX3CR1− CD8α+ cDC were ablated by this procedure, whereas the percentage of CX3CR1+ CD8α+ DC remained unchanged (Fig. 2D). This result suggests that CX3CR1+ CD8α+ DC lack characteristic cytosolic export mechanisms assigned to classical CD8α+ DC that allow them to cross-present antigens.
Classical CD8α+ DC are expanded preferentially upon systemic exposure to the growth factor Fms-related tyrosine kinase 3 ligand (Flt3L) (32). Interestingly, however, CX3CR1+ CD8α+ DC were significantly underrepresented among CD11chi cells of mice bearing an Flt3L-secreting tumor (Fig. 2E). This finding suggests that CX3CR1+ CD8α+ DC arise from developmental pathways distinct from those of classical CD8α+ DC. Development of the latter has been shown to require the basic leucine zipper transcription factor BatF3 (20). However, closer examination revealed that Batf3−/− mice retain a sizable population of CD8α+ DC. Moreover, subsequent flow cytometric analysis of Cx3cr1gfp Batf3−/− mice showed that these residual CD8α+ DC almost uniformly expressed CX3CR1 (Fig. 2F). Together with the results of the Flt3L exposure, this result indicates that developmental requirements of CX3CR1+ CD8α+ DC are distinct from those of classical CD8α+ DC. Collectively these findings establish that CD11chi CX3CR1+ CD8α+ cells are DC but lack hallmark features assigned to classical CD8α+ DC, such as IL-12 production, the ability to cross-present, and developmental BatF3 dependence.
Gene Expression Profiling of Splenic DC Subsets.
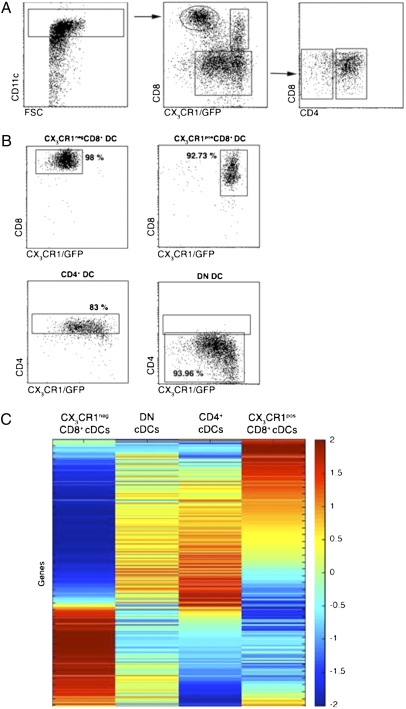
To compare the expression profiles of CX3CR1+ and CX3CR1 CD8α+ DC and to study their relationship to the CD8α− cDC subsets, we next isolated CD11chi DC from spleens of heterozygous mutant Cx3cr1gfp/+ C57BL/6 mice and sorted the two CD8α+ DC populations, as well as the CD4+ and DN cDC, to purity (Fig. 3 A and B). RNA was extracted from the four samples and subjected to gene-expression profiling using Mouse Genome 430.2 Affymetrix GeneChip arrays. Notably, all samples expressed equal levels of mRNAs encoding CD11c, MHC class II (I-Ab), Flt3, and CD40 (Table S1) but lacked expression of B- and T-cell markers such as CD19 and CD90. Moreover CX3CR1, CD4, and CD8α mRNAs were all expressed by the appropriate populations; the expression of CD8α mRNA establishes that the CD8α molecules on the CX3CR1+ population result from cell-intrinsic expression and not from passive acquisition from CD8+ T cells (33). Microarray results of selected genes were validated by RT-PCR analysis (Fig. S1).
Fig. 3.
Sorting strategy and gene-expression matrix of splenic cDC subsets. (A) Flow cytometric analysis of magnetic bead-enriched CD11chi cells isolated from Cx3cr1gfp/+ mice indicating sorting gates. (B) Analysis of sorted DC subsets. Percentages indicate purity of the respective cDC populations. (C) Expression matrix of the 500 modulated genes. Rows represent individual genes, and columns represent splenic cDC subsets. Colors indicate the relative expression levels of the genes in the different subsets, according to the code shown on the right.
Four-way comparison of all splenic DC subsets revealed only a few genes preferentially expressed in both CX3CR1+ and CX3CR1− CD8α+ DC as compared with CD8α− DC, including CD8α, CD24, and Sca1 (Table S2). Interestingly, however, expression of many hallmark proteins of classical CD8α+ DC, such as CD103, CD205, TLR3, XCR1, IL-15, and IL-12 (34), was absent from CX3CR1+ CD8α+ DC (Table S3), supporting the notion that CX3CR1+ CD8α+ DC are distinct entities. Rather, the microarray expression profile of CX3CR1+ CD8α+ DC revealed a significant overlap with the profile of both CD4+ and DN cDC (Fig. 3C). Thus, molecules that have been associated with the DN cDC, including Sirpβ, Notch3, TLR5, TLR7, DCIR2, and CD209 (34), were all expressed at >3-fold higher levels in CX3CR1+ than in CX3CR1− CD8α+ DC (Table S4). Notably, the expression of the transcription factors IRF4 and RelB, which are critical for the generation of CD4+ cDC but reportedly are dispensable for CD8α+ cDC production (8, 9), was also elevated (Table S4). Conversely, IRF8/ICSBP and Id2, reported to be essential for CD8α+ cDC development (16, 18, 19, 17), were prominently expressed in CX3CR1− CD8α+ cDC but were absent from CX3CR1+ CD8α+ DC (Table S3). Collectively, these results establish that, according to their gene-expression profile, CX3CR1+ CD8α+ DC are more closely related to CD4+ and DN cDC than to CD8α+ cDC.
CX3CR1+ CD8α+ DC Share Gene Expression and Somatic Rearrangements with Plasmacytoid Dendritic Cells.
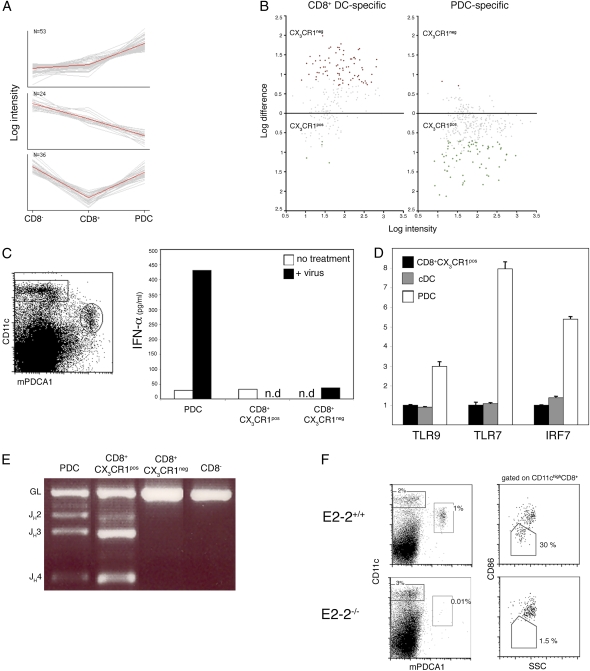
We next used the microarray data to define gene sets overexpressed in each population. These high-confidence sets then were compared with published expression profiles of DC populations (35). Principal component analysis (PCA) of the genes overexpressed specifically in the CD8+ CX3CR1+ population revealed a striking enrichment for PDC-specific genes (Fig. 4A and Table S5), including the surface markers Klra17/Ly-49Q, SiglecH, Bst2/mPDCA-1, and Ly-6C, as well as the transcription factors Tcf4/E2-2 and SpiB. Thus, PDC-specific genes were highly enriched in the CX3CR1+ subset, whereas classical CD8+ DC-specific genes were enriched only in the CX3CR1− subset (Fig. 4B). In further support of a potential link between PDC and CX3CR1+ CD8α+ DC, we identified a significant list of genes that were silent in these two populations but were expressed in both CD8α+ and CD8α− cDC (Table S6).
Fig. 4.
CX3CR1+ CD8α+ DC share expression profile, somatic Ig gene rearrangements, and E2-2 dependence with PDC. (A) PCA of the genes overexpressed in CX3CR1+ CD8α+ cells compared with CD8α+ cDC. Shown are the three principal component sets of genes analyzed against the expression data of Robbins et al. (35). Note the preferential expression in PDC (Top), in CD8α− DC (Middle), and in both PDC and CD8α− DC (Bottom). (B) Distribution of genes specific for CD8α+ DC or PDC in the two CD8α+ DC subsets. Shown is pairwise comparison of average probe intensities in CX3CR1+ and CX3CR1− CD8α+ DC; genes specific for CD8α+ DC or PDC were selected based on reference 35. (C) IFN-α production by sorted splenic DC subsets after incubation for 20 h in the presence or absence of 400 HAU/mL influenza virus. (D) Expression of the TLR/IFN signaling genes in sorted CD8α− cDC, PDC, and CX3CR1+ CD8α+ DC as determined by quantitative RT-PCR (mean ± SD of triplicate reactions). Expression levels are normalized relative to the CX3CR1+CD8α+ subset. (E) D–J rearrangement of IgH gene in sorted DC subsets. IgH D–J rearrangements were detected by genomic PCR using primer sets for the DHQ52 element. Data are representative of three experiments. (F) Absence of CX3CR1+ CD8α+ DC and PDC in absence of the transcription factor E2-2. (Left) Gated donor-derived (CD45.2+) splenocytes from E2-2−/− chimeras or control E2-2+/+ chimeras mice were analyzed for the presence of CD11cint Bst2+ PDC. (Right) Gated CD11chi CD8α+ DC comprise the CD86lo SSClo and the CD86hi SSChi subsets (corresponding to CX3CR1+ and CX3CR1− populations, respectively). Data are representative of three independent experiments.
A prime function of PDC is their production of type I interferons in antiviral responses (36). To probe for a functional overlap of CX3CR1+ CD8α+ DC and PDC, we tested their response to viral challenge. As seen in Fig. 4C, only PDC, but not CX3CR1+ CD8α+ DC, produced type I IFN upon in vitro exposure to influenza virus. In accordance with this finding, quantitative PCR analysis revealed that CX3CR1+ CD8α+ DC express lower levels of TLR9, TLR7, and IRF-7 than do PDC (Fig. 4D). Moreover, CX3CR1+ CD8α+ DC lacked surface expression of classical PDC markers such as B220 and mPDCA (Fig. S2). These findings establish that CX3CR1+ CD8α+ DC are phenotypically and functionally distinct from PDC.
As a result of the activation of a unique “lymphoid” gene-expression program, PDC, unlike cDC, harbor Ig heavy-chain (IgH) D–J rearrangements that can serve as distinctive and stable genetic lineage markers (37, 26). To probe for a developmental connection between CX3CR1+ CD8α+ DC and PDC, we investigated whether these two populations share this genetic feature by probing the rearrangement status of their IgH loci using a genomic PCR assay. As shown in Fig. 4E, D–J rearrangements were detected readily in both PDC and CX3CR1+ CD8α+ DC, whereas classical CD8α+ DC and CD8α− DC harbored only germline IgH alleles. This finding suggests that CX3CR1+ CD8α+ DC are developmentally related to PDC.
We next decided to probe for potential developmental connections between the two cell populations. Because the transcription factor E2-2 is required for PDC generation (27), we tested whether CX3CR1+ CD8α+ DC develop in the absence of E2-2. We found both PDC and CX3CR1+ CD8α+ DC were absent in chimeras established from E2-2–deficient fetal livers (38) (Fig. 4F). We also examined a conditional strain in which E2-2 was deleted using a DC/PDC-specific CD11c-Cre transgene (10, 27). In these animals, some PDC develop in the bone marrow but disappear from the spleen because of spontaneous differentiation. In these animals, splenic PDC were absent, but SSClow CD86lo CD8α+ DC were present at the same frequencies as in the control animals (Fig. S3). These data suggest that CX3CR1+ CD8α+ DC may be generated independently of mature PDC, possibly from a common bone marrow progenitor.
Collectively, these results show that CX3CR1+ CD8α+ DC share with PDC a significant expression signature, the presence of unique Ig rearrangements, and the dependence on the transcription factor E2-2. However, they are functionally distinct from PDC in that they are not specialized in IFN-α production.
Discussion
Here we report the identification and characterization of a CX3CR1+ CD8α+ DC subpopulation that coexists with cDC in lymphoid tissues of naïve mice and can comprise a major fraction of splenic and lymph node CD8α+ DC. CX3CR1+ CD8α+ DC lacked hallmark features assigned to classical CD8α+ DC, such as the ability to produce IL-12 and cross-present, as well as the developmental dependence on the transcription factor BatF3. Moreover, their gene-expression profile also confirmed that CX3CR1+ CD8α+ DC are distinct from CD8α+ cDC but rather resemble CD8α− cDC. Comparative microarray analysis and the presence of characteristic Ig gene rearrangements established that CX3CR1+ CD8α+ DC are ontologically related to PDC. This notion is supported further by the fact that development of both PDC and CX3CR1+ CD8α+ DC depends on the transcription factor E2-2. However, conditional ablation of E2-2 using CD11c-Cre revealed that, in contrast to PDC, the maintenance of CX3CR1+ CD8α+ DC does not require continuous E2-2 expression. Analysis of these conditional E2-2–deficient animals furthermore established that CX3CR1+ CD8α+ DC can emerge without an obligatory PDC intermediate and that their numbers remain unaffected by the lack of splenic PDC. Notably, PDC have been reported to give rise to cells with phenotypic and functional cDC features upon microbial challenge in vitro and in vivo (25, 39, 40). Specifically, these PDC-derived DC were shown to express CD8α, but, unlike classical CD8α+ DC, to lack CD205 expression (25). However, our data argue that steady-state CX3CR1+ CD8α+ DC arise from Rag-expressing immature bone marrow precursors shared with PDC, possibly the CD11c− Ly-6C+ B220− subset (27) (Fig. S4). Supporting this notion, Irf8−/−-deficient mice that lack both classical CD8α+ cDC and PDC (16, 8, 41) were reported to retain a residual population of CD8α+ DC. Moreover, these cells share striking similarity with the CX3CR1+ CD8α+ DC reported in this study, including low-level expression of CD86, absence of TLR3, and the inability to produce IL-12 (8). Together with our results of the constitutive and conditional E2-2 animals, this finding strongly suggests that CX3CR1+ CD8α+ DC share developmental pathways with PDC but develop in absence of the latter.
Importantly, our analysis revealed that the expression of a number of genes previously assigned to classical CD8α+ DC (34) are, in fact, restricted to the CX3CR1+ CD8α+ subset. By removing contaminating PDC-related CX3CR1+ CD8α+ DC from the CX3CR1− CD8α+ cDC population, our study thus significantly refines the resolution of previous expression profile-based DC definitions (5, 34) and sharpens the border between classical CD8α+ and CD8α− DC. Our findings should assist the identification of human correlates of murine DC subsets as exemplified by the recent reports on the human equivalent of classical mouse CD8α+ DC (42). Taken together, we identified and molecularly defined an additional steady-state DC subset in the mouse that is developmentally related to PDC but is functionally distinct. Future studies should reveal potential unique roles of CX3CR1+ CD8α+ DC in pathogen recognition and immunostimulation.
Materials and Methods
Mice.
The following mice were used in this study: WT C57BL/6 mice; heterozygous and homozygous mutant Cx3cr1gfp C57BL/6 mice, Cx3cr1gfp BALB/c mice (28), and E2-2+/− (38) and Batf3−/− mice (20) crossed with Cx3cr1gfp mice. Fetal liver chimeras were established as described (10, 27) by intercrossing E2-2+/− parents, one of which was Cx3cr1gfp/WT. All animals were maintained under specific pathogen-free conditions and handled according to protocols approved by each of the investigator's institutions in accordance with international guidelines.
Microarrays.
After collagenase D digestion, spleens from Cx3cr1gfp/+ C57BL/6 mice were enriched for CD11c+ cells by magnetic separation according to the manufacturer's protocol (Miltenyi Biotec GmbH). Splenic CD11chi cells were isolated using the FACS ARIA high-speed sorter (Becton-Dickinson). Total RNA was extracted and subjected to gene-expression profiling using the Mouse Genome 430.2 Affymetrix GeneChip. We then applied the Sorting Points into Neighborhoods (SPIN) algorithm, an unsupervised analysis tool for organization and visualization of the data. For pairwise comparison of the two CD8α+ DC subsets, microarray data from two independently sorted samples were combined and analyzed using NIA Array software (43).
Flow Cytometric Analysis.
Staining reagents used in this study included the phycoerythrin-coupled antibodies anti-MHC II, CD8, CD24, CD11c, CD103, and mPDCA1, the biotinylated antibodies anti-Ly6c, CD4, and CD8, the Allophycocyanin (APC)-coupled antibodies anti-CD11c, CD4, and CD8, the APC-Alexa750–coupled antibody anti-B220, and peridinin chlorophyll protein complex (PerCP)-coupled streptavidin. Unless indicated otherwise, the reagents were obtained from eBioscience, Biolegend, BD Biosciences, and Caltag. For CX3CR1 surface staining, cells were incubated with an CX3CL1-Fc fusion peptide (NTN-Fc; kindly provided by Millennium Biotherapeutics) and subsequently stained with Cy5-conjugated F(ab)2 goat anti-human Ig G1 (IgG1; Jackson ImmunoResearch Laboratories). The cells were analyzed on a FACSCalibur cytometer (Becton-Dickinson) using CellQuest software (Becton-Dickinson).
Mixed Leukocyte Reactions.
We cultured 104 and 5 × 104 splenic DC (C57BL/6) with 105 responder CD4+ T cells (BALB/c). Cultures were pulsed after 72 h with 1 μCi of [H3] thymidine, and incorporation was measured 16 h later.
In Vivo Exposure to FLT3L.
To test the impact of Flt3L exposure on the distribution of CD8α+ DC subsets, Cx3cr1gfp/+ C57BL/6 mice were inoculated with B16 tumor cells (3 × 106) that had been manipulated to overexpress Flt3L (44).
In Vitro Cytokine Secretion Assay.
We cultured 5 × 104 sorted DC subsets in 200 uL RPMI (Gibco/BRL) with CpG (5 ug/mL) for 24 h. Supernatant IL-12 levels were measured using IL-12 p40/p70 ELISA Abs (554476; BD Pharmingen) or the IL-12 p70 ELISA kit (88–7121-22; eBioscience). For IL-12 p70 production, the stimulation consisted of the cytokines IL-4 (200 ng/mL), GM-CSF (40 ng/mL), and IFN-γ (20 ng/mL).
Ablation of Cross-Presenting Cells.
To ablate cross-presenting cells, mice were injected i.v. with 7.5 mg equine CytC dissolved in PBS (C2506; Sigma) (31). Animals were analyzed 24 h after the CytC injection.
Diagnostic Genomic PCR for Ig Loci Rearrangements.
Genomic DNA was extracted from 2 × 105 sorted cells by incubation with proteinase K at 55 °C for 2 h followed by heat inactivation for15 min at 95 °C. PCR primers specific for the DHQ52 element were used to amplify rearranged IgH D–J as previously described (45).
Type I IFN Assay.
Sorted cells were cultured at 2 × 106 cells/mL in complete RPMI 1640 for 20 h, with 400 hemagglutinin (HA) units/mL influenza virus A/Texas/1/77 (kindly provided by R. Arnon, The Weizmann Institute, Rehovot, Israel). Supernatants were assayed using an IFN-α ELISA kit (Performance Biomedical Laboratories).
Real-Time PCR.
Splenocytes from Cx3cr1gfp mice were stained with antibody conjugates, including Bst2-APC (Miltenyi Biotec) and sorted by a FACSAria flow sorter (BD Immunocytometry Systems) into CD11cint Bst2+ (PDC), CD11chi CD8α− CD11b+ GFP+ (CD8α− DC), and CD11chi CD8α+ CD11b− GFP+ (CX3CR1+ CD8α+ DC) fractions. Cells were sorted directly into TRIzol LS reagent (Invitrogen). Total RNA from the sorted cells was extracted and reverse transcribed. Gene-expression levels were assayed by SYBR Green-based real-time PCR on an MX3000P instrument (Stratagene). All expression was normalized to β-actin and is presented relative to the CX3CR1+ CD8α+ DC sample using the ΔΔCT method. All primers were validated for linear amplification (sequences are available on request).
Supplementary Material
Acknowledgments
We thank Hilah Gal, Elad Bar-On, and Einat Zucker. This work was supported by the Israel Science Foundation and research grants from the estate of Edith F. Goldensohn and the Kekst Center (to S.J.), the United States–Israel Binational Science Foundation (S.J. and B.R.), the Howard Hughes Medical Institute (K.M.M), the Emmy Noether Program of the German Research Foundation (K.H.), National Institutes of Health Grant AI072571 (to B.R.), and National Institutes of Health Training Grant AI007161 (to K.L.L.). B.T.E. is the recipient of a Burroughs Wellcome Fund Career Award for Medical Scientists.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Microarray data reported in this paper have been deposited in the Gene Expression Omnibus (accession number GSE23212).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1001562107/-/DCSupplemental.
References
- 1.Sapoznikov A, Jung S. Probing in vivo dendritic cell functions by conditional cell ablation. Immunol Cell Biol. 2008;86:409–415. doi: 10.1038/icb.2008.23. [DOI] [PubMed] [Google Scholar]
- 2.Steinman RM, Witmer MD. Lymphoid dendritic cells are potent stimulators of the primary mixed leukocyte reaction in mice. Proc Natl Acad Sci USA. 1978;75:5132–5136. doi: 10.1073/pnas.75.10.5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu K, et al. In vivo analysis of dendritic cell development and homeostasis. Science. 2009;324:392–397. doi: 10.1126/science.1170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shortman K, Naik SH. Steady-state and inflammatory dendritic-cell development. Nat Rev Immunol. 2007;7:19–30. doi: 10.1038/nri1996. [DOI] [PubMed] [Google Scholar]
- 5.Dudziak D, et al. Differential antigen processing by dendritic cell subsets in vivo. Science. 2007;315:107–111. doi: 10.1126/science.1136080. [DOI] [PubMed] [Google Scholar]
- 6.Pulendran B, Tang H, Denning TL. Division of labor, plasticity, and crosstalk between dendritic cell subsets. Curr Opin Immunol. 2008;20:61–67. doi: 10.1016/j.coi.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu L, et al. RelB is essential for the development of myeloid-related CD8alpha- dendritic cells but not of lymphoid-related CD8alpha+ dendritic cells. Immunity. 1998;9:839–847. doi: 10.1016/s1074-7613(00)80649-4. [DOI] [PubMed] [Google Scholar]
- 8.Schiavoni G, et al. ICSBP is essential for the development of mouse type I interferon-producing cells and for the generation and activation of CD8alpha(+) dendritic cells. J Exp Med. 2002;196:1415–1425. doi: 10.1084/jem.20021263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tamura T, Nagamura-Inoue T, Shmeltzer Z, Kuwata T, Ozato K. ICSBP directs bipotential myeloid progenitor cells to differentiate into mature macrophages. Immunity. 2000;13:155–165. doi: 10.1016/s1074-7613(00)00016-9. [DOI] [PubMed] [Google Scholar]
- 10.Caton ML, Smith-Raska MR, Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J Exp Med. 2007;204:1653–1664. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iyoda T, et al. The CD8+ dendritic cell subset selectively endocytoses dying cells in culture and in vivo. J Exp Med. 2002;195:1289–1302. doi: 10.1084/jem.20020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.den Haan JM, Lehar SM, Bevan MJ. CD8(+) but not CD8(-) dendritic cells cross-prime cytotoxic T cells in vivo. J Exp Med. 2000;192:1685–1696. doi: 10.1084/jem.192.12.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pulendran B, et al. Distinct dendritic cell subsets differentially regulate the class of immune response in vivo. Proc Natl Acad Sci USA. 1999;96:1036–1041. doi: 10.1073/pnas.96.3.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reis e Sousa C, et al. In vivo microbial stimulation induces rapid CD40 ligand-independent production of interleukin 12 by dendritic cells and their redistribution to T cell areas. J Exp Med. 1997;186:1819–1829. doi: 10.1084/jem.186.11.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamazaki T, et al. CCR6 regulates the migration of inflammatory and regulatory T cells. J Immunol. 2008;181:8391–8401. doi: 10.4049/jimmunol.181.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aliberti J, et al. Essential role for ICSBP in the in vivo development of murine CD8alpha + dendritic cells. Blood. 2003;101:305–310. doi: 10.1182/blood-2002-04-1088. [DOI] [PubMed] [Google Scholar]
- 17.Tamura T, et al. IFN regulatory factor-4 and -8 govern dendritic cell subset development and their functional diversity. J Immunol. 2005;174:2573–2581. doi: 10.4049/jimmunol.174.5.2573. [DOI] [PubMed] [Google Scholar]
- 18.Hacker C, et al. Transcriptional profiling identifies Id2 function in dendritic cell development. Nat Immunol. 2003;4:380–386. doi: 10.1038/ni903. [DOI] [PubMed] [Google Scholar]
- 19.Kusunoki T, et al. TH2 dominance and defective development of a CD8+ dendritic cell subset in Id2-deficient mice. J Allergy Clin Immunol. 2003;111:136–142. doi: 10.1067/mai.2003.29. [DOI] [PubMed] [Google Scholar]
- 20.Hildner K, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colonna M, Trinchieri G, Liu YJ. Plasmacytoid dendritic cells in immunity. Nat Immunol. 2004;5:1219–1226. doi: 10.1038/ni1141. [DOI] [PubMed] [Google Scholar]
- 22.Naik SH, et al. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat Immunol. 2007;8:1217–1226. doi: 10.1038/ni1522. [DOI] [PubMed] [Google Scholar]
- 23.Onai N, et al. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat Immunol. 2007;8:1207–1216. doi: 10.1038/ni1518. [DOI] [PubMed] [Google Scholar]
- 24.Liu K, et al. Origin of dendritic cells in peripheral lymphoid organs of mice. Nat Immunol. 2007;8:578–583. doi: 10.1038/ni1462. [DOI] [PubMed] [Google Scholar]
- 25.O'Keeffe M, et al. Mouse plasmacytoid cells: Long-lived cells, heterogeneous in surface phenotype and function, that differentiate into CD8(+) dendritic cells only after microbial stimulus. J Exp Med. 2002;196:1307–1319. doi: 10.1084/jem.20021031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shigematsu H, et al. Plasmacytoid dendritic cells activate lymphoid-specific genetic programs irrespective of their cellular origin. Immunity. 2004;21:43–53. doi: 10.1016/j.immuni.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 27.Cisse B, et al. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell. 2008;135:37–48. doi: 10.1016/j.cell.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jung S, et al. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol. 2000;20:4106–4114. doi: 10.1128/mcb.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dakic A, et al. Development of the dendritic cell system during mouse ontogeny. J Immunol. 2004;172:1018–1027. doi: 10.4049/jimmunol.172.2.1018. [DOI] [PubMed] [Google Scholar]
- 30.Yarovinsky F, et al. TLR11 activation of dendritic cells by a protozoan profilin-like protein. Science. 2005;308:1626–1629. doi: 10.1126/science.1109893. [DOI] [PubMed] [Google Scholar]
- 31.Lin ML, et al. Selective suicide of cross-presenting CD8+ dendritic cells by cytochrome c injection shows functional heterogeneity within this subset. Proc Natl Acad Sci USA. 2008;105:3029–3034. doi: 10.1073/pnas.0712394105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Keeffe M, et al. Effects of administration of progenipoietin 1, Flt-3 ligand, granulocyte colony-stimulating factor, and pegylated granulocyte-macrophage colony-stimulating factor on dendritic cell subsets in mice. Blood. 2002;99:2122–2130. doi: 10.1182/blood.v99.6.2122. [DOI] [PubMed] [Google Scholar]
- 33.Morón G, Rueda P, Casal I, Leclerc C. CD8alpha- CD11b+ dendritic cells present exogenous virus-like particles to CD8+ T cells and subsequently express CD8alpha and CD205 molecules. J Exp Med. 2002;195:1233–1245. doi: 10.1084/jem.20011930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edwards AD, et al. Relationships among murine CD11c(high) dendritic cell subsets as revealed by baseline gene expression patterns. J Immunol. 2003;171:47–60. doi: 10.4049/jimmunol.171.1.47. [DOI] [PubMed] [Google Scholar]
- 35.Robbins SH, et al. Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome-wide expression profiling. Genome Biol. 2008;9:R17. doi: 10.1186/gb-2008-9-1-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cervantes-Barragan L, et al. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood. 2007;109:1131–1137. doi: 10.1182/blood-2006-05-023770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corcoran L, et al. The lymphoid past of mouse plasmacytoid cells and thymic dendritic cells. J Immunol. 2003;170:4926–4932. doi: 10.4049/jimmunol.170.10.4926. [DOI] [PubMed] [Google Scholar]
- 38.Zhuang Y, Cheng P, Weintraub H. B-lymphocyte development is regulated by the combined dosage of three basic helix-loop-helix genes, E2A, E2-2, and HEB. Mol Cell Biol. 1996;16:2898–2905. doi: 10.1128/mcb.16.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schlecht G, et al. Murine plasmacytoid dendritic cells induce effector/memory CD8+ T-cell responses in vivo after viral stimulation. Blood. 2004;104:1808–1815. doi: 10.1182/blood-2004-02-0426. [DOI] [PubMed] [Google Scholar]
- 40.Zuniga EI, McGavern DB, Pruneda-Paz JL, Teng C, Oldstone MB. Bone marrow plasmacytoid dendritic cells can differentiate into myeloid dendritic cells upon virus infection. Nat Immunol. 2004;5:1227–1234. doi: 10.1038/ni1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsujimura H, Tamura T, Ozato K. Cutting edge: IFN consensus sequence binding protein/IFN regulatory factor 8 drives the development of type I IFN-producing plasmacytoid dendritic cells. J Immunol. 2003;170:1131–1135. doi: 10.4049/jimmunol.170.3.1131. [DOI] [PubMed] [Google Scholar]
- 42.Villadangos JA, Shortman K. Found in translation: The human equivalent of mouse CD8+ dendritic cells. J Exp Med. 2010;207:1131–1134. doi: 10.1084/jem.20100985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharov AA, Dudekula DB, Ko MS. A web-based tool for principal component and significance analysis of microarray data. Bioinformatics. 2005;21:2548–2549. doi: 10.1093/bioinformatics/bti343. [DOI] [PubMed] [Google Scholar]
- 44.Mach N, et al. Differences in dendritic cells stimulated in vivo by tumors engineered to secrete granulocyte-macrophage colony-stimulating factor or Flt3-ligand. Cancer Res. 2000;60:3239–3246. [PubMed] [Google Scholar]
- 45.ten Boekel E, Melchers F, Rolink A. The status of Ig loci rearrangements in single cells from different stages of B cell development. Int Immunol. 1995;7:1013–1019. doi: 10.1093/intimm/7.6.1013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.