Abstract
The ternary complex of cadherin, β-catenin, and α-catenin regulates actin-dependent cell–cell adhesion. α-Catenin can bind β-catenin and F-actin, but in mammals α-catenin either binds β-catenin as a monomer or F-actin as a homodimer. It is not known if this conformational regulation of α-catenin is evolutionarily conserved. The Caenorhabditis elegans α-catenin homolog HMP-1 is essential for actin-dependent epidermal enclosure and embryo elongation. Here we show that HMP-1 is a monomer with a functional C-terminal F-actin binding domain. However, neither full-length HMP-1 nor a ternary complex of HMP-1–HMP-2(β-catenin)–HMR-1(cadherin) bind F-actin in vitro, suggesting that HMP-1 is auto-inhibited. Truncation of either the F-actin or HMP-2 binding domain of HMP-1 disrupts C. elegans development, indicating that HMP-1 must be able to bind F-actin and HMP-2 to function in vivo. Our study defines evolutionarily conserved properties of α-catenin and suggests that multiple mechanisms regulate α-catenin binding to F-actin.
Keywords: cell–cell adhesion, HMP-1, HMP-2, HMR-1
Cadherin-mediated cell–cell adhesion is critical for normal development and tissue organization in metazoans (1). Cadherins bind cytoplasmic β-catenin and p120 directly, and strengthening of cell–cell adhesion involves local reorganization of the actin cytoskeleton (2–4). α-Catenin binds β-catenin, can bundle F-actin (5), and associates with actin-regulatory proteins (6, 7). Thus, the classical model of the cadherin–catenin complex posits that α-catenin forms a static bridge between the cadherin–catenin complex and the actin cytoskeleton.
In vitro studies revealed a more complex regulation of these protein interactions. Mammalian αE-catenin forms monomers or homodimers (8–10). Association of αE-catenin monomer with β-catenin significantly weakens the affinity of αE-catenin for F-actin, whereas αE-catenin homodimer binds strongly to F-actin. It is unknown if this conformational regulation is evolutionarily conserved by other α-catenins.
The only α-catenin homolog in Caenorhabditis elegans is HMP-1. Mutations in hmp-1 cause the detachment of circumferential actin filament bundles from adherens junctions, which are required for embryo elongation during epidermal morphogenesis, and results in dorsal folds in the epidermis (11). Nothing, however, is known about the molecular properties of HMP-1: Does HMP-1 bind F-actin directly, does the ternary HMR-1–HMP-2–HMP-1 (cadherin-β–catenin-αE—catenin) complex bind F-actin, and is HMP-1 function regulated by homodimerization?
Here we show that HMP-1 is a monomer that does not bind directly to F-actin in vitro despite a functional C-terminal F-actin binding domain. However, both the HMP-2/β-catenin and F-actin binding regions are necessary for HMP-1 function during embryogenesis, suggesting additional factors regulate HMP-1 activity in vivo. Our study is a detailed analysis of an invertebrate α-catenin and provides unique insights into the molecular properties and evolution of α-catenin.
Results and Discussion
HMP-1 Is a Bona Fide α-Catenin that Binds Directly to HMP-2.
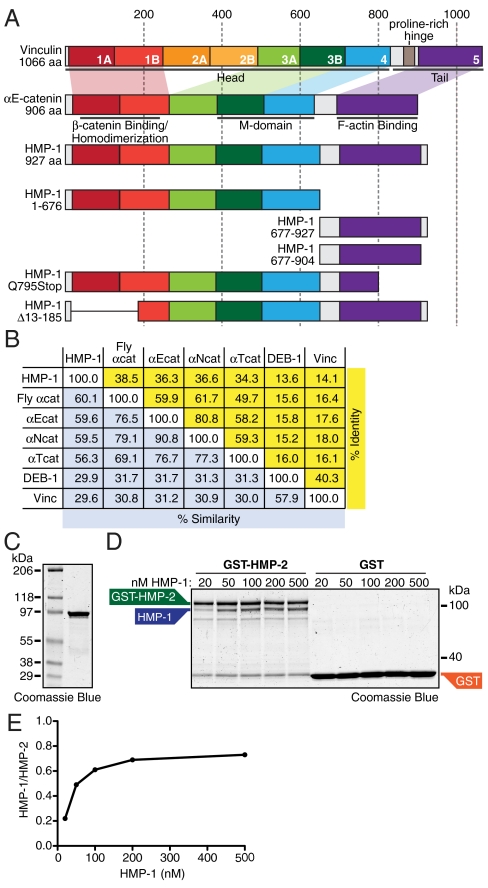
Crystal structures of αE-catenin domains (12–14) and vinculin (15–17) show that these proteins are a series of four-helix bundles (Fig. 1A). The N-terminal domain of αE-catenin, comprising two four-helix bundles, has overlapping sites for β-catenin binding and homodimerization, making these interactions mutually exclusive (5, 8, 9, 14). The middle (M) domain consists of two flexibly linked four-helix bundles (12, 13). The C-terminal “tail” region of both αE-catenin and vinculin is a five-helix bundle that binds F-actin (17). Vinculin includes an additional pair of helical bundles between the αE-catenin N-terminal and M domains (Fig. 1A, orange boxes 2a and 2b). In vinculin, the N-terminal “head” region binds intramolecularly to the tail to inhibit actin binding. This auto-inhibition is relieved upon binding to talin, and is considered critical for regulating vinculin function (18–20). Despite the similarities between vinculin and α-catenin, there is no evidence for a head to tail interaction in α-catenin (6, 21).
Fig. 1.
Recombinant HMP-1 binds HMP-2. (A) Vinculin is composed of an array of 7 four-helix bundles (first 6 paired), a hinge region and C-terminal five-helix bundle. Matching helical domains in αE-catenin and HMP-1 are color-coded to mark homology. Amino acid scale at top. Head and tail regions of vinculin are marked, as are the β-catenin/dimerization, M-domain, and F-actin binding regions in αE-catenin. All HMP-1 constructs used in this study are defined. (B) Percent identity (yellow) and similarity (blue) between HMP-1, fly α-catenin, mouse α-catenins (αE-, αN-, and αT-catenin), mouse vinculin, and worm vinculin (DEB-1). (C) Recombinant FL HMP-1 run on an SDS-PAGE gel and stained with Coomassie blue. (D) Increasing concentrations of HMP-1 were incubated with 300 nM GST-HMP-2 or 1 μM GST bound to glutathione-agarose beads for 1 hr at RT, washed, and then analyzed by SDS-PAGE. (E) Amounts of precipitated HMP-1 were measured, normalized to GST-HMP-2, and plotted.
We compared the amino acid (aa) sequence of C. elegans HMP-1, Drosophila melanogaster α-catenin, mouse α-catenins (αE-, αN-, and αT-catenin), and vinculin from C. elegans (DEB-1) and mouse (Fig. 1B). Based on sequence homology and domain organization, HMP-1 is a bona fide member of the α-catenin family.
We first tested whether recombinant full-length (FL) HMP-1 (Fig. 1C) binds HMP-2, a C. elegans β-catenin homolog (11). HMP-1 bound to GST-HMP-2, but not GST, at a stoichiometry of ∼1∶1 at saturating amounts of HMP-1 (Fig. 1D), similar to the mammalian αE-catenin–β-catenin complex (9, 14). HMP-2 also interacted robustly with FL HMP-1 and HMP-1 aa 1–676 (head) in a yeast two-hybrid assay (Fig. S1). Saturated binding of HMP-1 to HMP-2 was reached at approximately 200 nM HMP-1, and the resulting binding curve (Fig. 1E) indicates the affinity between HMP-1 and HMP-2 is comparable to the affinity of αE-catenin for β-catenin (22).
HMP-1 Is a Compact Monomer.
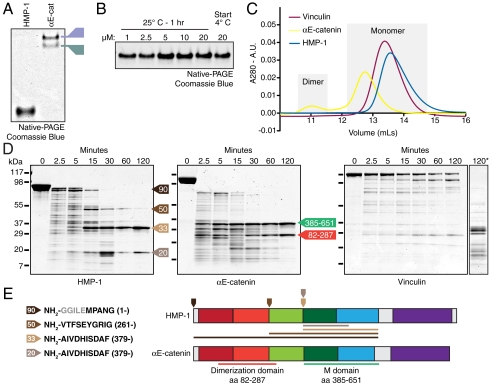
Homodimerization is an intrinsic property of mammalian αE-catenin (9). However, HMP-1 migrated as a single band by native-PAGE, faster than either αE-catenin monomer or homodimer (Fig. 2A), indicating that recombinant HMP-1 is a monomer in solution; neither temperature nor protein concentration affected the monomeric state of HMP-1 (Fig. 2B). HMP-1 aa 1–676 (head) also migrated as a single band by native-PAGE, and no interaction between FL HMP-1 proteins was observed in a yeast two-hybrid assay (Fig. S2 and Fig. S1), indicating that the N-terminal region of HMP-1, unlike that of αE-catenin, does not mediate homodimerization.
Fig. 2.
HMP-1 is a monomer in solution. (A) FL HMP-1 and mouse αE-catenin were separated by native-PAGE. Monomer (M) and dimer (D) species of αE-catenin noted. (B) HMP-1 was incubated at concentrations ranging from 1 to 20 μM for 1 h at 25 °C and separated by native-PAGE. (C) Superdex 200 (S200) gel filtration chromatography of recombinant HMP-1, mouse αE-catenin, and mouse vinculin. (D) Proteolytic sensitivity of recombinant HMP-1, αE-catenin monomer, and purified chicken vinculin. Coomassie-stained SDS-PAGE of proteins incubated for 0, 2.5, 5, 15, 30, 60, and 120 min at RT with 0.05 mg/mL of trypsin. 120* is vinculin digested for 120 min at 0.2 mg/mL trypsin. M-domain (385–651) and dimerization domain (82–287) αE-catenin fragments marked with arrows. HMP-1 fragments of 90, 50, 33, and 20 kDas noted with arrows. (E) Edman sequencing results (Left, amino acid start in HMP-1 sequence listed) and position of NH2-termini marked on HMP-1 schematic (Right, downward-facing arrows). Linker region retained after GST cleavage colored gray. Projected fragment size shown as color-coded lines. αE-catenin diagram provided as reference.
The difference in electrophoretic migration between HMP-1 and αE-catenin monomers was surprising given the similarities in their molecular weights (104 kDa vs. 100 kDa) and overall net charge (calculated pI 5.6 vs. 5.8). The faster migration of HMP-1 suggests that it is more compact than αE-catenin monomer. We compared the gel filtration elution profiles of HMP-1, mouse αE-catenin and mouse vinculin. αE-catenin eluted in two distinct peaks corresponding to a homodimer and monomer (Fig. 2C) (9, 10). Vinculin, a compact, folded monomer in solution (9, 23), eluted after αE-catenin monomer in a single peak. HMP-1 eluted in a single peak after αE-catenin monomer, consistent with its faster migration in native-PAGE.
Small-angle X-ray scattering (SAXS) provides information about molecular dimensions of proteins in solution (a mass-weighted average radius termed the radius of gyration Rg, and the maximum interatomic distance, Dmax) that are independent of hydrodynamic properties (Table 1). Vinculin has the smallest Rg value as well as the shortest Dmax, which corresponds to its highly compact structure in the auto-inhibited state. In contrast, αE-catenin homodimer has much larger Rg and Dmax values. αE-catenin monomer preparations always contain small amounts of homodimer that contribute strongly to the X-ray scattering, preventing accurate determination of SAXS parameters. The β-αE-catenin chimera [α-catenin binding region of β-catenin fused to αE-catenin to block homodimerization (9, 14)] is a surrogate for monomeric αE-catenin and has Rg and Dmax values that are considerably larger than those of vinculin despite having fewer residues (891 vs. 1066, respectively), consistent with the chimera having a more extended conformation than vinculin. HMP-1 has Rg and Dmax values that are closest to, albeit slightly larger than, vinculin. Given the larger number of residues in vinculin than HMP-1 (1066 vs. 926), these data indicate that HMP-1 adopts a relatively compact conformation in solution compared to β-αE-catenin chimera. The compact nature of HMP-1 explains the difference in electrophoretic migration between HMP-1 and αE-catenin monomers (Fig. 2A).
Table 1.
SAXS parameters, Rg and Dmax of HMP-1, vinculin, αE-catenin dimer and β-αE-catenin
| Protein | # aa | Rg [Å] | Dmax [Å] |
| HMP-1 | 932 | 37.2 | 123 |
| Vinculin | 1069 | 36.7 | 121 |
| αE-catenin dimer | 1820 | 59.6 | 193 |
| β-αE-catenin | 893 | 44.6 | 150 |
The Rg of vinculin calculated from the crystal structure (PDB ID 1ST6) using Crysol (31) is 33.7 Å. The number of amino acids (# aa) includes residues left after cleavage of the GST-tag (or His-tag in the case of vinculin). The estimated uncertainty in Dmax is ± 5 Å.
To determine if HMP-1 adopts a conformation similar to α-catenin, we used limited proteolysis to probe for stable subdomains of HMP-1 and compare them to αE-catenin and vinculin. Tryptic digests of HMP-1 revealed four prominent fragments of 90, 50, 33, and 20 kDa (Fig. 2D). Edman sequencing revealed that the 33 kDa fragment is similar to the M domain in αE-catenin (aa 385–651), and that the 50 and 20 kDa fragments are larger and smaller fragments, respectively, of this region (Fig. 2E). Notably, a protease-resistant dimerization domain was not found (aa 82–287), suggesting the N-terminus of HMP-1 adopts a conformation different from that of αE-catenin (14). The 90 kDa fragment may result from loss of the C-terminal tail, and thus represents the head region (Fig. 2E and Fig. S2). Importantly, the tryptic digest pattern of vinculin was distinct from both HMP-1 and αE-catenin (Fig. 2D, right panel), reflecting intrinsic differences in the conformation of these proteins.
Together, these results demonstrate that HMP-1 is a compact, monomeric α-catenin. Whereas we cannot rule out the possibility that HMP-1 homodimerizes in vivo, the absence of HMP-1 dimers observed in vitro distinguishes it from mammalian αE-catenin (9, 22) and suggests that homodimerization is not a property common to all α-catenins.
FL HMP-1 Does Not Bind F-actin In Vitro.
Mammalian αE-catenin monomer binds weakly to F-actin, whereas αE-catenin homodimers bind strongly (8, 9). HMP-1 monomer failed to bind F-actin (Fig. 3A and Fig. 4C) above background (BSA) (Fig. 3B). The lack of F-actin binding activity could be due to HMP-1 misfolding or the absence of a functional F-actin-binding domain. The former possibility is unlikely because HMP-1 bound to HMP-2 with high affinity and stoichiometry, and the SAXS data indicates that it was folded compactly with no aggregation. Therefore, we examined whether HMP-1 has a functional F-actin binding domain.
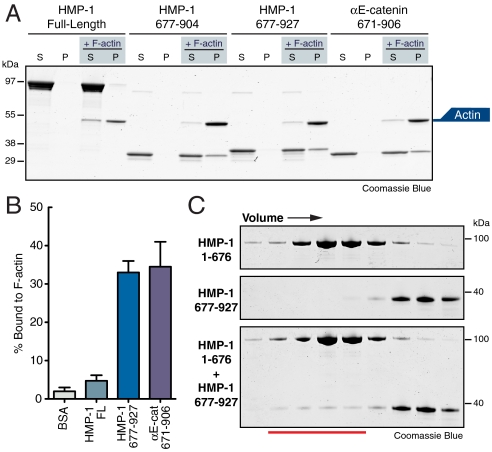
Fig. 3.
The F-actin binding region in FL HMP-1 is masked. (A) High-speed supernatant (S) and pellet (P) fractions of 5 μM of FL HMP-1, HMP-1 aa 677–904, HMP-1 aa 677–927, and mouse αE-catenin aa 671–906 incubated with 5 μM F-actin. (B) Percentage of BSA, FL HMP-1, HMP-1 aa 677–927, and mouse αE-catenin aa 671–906 pelleting with F-actin was measured and plotted. Errors bars are standard error of the mean (SEM) from at least two independent experiments. (C) S200 gel filtration chromatography fractions of 50 μM HMP-1 aa 1–676 (head, Upper) and 50 μM HMP-1 aa 677–927 (tail, Center). When mixed before gel filtration (50 μM final concentration for both fragments), a small amount of tail domain elutes with the head domain (red line, Lower).
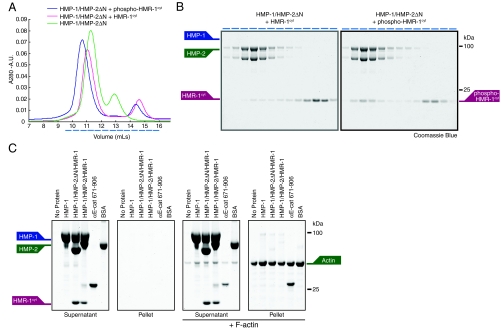
Fig. 4.
The HMR-1/HMP-2/HMP-1 ternary complex does not bind F-actin. (A, B) S200 gel filtration chromatography (A) of 100 μM HMP-1/HMP-2ΔN complex alone (green line) and incubated with 150 μM HMR-1cyt (pink line) or 150 μM CK1 phoshorylated HMR-1cyt (blue line). Fractions (represented by dashed blue lines below chromatogram in A) were analyzed by SDS-PAGE (B). HMP-1, HMP-2ΔN and HMR-1cyt proteins are marked. (C) Sedimentation of 8 μM HMP-1, HMP-1/HMP-2ΔN/HMR-1cyt, HMP-1/HMP-2/HMR-1cyt and BSA in the absence or presence of 8 μM F-actin. Supernatant (S) and pellet (P) fractions were analyzed by SDS-PAGE and stained with Coomassie blue. HMP-1, HMP-2, HMR-1cyt and actin proteins marked.
The C-terminal domain of HMP-1 (aa 677–904) is 50% identical and 75% similar to the comparable domain of αE-catenin (aa 671–906, Fig. S3). We expressed two C-terminal fragments of HMP-1: aa 677–904, which contains the putative F-actin binding domain, and aa 677–927, which has an additional 23 aa C-terminal tail absent in vertebrate and Drosophila α-catenin (Fig. S3). Both C-terminal HMP-1 proteins bound F-actin (Fig. 3A) at levels similar to the corresponding C-terminal F-actin-binding domain of mammalian αE-catenin (Fig. 3 A and B). Therefore, HMP-1 has a functional F-actin binding domain that is masked in the FL protein, indicating auto-inhibition.
The tail and head domains of vinculin bind in the auto-inhibited state, preventing F-actin association (18). We tested whether the tail (aa 677–927) and head (aa 1–676) domains of HMP-1 bound to each other. When the two domains were mixed before gel filtration, we detected a small amount of tail domain eluting with the head domain (Fig. 3C), indicating a weak interaction between these regions in trans. This interaction, however, was below the threshold of detection by yeast two-hybrid analysis (Fig. S1). This is consistent with a weak affinity (Kd > 1 μM) compared to vinculin head to tail binding in trans (Kd < 50 nM) (19). Thus, the mechanism of HMP-1 auto-inhibition may be distinct from that of vinculin.
This analysis of HMP-1 is the first to examine F-actin binding to a pure α-catenin monomer, because analysis of F-actin binding to mammalian αE-catenin monomers is complicated by their rapid homodimerization. We note, however, when if homodimerization is blocked by β-catenin binding, as in the β–αE-catenin chimera, αE-catenin “monomer” binds weakly, if at all, to F-actin (9). We speculate that auto-inhibition of F-actin binding is a feature common to α-catenins.
The HMR-1/HMP-2/HMP-1 Ternary Complex Does Not Associate with F-actin.
We next examined whether binding of HMP-1 in the ternary complex of HMR-1 and HMP-2 relieved the apparent auto-inhibition of HMP-1 binding to F-actin. First, we attempted to form the HMR-1–HMP-2–HMP-1 ternary complex in vitro using the C-terminal cytoplasmic domain of HMR-1 (HMR-1cyt). To improve HMP-2 stability, we removed 35 aa from the N-terminus of HMP-2 (HMP-2ΔN), which did not affect stoichiometric binding of HMP-2 to HMP-1 or HMR-1cyt (Fig. 4C).
Gel filtration of a mixture of HMR-1cyt, HMP-2ΔN and HMP-1 resulted, surprisingly, in the elution of HMR-1cyt separately from the HMP-2ΔN–HMP-1 complex (Fig. 4 A and B). Binding between mammalian cadherin cytoplasmic domain and β-catenin is increased by serine/threonine phosphorylation (24), and HMR-1cyt contains consensus sites for casein kinase 1 (CK1) phosphorylation. When phosphorylated by CK1 in vitro, phospho-HMR-1cyt bound readily to HMP-2ΔN/HMP-1 (Fig. 4 A and B), indicating CK1-dependent phosphorylation regulates formation of the ternary complex from C. elegans. Significantly, neither the HMP-2ΔN–HMP-1 complex nor phospho-HMR-1cyt–HMP-2ΔN–HMP-1 ternary complex bound F-actin (Fig. 4C). Thus, neither HMP-2 binding nor the formation of the ternary complex increased HMP-1 binding to F-actin in vitro.
In summary, our in vitro studies demonstrate HMP-1 is a monomer that does not bind F-actin, even when bound to HMP-2 or in the HMR-1cyt–HMP-2 complex. The C-terminus of HMP-1, like that of other α-catenins, can bind F-actin, but this site is blocked in the FL protein. F-actin binding by mammalian αE-catenin is activated by homodimerization, although the molecular basis for this change is not understood. Because HMP-1 does not homodimerize by itself, its F-actin binding activity must be activated by a different mechanism.
N- and C-Terminal Domains Are Required for HMP-1 Function In Vivo.
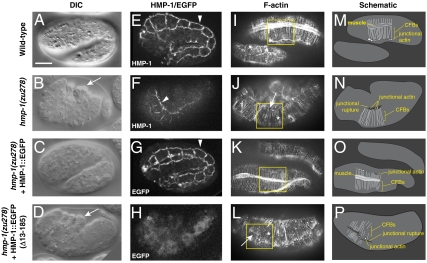
We tested in vivo requirements for selected molecular properties of HMP-1 uncovered in our in vitro experiments. In elongating wild-type embryos, actin accumulates at cell–cell junctions in the epidermis, and a series of thick actin bundles (circumferential filament bundles, or CFBs) form that insert orthogonally to the junctional actin (Fig. 5 I and M). A putative null allele of hmp-1, zu278, was identified in a screen for mutants defective in embryonic elongation (11). Zygotic loss of wild-type HMP-1 in zu278 mutants results in two major defects to these actin structures: loss of junctional proximal actin, and detachment of CFBs (Fig. 5 J and N; for further details see ref. 11). We found, however, that hmp-1(zu278) has a point mutation that creates a premature stop codon (Q795Stop) in the F-actin-binding domain (Fig. 1A). Using an antibody against FL HMP-1, we observed HMP-1 staining at cell–cell contacts in homozygous mutants similar to wild-type (wt), although the amount appeared to be reduced compared to controls (Fig. 5 E and F), indicating that the truncated protein associates with junctions. Importantly, hmp-1(zu278) homozygous mutants displayed a strong Humpback phenotype (Fig. 5B, white arrow; compare to wt in Fig. 5A), demonstrating that F-actin binding is critical for HMP-1 function during embryogenesis.
Fig. 5.
HMP-1 N- and C-termini are required for function in vivo. (A–D) DIC images. Wild-type embryo (A); homozygous hmp-1(zu278) embryo (B) with characteristic dorsal humps (arrow); homozygous hmp-1(zu278) embryo rescued by expression of HMP-1::EGFP (C); and homozygous hmp-1(zu278) embryo expressing HMP-1∷EGFP(Δ13–185) (D) with dorsal humps as in (B) (arrow). (E, F) HMP-1 staining. Wild-type embryo (E) with HMP-1 staining at cell–cell junctions (arrowhead) and homozygous hmp-1(zu278) embryo (F) with reduced HMP-1 staining at cell–cell junctions (arrowhead). (G, H) EGFP localization. (G) Embryo in (C) showing localization of HMP-1∷EGFP at cell–cell junctions (arrowhead). (H) Embryo in (D) showing cytoplasmic localization of HMP-1∷EGFP(Δ13–185). (I–L) Phalloidin staining. Wild-type embryo (I) with typical parallel CFBs; homozygous hmp-1(zu278) embryo (J) with gaps between F-actin and junctions (arrow) and larger than normal gaps between CFBs (asterisk); homozygous hmp-1(zu278) embryo rescued by expression of HMP-1∷EGFP (K) with parallel CFBs similar to wt in (I); and homozygous hmp-1(zu278) embryo expressing HMP-1∷GFP(Δ13–185) (L) with gaps between F-actin and junctions (arrow) and larger than normal gaps between CFBs (asterisks) similar to homozygous hmp-1(zu278) embryo in (J). (M–P) Schematics of phalloidin-stained regions in (I–L). Only the boxed regions in (I–L) are drawn in the schematics. CFBs, junctional actin, junctional tears and muscle are illustrated. Note F-actin stained muscle is below the epidermis. All embryos are at 2-fold elongation or terminal, positioned anterior left and dorsal up. Scale bar is 10 μm.
We determined if hmp-1(zu278) could be rescued by expression of FL HMP-1∷EGFP driven by its endogenous promoter. HMP-1∷EGFP localized correctly to cell–cell junctions and, importantly, rescued the developmental defects in zu278 homozygous mutant embryos to viability (Fig. 5 C, G, K and O). Rescued adults exhibited a slightly uncoordinated (Unc) phenotype, but were otherwise indistinguishable from wt.
We next assessed the ability of a hmp-1 mutant lacking the N-terminal HMP-2 binding site to rescue the hmp-1(zu278) mutation. The predicted HMP-2 binding site—aa 13–185—was deleted from HMP-1 (Fig. 1A). Though expressed at levels comparable to HMP-1∷EGFP, HMP-1∷EGFP(Δ13–185) failed to localize to cell–cell junctions in hmp-1(zu278) homozygous mutants (Fig. 5 H and I) and did not rescue the hmp-1 mutant phenotype (Fig. 5D). Thus, HMP-2 binding is required to localize HMP-1 to cell–cell junctions and to rescue the hmp-1(zu278) mutant phenotype.
Because in vitro binding studies showed that HMP-1, either alone or in a complex with HMR-1 and HMP-2, did not bind F-actin, how is the C-terminal F-actin-binding domain required for function in vivo? One possibility is that a yet unidentified factor, such as another interacting protein or a posttranslational modification of HMP-1, is required to relieve the auto-inhibited state and activate HMP-1 binding to F-actin. Recently, it was proposed that E-cadherin is a mechanosensor that transmits force to the actin cytoskeleton, and that vinculin interacts with the cadherin complex to potentiate this activity (25). Additionally, a recent independent study suggested that αE-catenin is a force transducer at adherens junctions, and recruits vinculin upon mechanical strain to increase F-actin binding and adhesive strength (26). However, it is unlikely that DEB-1/vinculin promotes HMP-1 association with the actin cytoskeleton as DEB-1 is not expressed in C. elegans epithelia (27). A putative HMP-1 binding protein could induce HMP-1 homodimerization in vivo. Alternatively HMP-1, like vinculin, may function exclusively as a monomer. Because the N-terminal HMP-2 binding domain is also required for HMP-1 function in vivo, HMP-1 may link the cadherin-catenin complex to the actin cytoskeleton upon activation by an additional protein. Indeed, given that cadherin-complex proteins are conserved from worms to mammals, a similar protein could also regulate the F-actin binding activity of αE-catenin in vertebrates. Further studies are required to identify this putative α-catenin modulator.
Materials and Methods
Protein Expression and Purification.
GST-tagged proteins were expressed in BL21 (DE3) Codon Plus Escherichia coli cells and purified as described (14). GST-tagged proteins bound to glutathione-agarose were equilibrated in cleavage/elution buffer (20 mM Tris pH 8.0, 150 mM NaCl, 2 mM EDTA, 1 mM DTT and 10% glycerol) and then incubated with tobacco etch virus (TEV) protease overnight at 4 °C to remove protein from GST tag. His-tagged full-length vinculin was purified using Ni-NTA agarose beads (Qiagen). After cleavage of his-tag by thrombin, the his-peptide was removed with Ni-NTA agarose beads. All proteins were further purified by FPLC using an anion exchange MonoQ column, and when necessary, a Superdex200 (S200) preparative gel filtration column. When required, eluted protein was concentrated to 20–100 μM working concentrations using a Millipore column concentrator. To analyze oligomerization, FPLC-purified proteins were run over an analytical S200 column.
HMR-1cyt Phosphorylation.
Bacterially expressed HMR-1cyt was phosphorylated by recombinant CKI (New England Biolabs) using the manufacturer’s recommended buffer. The reaction was carried out for 12 h at 30 °C, and stopped by the addition of 4 mM EDTA. Phosphorylated and nonphosphorylated HMR-1cyt were separated over a MonoQ column. Phosphorylation was confirmed by mass spectrometry. HMR-1cyt molecules bearing 4 or 5 phosphate groups gave equivalent binding to HMP-2.
Native-PAGE.
1 μg of FPLC-purified protein was diluted into ice-cold CSK buffer (10 mM Pipes, pH 6.8, 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose) plus 100 mM DTT and 0.02% bromophenol blue (for color) and immediately loaded onto a 5% native gel (running gel: 0.4 M Tris pH 8.8, 5% acrylamide; stacking gel: 0.1 M Tris pH 6.8, 5% acrylamide). Gels were run at 80 V for 4–5 h at 4 °C, stained with Coomassie blue, destained and imaged on a LI-COR scanner.
Actin Pelleting Assay.
Chicken G-actin was incubated in polymerization buffer (200 mM Imidazole pH 7, 1 M KCl, 20 mM MgCl2, 5 mM ATP, 10 mM EGTA) for 1 h at room temperature (RT) to promote filament assembly. HMP-1 and control proteins were diluted to 5 μM in reaction buffer (200 mM Imidazole pH 7.0, 1.5 M NaCl, 20 mM MgCl2, 5 mM ATP, 10 mM EGTA) with and without 5 μM F-actin and incubated for 30 min at RT. Samples were then centrifuged at 100,000 rpm for 20 min in TLA-120.1 rotor. Supernatant and pellet samples were diluted into equal volumes of Laemmli sample buffer, loaded onto an SDS-PAGE gel and stained with Coomassie blue. Gels were imaged on a LI-COR scanner and band intensity measured and quantified in ImageJ.
Tryptic Digest and Edman Sequencing.
FL HMP-1 (12 μM), murine αE-catenin monomer (12 μm) and chicken vinculin (8 μM) were incubated in 0.05 mg/mL sequence-grade trypsin (Roche) in 20 mM Tris pH 8.0, 150 mM NaCl and 1 mM DTT. Reactions were stopped with 2X Laemmli buffer at the indicated times and samples analyzed by SDS-PAGE. For Edman degradation sequencing, digested peptides were blotted onto PVDF membrane, stained with 0.1% Coomassie R-250/40% methanol/1% acetic acid, destained and dried. Individual bands were excised and sequenced.
Strains and Alleles.
C. elegans strains were cultured using standard protocols (28). Bristol strain N2 was used as wild type. The putative null allele hmp-1(zu278) was originally isolated from an ethane methyl sulfonate (EMS) mutagenesis screen (11). To sequence the allele, homozygous hmp-1(zu278) embryos were isolated based on phenotype and treated briefly with a 1% chitinase solution before lysis with standard single worm lysis buffer. Embryonic lysate was used to PCR amplify hmp-1 and the product was cloned using the TOPO-TA kit (Invitrogen). Subsequent sequencing revealed a nonsense mutation, Gln795Ochre. SU307 (skn-1(zu67)/nT1[qIsI] IV; +/nT1[qIsI] V) males were mated to SU290 (hmp-1(zu278)/lon-3(e2175) V) hermaphrodites to obtain SU370 (+/nT1[qIsI] IV; hmp-1(zu278)/nT1[qIsI] V).
For the in vivo functional analysis, hmp-1:egfp constructs were microinjected (29) at 1 ng/μL along with rol-6(su1006) (79 ng/μL) and noncoding DNA (F35D3, 20 ng/μL) into the gonads of either N2 or SU370 hermaphrodites. A targeted in-frame deletion of the N-terminus was created in the hmp-1:egfp construct by site-directed mutagenesis using the following primers: hmp-1:egfpΔ13-185 (pSM21), SM23REV (5′-GTT GAA ATA CGC ATG AGA ATT GCC-3′) and SM6 (5′-GTT CGA CGA CGA GCC ATT GAT TTG-3′). Extrachromosomal arrays maintained in N2 were subsequently crossed into SU370 hermaphrodites later. Homozygous hmp-1(zu278) embryos rescued to viability were maintained as a separate line. The following strains were created: SU401 (+/nT1(qIsI) IV; hmp-1(zu278)/nT1(qIsI) V; jcEx110 (pJS434(hmp-1:egfp), pRF4(rol-6(su1006)))), SU402 (hmp-1(zu278)/hmp-1(zu278) V; jcEx110), SU419 (jcEx123 (pSM21(hmp-1:egfpΔ13-185), pRF4(rol-6(su1006))), SU446 (+/nT1(qIsI) IV; hmp-1(zu278)/nT1(qIsI) V; jcEx123), SU449 (+/nT1(qIsI) IV; hmp-1(zu278)/nT1(qIsI) V; jcEx124.
Phalloidin and Antibody Staining.
Transgenic embryos were isolated from gravid hermaphrodites with 0.5% NaOCl in 250 mM NaOH for 5 min followed by three washes in M9 buffer. Embryos were aged in M9 for 5 h at RT, washed three times with water, and mounted on poly-L-lysine coated ring slides. For phalloidin staining, embryos were fixed with 4% paraformaldehyde, 0.1 mg/mL lysolecithin, 48 mM Pipes pH 6.8, 25 mM Hepes pH 6.8, 2 mM MgCl2, and 10 mM EGTA for 20 min then washed three times with PBS. Embryos were incubated in the dark with 1∶20 Alexa 455 phalloidin overnight at 4 °C then washed three times with PBS before being covered with SlowFade antifade reagent (Invitrogen) and a coverslip. For antibody staining, embryos were freeze-cracked as described previously (30) and incubated with a rabbit α-HMP-1 polyclonal antibody (1∶4000) overnight at 4 °C, washed 3 times in PBST, then incubated at room temperature with an α-rabbit FITC antibody (1∶50). Slides were washed three times with PBST before being covered in SlowFade and a coverslip.
Supplementary Material
Acknowledgments.
We thank Thomas Weiss and Hiro Tsuruta for assistance with the SAXS measurements, Allyson O’Donnell for help with the yeast two-hybrid analysis, Dan Dickinson for assistance with the F-actin pelleting assays, and members of the Nelson lab for critical feedback on the manuscript. A.V.K was supported by Ruth L. Kirschstein National Research Service Award 5T32 CA09302. S.L.M. was supported by an American Heart Association Predoctoral Fellowship (0815662G). A.M.L. was supported by a Genetics predoctoral training grant [National Institutes of Health (NIH) Grant 5T32 GM07133]. This work was supported by NIH Grants GM058038 (J.H.), GM035527 (W.J.N.), and GM56169 (W.I.W.). Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, supported by the US Department of Energy and the National Institute of General Medical Sciences (NIGMS).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007349107/-/DCSupplemental.
References
- 1.Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol. 2005;6(8):622–634. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- 2.Krendel MF, Bonder EM. Analysis of actin filament bundle dynamics during contact formation in live epithelial cells. Cell Motil Cytoskeleton. 1999;43(4):296–309. doi: 10.1002/(SICI)1097-0169(1999)43:4<296::AID-CM3>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 3.Ehrlich JS, Hansen MD, Nelson WJ. Spatio-temporal regulation of Rac1 localization and lamellipodia dynamics during epithelial cell–cell adhesion. Dev Cell. 2002;3(2):259–270. doi: 10.1016/s1534-5807(02)00216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamada S, Nelson WJ. Localized zones of Rho and Rac activities drive initiation and expansion of epithelial cell–cell adhesion. J Cell Biol. 2007;178(3):517–527. doi: 10.1083/jcb.200701058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rimm DL, Koslov ER, Kebriaei P, Cianci CD, Morrow JS. Alpha 1(E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc Natl Acad Sci USA. 1995;92(19):8813–8817. doi: 10.1073/pnas.92.19.8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobielak A, Fuchs E. Alpha-catenin: At the junction of intercellular adhesion and actin dynamics. Nat Rev Mol Cell Biol. 2004;5(8):614–625. doi: 10.1038/nrm1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abe K, Takeichi M. EPLIN mediates linkage of the cadherin catenin complex to F-actin and stabilizes the circumferential actin belt. Proc Natl Acad Sci USA. 2008;105(1):13–19. doi: 10.1073/pnas.0710504105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamada S, Pokutta S, Drees F, Weis WI, Nelson WJ. Deconstructing the cadherin–catenin–actin complex. Cell. 2005;123(5):889–901. doi: 10.1016/j.cell.2005.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drees F, Pokutta S, Yamada S, Nelson WJ, Weis WI. Alpha-catenin is a molecular switch that binds E-cadherin–beta-catenin and regulates actin-filament assembly. Cell. 2005;123(5):903–915. doi: 10.1016/j.cell.2005.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benjamin JM, et al. AlphaE-catenin regulates actin dynamics independently of cadherin-mediated cell–cell adhesion. J Cell Biol. 2010;189(2):339–352. doi: 10.1083/jcb.200910041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa M, et al. A putative catenin–cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J Cell Biol. 1998;141(1):297–308. doi: 10.1083/jcb.141.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang J, Dokurno P, Tonks NK, Barford D. Crystal structure of the M-fragment of alpha-catenin: Implications for modulation of cell adhesion. EMBO J. 2001;20(14):3645–3656. doi: 10.1093/emboj/20.14.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pokutta S, Drees F, Takai Y, Nelson WJ, Weis WI. Biochemical and structural definition of the l-afadin- and actin-binding sites of alpha-catenin. J Biol Chem. 2002;277(21):18868–18874. doi: 10.1074/jbc.M201463200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pokutta S, Weis WI. Structure of the dimerization and beta-catenin-binding region of alpha-catenin. Mol Cell. 2000;5(3):533–543. doi: 10.1016/s1097-2765(00)80447-5. [DOI] [PubMed] [Google Scholar]
- 15.Borgon RA, Vonrhein C, Bricogne G, Bois PR, Izard T. Crystal structure of human vinculin. Structure. 2004;12(7):1189–1197. doi: 10.1016/j.str.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 16.Bakolitsa C, et al. Structural basis for vinculin activation at sites of cell adhesion. Nature. 2004;430(6999):583–586. doi: 10.1038/nature02610. [DOI] [PubMed] [Google Scholar]
- 17.Bakolitsa C, de Pereda JM, Bagshaw CR, Critchley DR, Liddington RC. Crystal structure of the vinculin tail suggests a pathway for activation. Cell. 1999;99(6):603–613. doi: 10.1016/s0092-8674(00)81549-4. [DOI] [PubMed] [Google Scholar]
- 18.Johnson RP, Craig SW. F-actin binding site masked by the intramolecular association of vinculin head and tail domains. Nature. 1995;373(6511):261–264. doi: 10.1038/373261a0. [DOI] [PubMed] [Google Scholar]
- 19.Johnson RP, Craig SW. An intramolecular association between the head and tail domains of vinculin modulates talin binding. J Biol Chem. 1994;269(17):12611–12619. [PubMed] [Google Scholar]
- 20.Humphries JD, et al. Vinculin controls focal adhesion formation by direct interactions with talin and actin. J Cell Biol. 2007;179(5):1043–1057. doi: 10.1083/jcb.200703036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rudiger M. Vinculin and alpha-catenin: Shared and unique functions in adherens junctions. Bioessays. 1998;20(9):733–740. doi: 10.1002/(SICI)1521-1878(199809)20:9<733::AID-BIES6>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 22.Koslov ER, Maupin P, Pradhan D, Morrow JS, Rimm DL. Alpha-catenin can form asymmetric homodimeric complexes and/or heterodimeric complexes with beta-catenin. J Biol Chem. 1997;272(43):27301–27306. doi: 10.1074/jbc.272.43.27301. [DOI] [PubMed] [Google Scholar]
- 23.Geiger B. A 130 K protein from chicken gizzard: Its localization at the termini of microfilament bundles in cultured chicken cells. Cell. 1979;18(1):193–205. doi: 10.1016/0092-8674(79)90368-4. [DOI] [PubMed] [Google Scholar]
- 24.Lickert H, Bauer A, Kemler R, Stappert J. Casein kinase II phosphorylation of E-cadherin increases E-cadherin/beta-catenin interaction and strengthens cell–cell adhesion. J Biol Chem. 2000;275(7):5090–5095. doi: 10.1074/jbc.275.7.5090. [DOI] [PubMed] [Google Scholar]
- 25.le Duc Q, et al. Vinculin potentiates E-cadherin mechanosensing and is recruited to actin-anchored sites within adherens junctions in a myosin II-dependent manner. J Cell Biol. 2010;189(7):1107–1115. doi: 10.1083/jcb.201001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yonemura S, Wada Y, Watanabe T, Nagafuchi A, Shibata M. alpha-Catenin as a tension transducer that induces adherens junction development. Nat Cell Biol. 2010;12(6):533–542. doi: 10.1038/ncb2055. [DOI] [PubMed] [Google Scholar]
- 27.Barstead RJ, Waterston RH. Vinculin is essential for muscle function in the nematode. J Cell Biol. 1991;114(4):715–724. doi: 10.1083/jcb.114.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- 30.Williams-Masson EM, Malik AN, Hardin J. An actin-mediated two-step mechanism is required for ventral enclosure of the C. elegans hypodermis. Development. 1997;124(15):2889–2901. doi: 10.1242/dev.124.15.2889. [DOI] [PubMed] [Google Scholar]
- 31.Svergun D, Barberato C, Koch MHJ. CRYSOL—A program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr. 1995;28(6):768–773. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.