Abstract
Aminoacyl-tRNA synthetases (aaRSs) are ancient and evolutionary conserved enzymes catalyzing the formation of aminoacyl-tRNAs, that are used as substrates for ribosomal protein biosynthesis. In addition to full length aaRS genes, genomes of many organisms are sprinkled with truncated genes encoding single-domain aaRS-like proteins, which often have relinquished their canonical role in genetic code translation. We have identified the genes for putative seryl-tRNA synthetase homologs widespread in bacterial genomes and characterized three of them biochemically and structurally. The proteins encoded are homologous to the catalytic domain of highly diverged, atypical seryl-tRNA synthetases (aSerRSs) found only in methanogenic archaea and are deprived of the tRNA-binding domain. Remarkably, in comparison to SerRSs, aSerRS homologs display different and relaxed amino acid specificity. aSerRS homologs lack canonical tRNA aminoacylating activity and instead transfer activated amino acid to phosphopantetheine prosthetic group of putative carrier proteins, whose genes were identified in the genomic surroundings of aSerRS homologs. Detailed kinetic analysis confirmed that aSerRS homologs aminoacylate these carrier proteins efficiently and specifically. Accordingly, aSerRS homologs were renamed amino acid:[carrier protein] ligases (AMP forming). The enzymatic activity of aSerRS homologs is reminiscent of adenylation domains in nonribosomal peptide synthesis, and thus they represent an intriguing link between programmable ribosomal protein biosynthesis and template-independent nonribosomal peptide synthesis.
Keywords: aaRS homologs, amino acid:[carrier protein] ligase, noncanonical functions of aaRS, seryl-tRNA synthetase
Aminoacyl-tRNA synthetases (aaRSs) are essential multidomain enzymes that catalyze the specific ATP-dependent aminoacylation of tRNAs. Charged tRNAs then act as substrates for the ribosome during protein biosynthesis (1). In addition to canonical aminoacylation activity, there is increasing evidence that aaRSs are also involved in processes not directly related to the protein synthesis (2). Analysis of completed genomes reveals that many organisms, which possess full length functional aaRSs genes, also display additional ORFs encoding single-domain aaRS-like proteins (3, 4). Some of these enzymes have developed unexpected aminoacylation activity. YadB, a homolog of glutamyl-tRNA synthetase deprived of tRNA-binding domain catalyzes modification of tRNAAsp by transfer of activated glutamate to the anticodon loop (5). A truncated cysS-like gene, identified in actinomycetes (6) encodes an ATP-dependent cysteine-specific ligase MshC, catalyzing attachment of cysteine to carbohydrate intermediates in the mycothiol biosynthesis pathway. The archaeal Asn synthetase, a paralog of AsnRS does not form Asn-tRNAAsn, but produces Asn by amidation of the β-carboxyl group of aspartate with ammonia (7). This Asn paralog lacks the anticodon binding domain and it is structurally and functionally related to bacterial Asn synthetase A (AsnA).
Additional SerRSs, encoded by duplicated serS genes in two Streptomyces species, are required for antibiotic production (8) or resistance (9). These duplicated SerRSs are significantly divergent from canonical housekeeping bacterial SerRSs, but in both cases they have retained their canonical tRNA aminoacylating activity. Herein, we describe and characterize a new class of SerRS-like proteins, whose genes were identified in numerous bacterial genomes. These truncated SerRS homologs are structurally related to the catalytic core of highly diverged, atypical seryl-tRNA synthetases (aSerRS) found only in methanogenic archaea. We have recently shown that methanogenic-type SerRSs display several idiosyncratic structural features and exhibit a different mode of substrate recognition in comparison with bacterial-type SerRSs (10–12). Truncated SerRS homologs are deprived of the tRNA-binding domain and lack canonical tRNA aminoacylating activity. Instead, truncated SerRS homologs catalyze ATP-dependent activation of selected amino acids and their transfer to the phosphopantetheine prosthetic group, covalently attached to carrier proteins of yet unknown function. Furthermore, the genes for these hypothetical carrier proteins colocalize with the genes for truncated SerRS homologs in numerous sequenced genomes. These results suggest that the newly discovered SerRS-like enzymes described here may participate in nonribosomal peptide synthesis (13), where activated precursors and intermediates of structurally diverse peptides assembled in template-independent fashion are tethered by thioester bond to dedicated carrier proteins. A weak thiol acylation activity has been detected for several other aaRSs, and interpreted as a vestige from a thioester world (14, 15) in which ancestral aaRSs provided aminoacyl thioesters for noncoded peptide synthesis. It is therefore tempting to speculate that single-domain SerRS-like proteins are ancestors of both thiol-acylating enzymes needed for nonribosomal peptide synthesis (or related processes) and of class II synthetases required for ribosomal protein synthesis.
Results
Truncated serS-Like Genes Encode Homologs of Methanogenic-Type SerRSs with Different and Relaxed Amino Acid Specificity.
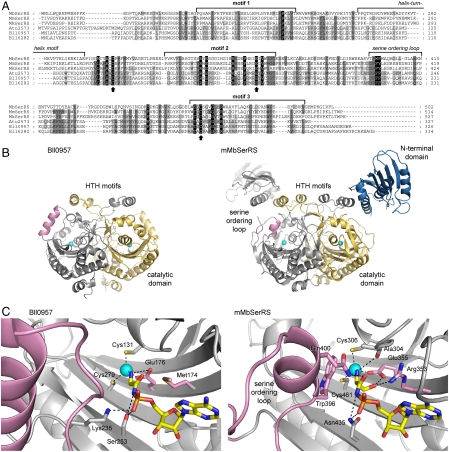
Along with canonical bacterial-like SerRS genes, putative ORFs encoding homologs of atypical archaeal SerRS (aSerRS) were found in the genomes of various bacterial species, predominantly those that belong to Alpha- and Betaproteobacteria, and in some Clostridium, Bacillus, and Streptomyces species (Fig. 1A). A few bacterial species such as Bradyrhizobium japonicum possess two different putative ORFs encoding homologs of aSerRSs. These hypothetical bacterial proteins are homologous to the catalytic domain of atypical SerRSs, but they are shorter and lack the N-terminal tRNA-binding domain. Although the sequence similarity between bacterial homologs and the catalytic core of methanogenic-type SerRS is rather low (15–21% sequence identity or less), the signature motifs of class II aminoacyl-tRNA synthetases are preserved in the sequence of truncated SerRS homologs. Residues that bind the zinc ion in the active site of aSerRS, required for serine binding (10), are strictly conserved in all inspected aSerRS-like sequences. There is a notable lack of sequence conservation in the region corresponding to the helix-turn-helix (HTH) motif of methanogenic-type SerRS from Methanosarcina barkeri (mMbSerRS) (Fig. 1A), which has an important architectural role in mMbSerRS and directs the positioning of tRNA-binding domain relative to the catalytic core (11).
Fig. 1.
Sequence and structure comparison of atypical SerRS from methanogenic archaea and their truncated bacterial homologs. (A) Sequence alignment of representative archaeal aSerRSs and their bacterial homologs characterized in this work. 12 aSerRS sequences and 33 sequences of their bacterial homologs were used to generate the alignment. For clarity, only selected sequences are displayed. Different shades denote positions with 100% (black), 80% (gray), and 60% (light gray) sequence conservation in the complete set. Signature motifs of class II aaRS (motif 1, 2, 3) and unique structural features of aSerRS (HTH motif and serine-ordering loop) are indicated. Black arrows denote strictly conserved residues that bind the zinc ion in the active site of aSerRSs. MbSerRS—Methanosarcina barkeri SerRS (YP_305637); MmSerRS—Methanococcus maripaludis SerRS (AAD03476); MkSerRS—Methanopyrus kandleri SerRS (NP_614743); Bll0957 (NP_767597); and Bll6282 (NP_772922)—two different aSerRS homologs from Bradyrhizobium japonicum (see Results); Atu2573—aSerRS homolog from Agrobacterium tumefaciens (NP_355511.1). National Center for Biotechnology Information Protein accession numbers are given in parenthesis. (B) Comparison of Bll0957 (Left) and mMbSerRS (Right) structures. Two subunits of Bll0957 and corresponding catalytic cores of mMbSerRS are displayed in gray and yellow. Zn2+ in the active site is shown as cyan sphere. The N-terminal domain (blue), which is in mMbSerRS involved in tRNA binding, is absent in Bll0957 structure. Serine-ordering loop in mMbSerRS and corresponding region in Bll0957 are colored pink. (C) Comparison of the active sites of Bll0957 (Left) and mMbSerRS (Right) occupied with aminoacyl-adenylate analogs. The substrate analogs and protein residues responsible for substrate and zinc coordination are shown in ball-and-stick representation and zinc ion as the cyan sphere. Major structural differences in amino acid binding pocket are emphasized in pink. Hydrogen bonds are presented by dashed lines. For clarity, residues interacting with adenylate moiety of the analogs are not shown (see Fig. S2).
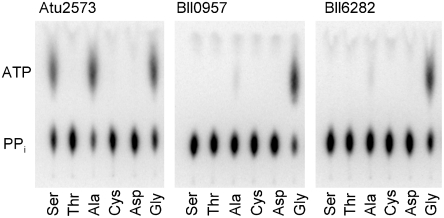
Three representatives of aSerRS homologs were recombinantly produced, purified, and their properties analyzed: hypothetical protein Atu2573 from Agrobacterium tumefaciens, as well as the Bll0957 and Bll6282 proteins from B. japonicum. In spite of weak, but evident sequence similarity to aSerRSs, only A. tumefaciens homolog was able to activate serine in the standard active-site titration assay (16), while the other two homologs from B. japonicum appeared catalytically inactive. This finding was quite puzzling, since all three homologs seemed to be properly folded as evaluated by circular dichroism spectroscopy and have dimeric structure as shown by size-exclusion chromatography. The activation of all 20 standard amino acids by the aSerRS homologs was therefore tested by a complementary method, the amino acid-depended ATP-pyrophosphate exchange assay (16). The results confirmed that only A. tumefaciens homolog is capable of serine activation, but revealed that alanine and glycine are also its substrates (Fig. 2). A strong enzymatic activity of B. japonicum homologs Bll0957 and Bll6282 was detected in the presence of glycine, while a very weak signal was observed upon alanine addition. Activation of amino acids other than serine was completely unexpected, since the amino acid specificity of aaRS is usually evolutionary strictly conserved (17).
Fig. 2.
Amino acid-dependent ATP-pyrophosphate exchange assay. The homologs of aSerRS from A. tumefaciens (Atu2573) and B. japonicum (Bll0957 and Bll6282) were incubated in the presence of ATP, 32P-labeled pyrophosphate (PPi), Mg2+, and different amino acids. Formation of enzyme bound aminoacyl-adenylate intermediate allows incorporation of 32P into ATP. Radioactive products were separated on polyethylenimine-cellulose TLC plates and visualized by PhosphoImager.
A detailed kinetic analysis (Table S1) revealed a strong preference of B. japonicum homologs for glycine activation. Alanine is activated approximately 300-fold less efficiently, while serine was not a substrate for B. japonicum homologs. The homolog from A. tumefaciens (Atu2573) is more relaxed in the amino acid recognition than the enzymes from B. japonicum; it prefers alanine over glycine or serine about 30–40-fold, and activates Glu and Pro (Table S1) albeit more than three orders of magnitude less efficiently than alanine. Thus, the amino acid specificity of the homologs is less stringent than observed for aaRSs. The enzymes were also tested for the activation of D-amino acids (D-Ala, D-Ser, D-Pro, and D-Glu), but no significant activity was detected.
The Structure of Bll0957.
The crystal structures of B. japonicum homolog Bll0957 in complex with AMP, ATP, and GlyAMS were determined and refined at resolutions of 2.15, 2.50, and 2.20 Å, respectively (PDB IDs: 3MF2, 3MEY and 3MF1). The electron density maps of the protein considered as apo-form revealed that both active sites were occupied by an AMP molecule which probably originates from hydrolysis of ATP present in expression host cells. Bll0957 is a homodimer with remarkable resemblance to the catalytic core domain of mMbSerRS structure (10) (rmsd 2.94 Å) despite of only 15% sequence identity (Fig. 1B). The structure shows all three motifs characteristic for class II aaRSs and an active-site zinc ion that is also present in aSerRSs. The motifs 1 and 3 are structurally conserved as in mMbSerRS, while motif 2 responsible for the binding of the 3′ end of tRNA is shorter and consists of two long β-strands (Fig. S1). The Zn2+ found in the active site is tetracoordinated by three conserved amino acids (Cys131, Glu176, and Cys279) and one water molecule in a way characteristic for methanogenic-type SerRS. In addition, a unique insertion found only in aSerRSs that forms a HTH motif is also structurally preserved.
The structure of Bll0957 in three binary complexes is nearly identical (rmsd of Cα atoms between Bll0957:AMP and Bll0957:ATP is 0.28 Å and between Bll0957:AMP and Bll0957:GlyAMS 0.27 Å). The Bll0957:ATP structure confirmed that ATP binds into the active site in the bent conformation in a manner characteristic for class II synthetases (Fig. S2).
In order to examine the recognition of aminoacyl moiety in the active site of Bll0957, a complex with a nonhydrolyzable analog of Gly-AMP, 5′-O-(N-glycyl-sulfamoyl)adenosine, Bll0957:GlyAMS, was prepared. The adenosine moiety of bound GlyAMS shows the same pattern of interactions with the protein as ATP in Bll0957:ATP complex (Fig. S2). The glycine amino group replaces the water molecule coordinated to Zn2+ keeping the metal coordination unchanged (Fig. 1C). The amino group is additionally stabilized by hydrogen bonding to Glu176, a residue that also coordinates the zinc ion. The arginine residue that helps to orient the serine hydroxyl-group in mMbSerRS (Arg353) is replaced by Met174, which has no direct contact with the glycine. Furthermore, Glu176 which coordinates the zinc ion in mMbSerRS (Glu355) but also makes a hydrogen bond with the serine hydroxyl-group has different orientation in Bll0957 (Fig. 1C). The lack of stabilizing interactions between Met174 and Glu176 in Bll0957 and serine hydroxyl-group could explain the shift of amino acid specificity of aSerRS homologs toward small aliphatic amino acids Gly and Ala.
In contrast to the mMbSerRS, no conformational change was observed after binding of GlyAMS analog in the active site of Bll0957. In mMbSerRS, a stretch of 15 amino acids (residues 394–410) completely disordered in the apo-enzyme, becomes ordered after the serine binding (10). This flexible loop, designated as “serine-ordering loop,” comprises two residues in direct contact with serine or Ser-AMS bound in the active site: Gln400 interacts with serine carbonyl oxygen and Trp396 packs above the amino group of serine and acts as a gatekeeper. In all three presented Bll0957 complex structures this segment is replaced with a loop followed by a long helix, which is located further away from the active site and apparently does not participate in amino acid binding (Fig. 1C).
Probing the Transfer of Activated Amino Acids to tRNA.
The ability of aSerRS homologs to activate amino acids, raises the question whether the activated amino acids can be transferred to tRNA. The tRNA aminoacylation activity was tested with bulk, unfractionated tRNA since it was unclear whether aSerRS homologs should charge tRNASer, or the switch in amino acid specificity was accompanied by the change in tRNA selectivity. However, no tRNA aminoacylation could be detected. Furthermore, in an attempt to restore their tRNA aminoacylation activity, N-terminal tRNA-binding domain from mMbSerRS was fused to Atu2573, Bll0957, and Bll6282. The fusion proteins were capable of amino acid activation, but they were still unable to transfer activated amino acid to tRNA.
Beside the absence of tRNA-binding domain, shortened motif 2 loop (fragment Arg159-Gln170 in Bll0957; Fig. 1) is in accordance with the inability of aSerRS homologs to aminoacylate tRNA. In mMbSerRS motif 2 loop plays an important role in the positioning of tRNA 3′-end into the active site (18).
Carrier Protein Thioesterification by aSerRS Homologs.
Inability of aSerRS homologs to aminoacylate tRNA prompted us to search for alternative amino acid acceptors. The genes for aSerRS homologs in numerous genomes show a strong synteny with the genes for small hypothetical proteins (80–100 amino acids; Mr ∼ 10, 000) (Fig. S3), carrying a putative 4′-phosphopantetheine attachment site. Such small proteins act as the carriers of activated precursors, attached to the prosthetic group by thioester bond, in fatty acids and nonribosomal peptide biosynthesis, the synthesis of polyketide antibiotics, etc. (19). Thus, we wondered whether these small hypothetical proteins might act as the carriers for amino acids activated by aSerRS homologs.
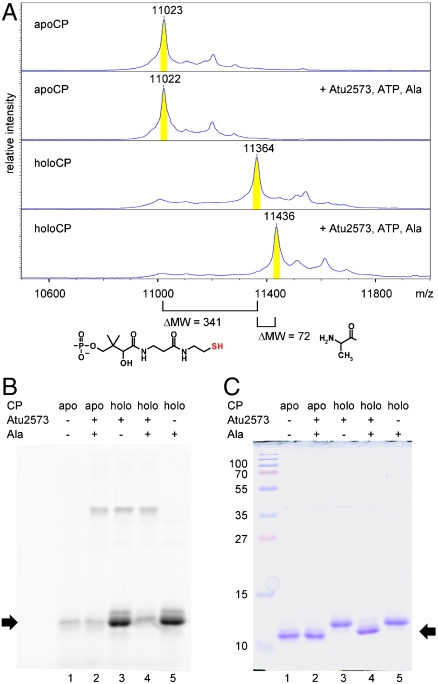
Three putative carrier proteins (CP) residing close to aSerRS homologs Bll0957, Bll6282, and Atu2573 (Bsr0959, Blr6284, and Atu2571, respectively) were cloned and overexpressed in Escherichia coli. The 4′-phosphopantetheinyl prosthetic group was attached to purified apoCPs using broad-specificity phosphopantetheinyl transferase Sfp from Bacillus subtilis (13, 19). Attachment of amino acids to CPs catalyzed by aSerRS homologs was monitored by mass spectrometry (Fig. 3A and Fig. S4). In all three cases, a shift in CP molecular weight corresponding to the Mr of the respective aminoacyl- moiety (58 for Gly, 72 for Ala) was observed when CPs were incubated with the enzyme, ATP, and appropriate amino acid (Fig. 3A, lanes 3 and 4; Fig S4). No change in molecular weight was observed when apoCPs without prosthetic group attached were incubated with the enzymes under the same conditions (Fig. 3A, lanes 1 and 3; Fig. S4), indicating that aSerRS homologs catalyze attachment of amino acids to the 4′-phosphopantetheine arm and thioester bond formation. Amino acid transfer to prosthetic group was further confirmed by treatment of CPs with fluorescein-5-maleimide, a chemical reagent selective for fluorescent labeling of sulfhydryl-containing molecules (20). Since CPs under investigation do not contain any cysteines in their sequence, only CPs bearing a free thiol group in 4′-phosphopantetheine prosthetic arm are fluorescently labeled (Fig. 3B, lanes 3 and 5; Fig. S5). Consequently, after incubation with the enzymes, ATP, and amino acids, aminoacylated CPs were protected from chemical labeling (Fig. 3B, lane 4). After fluorescence visualization, the same gels were stained with Coomassie Brilliant Blue (Fig. 3C and Fig. S5) to ensure that the protein load was comparable.
Fig. 3.
Transfer of alanine to CP catalyzed by A. tumefaciens aSerRS homolog Atu2573. ApoCP—CP Atu2571 with no prosthetic group attached; holoCP—CP with 4′-phosphopantetheine prosthetic group attached. (A) Mass spectrometric analysis of CP products after incubation of CP with A. tumefaciens aSerRS homolog Atu2573, ATP, and alanine (lanes 2 and 4). The mass difference corresponding to 4′-phosphopantetheine prosthetic group (lanes 1 and 3) and aminoacyl- residue (lanes 3 and 4) is readily observed. (B) Labeling of CP with fluorescein-5-maleimide after incubation with Atu2573. ApoCP (lanes 1, 2), or holoCP (lanes 3-5) were incubated under the same conditions, with or without addition of the enzyme (lanes 2-4) and alanine (lanes 2, 4, 5). Samples were treated with fluorescein-5-maleimide and subjected to SDS-PAGE. Labeled proteins were revealed after scanning of fluorescent bands. Black arrow marks position of fluorescently labeled CP. Attachment of amino acid protects CP from flurescein-5-maleimide labeling (lane 4). (C) Coomassie staining of SDS-PAGE gel shown in B. Fluorescent labeling affects electrophoretic mobility of labeled CP (black arrow). Molecular weight of protein markers is indicated on the left.
Determination of Kinetic Parameters for Carrier Protein Aminoacylation.
Detailed kinetic analysis of CP aminoacylation reveals that aSerRS homologs efficiently charge CPs. The affinity of aSerRS homologs for CPs in aminoacylation reaction is high, in micromolar range (Table 1) and the KM values are comparable to KM of aSerRSs for tRNASer (1.54–4.7 μM) (18, 21). These KM values are also in the same range as observed for structurally unrelated adenylation domains exemplified by DltA, the D-alanine:[D-alanyl CP] ligase, from B. subtilis, catalyzing attachment of D-Ala to CP Dcp (KM = 8 μM) (22), and EntE from E. coli (KM = 23 μM) (23), which acylates the CP domain of EntB in siderophore biosynthesis. The turnover numbers for aSerRS homologs (kcat = 0.35 s-1 for Bll0957, 0.1 s-1 for Bll6282, and 0.64 s-1 for Atu2573) determined under saturating amino acid concentrations are also comparable to kcat values reported for aSerRSs in tRNA aminoacylation (0.11–4.4 s-1), albeit they remain lower than turnover number for DltA (816 s-1) or EntE (5.9 s-1). Therefore, catalytic parameters for aSerRS homologs are within reasonable expectations and indicate that CPs may be biologically relevant substrates.
To ascertain that vicinal CPs are relevant biological substrates for aSerRS homologs, they were challenged with acyl CPs (ACPs) from fatty acid biosynthesis. ACPs are archetypal CPs, with similar overall topology and the same prosthetic group as for the CPs found in the genomic context of aSerRS homologs. Thus, ACPs represent a stringent test of CP aminoacylation specificity and proposed biological relevance. Each enzyme was challenged with ACP from its cognate organism and heterologous ACP from E. coli. All ACPs were poor substrates for aSerRS homologs (Table 1) and especially for Bll6282 whose activity in ACP acylation was too low for reliable quantification.
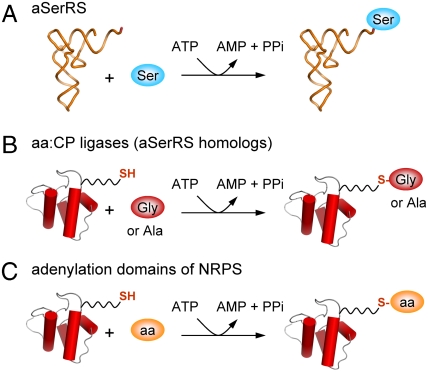
Based on detailed kinetic analysis, we propose that CPs found in genomic neighborhood are cognate and relevant substrates of aSerRS homologs. Therefore, we propose that homologs of atypical SerRS represent a previously uncharacterized family of amino acid:[carrier protein] ligases (AMP forming), abbreviated: aa:CP ligases. The aSerRS homologs catalyze thioester bond formation by amino acid attachment to 4′-phosphopantetheinyl prosthetic group and functionally closely resemble adenylation domains (13) found in nonribosomal peptide synthetases or as stand-alone activating enzymes such as DltA (D-alanine:[D-alanyl carrier protein] ligase) or EntE, catalyzing essentially the same reaction (Fig. 4).
Fig. 4.
Schematic representation of reactions catalyzed by atypical SerRSs (A), amino acid:[CP] ligases (B), and adenylation domains of nonribosomal peptide synthetases (C). aSerRSs aminoacylate tRNA, while aa:CP ligases and adenylation domains attach activated amino acids to prosthetic groups of CPs. Although aa:CP ligases (B) are homologous to aSerRSs (A), the reaction they catalyze resembles those catalyzed by evolutionary and structurally unrelated NRPS adenylation domains (C).
Discussion
Evolutionary Implications of aa:CP Ligases.
Functional differences between archaeal atypical SerRSs and their truncated bacterial homologs, amino acid:[CP] ligases (aa:CP ligases), are summarized in Fig. 4. It is remarkable that aa:CP ligases display both different amino acid and macromolecular acceptor specificity.
A switch in amino acid specificity is unprecedented in the world of aminoacyl-tRNA synthetases. The specificity of these ancient enzymes is thought to be established and fixed in the earliest stages of evolution (17, 24), with exception of GlnRS and AsnRS, which evolved from GluRS and AspRS, respectively. Even the aaRS homologs which do not participate in tRNA aminoacylation for protein biosynthesis, such as YadB [tRNA hypermodification (5)], AsnA [asparagine biosynthesis (7)], HisZ [histidine biosynthesis (25)], or MshC [mycothiol biosynthesis (6)] retained cognate amino acid selectivity. To our knowledge, aa:CP ligases described herein represent the only example of aaRS homologs with diverged amino acid specificity. In this context, it is notable that the two types of SerRSs (bacterial- and methanogenic-type) represent the only aaRSs with different mode of recognition of the same amino acid substrate (10, 26), reflecting their distinct evolutionary origin.
Differences in amino acid specificity of A. tumefaciens and B. japonicum aSerRS homologs (Fig. 2 and Table S1) compared to aSerRS, the lack of tRNA-binding domain, and absence of residual tRNA aminoacylation activity have interesting evolutionary implications. In agreement with the generally accepted view that modern aaRSs originated from primitive catalytic cores capable of amino acid activation by subsequent acquisition of additional domains (27), aa:CP ligases might represent direct descendants of primordial aaRS class II catalytic core of ambiguous amino acid specificity, before it acquired tRNA-binding domain. In another instance, possibly in archaeal lineage, it developed specificity for serine, followed by (or concomitantly with) acquisition of idiosyncratic tRNA-binding domain, giving rise to atypical SerRSs. Different and relaxed amino acid specificity of aa:CP ligases Bll0957, Bll6282, and especially Atu2573, intolerable in protein biosynthesis, could reflect ambiguous amino acid specificity of ancestral aaRS class II catalytic core since it was not imposed to selective pressure to achieve the high level of amino acid specificity characteristic for modern aaRS. On the other hand, it is possible, that aa:CP ligases arose by aSerRS gene duplication or horizontal gene transfer, followed by loss of tRNA-binding domain, change of amino acid selectivity and reassignment to a different biological role. However, this scenario seems less probable because it would require “a double switch”: the change in amino acid selectivity and the switch from tRNA to CP aminoacylation.
Possible Roles of aa:CP Ligases and Aminoacylated CPs.
The ultimate cellular fate of products of newly discovered aa:CP ligases is currently unknown. The identification of CPs as substrates for aSerRS homologs was fostered by their genomic surroundings (Fig. S3), but genetic context provides no clear indications about further fate of aminoacylated CPs. However, it is curious that aa:CP ligase (designated as ORF2 of unknown function), was found close to valinomycin biosynthetic gene cluster in Streptomyces tsusimaensis ATCC 15141 (28).
CPs participate as cofactors in diverse biological processes that require acyl transfer steps, such as fatty acid and polyketide synthesis, nonribosomal peptide synthesis, synthesis of membrane-derived oligosaccharides, etc (19). In rhizobia three different CPs (NodF, AcpXL, and RkpF) are involved in synthesis of cell-surface molecules and nodulation factors required for establishing a symbiotic relationship with their legume hosts (29). DltA, the D-alanine:[D-alanyl CP] ligase, catalyzes attachment of D-alanine to corresponding CP in initial step of D-alanylation of lipoteichoic acids in the cell walls of Gram-positive bacteria (22). Clearly, aa:CP ligases and their aminoacylated CP substrates could play similar roles.
It is worth mentioning that aminoacyl-tRNAs function as donors of activated amino acids outside the protein synthesis as well (e.g., in porphyrin or phospholipid synthesis, peptidoglycan crosslinking; 30–32). Recently, it was shown that VlmL, a dedicated seryl-tRNA synthetase, and Ser-tRNASer participate in the biosynthesis of valanimycin, azoxy antibiotic produced by Streptomyces viridifaciens (8). Aminoacyl-CPs synthesized by aa:CP ligases may represent a substitute for aa-tRNAs as amino acid donors in such processes outside the protein biosynthesis.
aa:CP Ligases—A Missing Link Between Coded Protein Synthesis and Thioester-Dependent Peptide Synthesis?
AaRSs and adenylation domains found in nonribosomal peptide synthetases (NRPS) catalyze a mechanistically similar two-step reaction, with aminoacyl-adenylate intermediates: the substrate amino acid is activated at the expense of ATP, and subsequently transferred to a suitable macromolecular carrier (13) (Fig. 4). Moreover, the function of aaRS and adenylation domains is analogous, because they provide activated precursors for protein synthesis or noncoded thioester-dependent peptide synthesis. In spite of mechanistical and functional analogies, no evolutionary or structural relation between aaRS and NRPS adenylation domains have been determined thus far (33).
Homologs of aSerRSs, described in this paper, possess all the structural features unique to class II aminoacyl-tRNA synthetases, share striking structural homology with mMbSerRS, and yet they have no tRNA aminoacylation activity. Instead, aSerRS homologs act as functional amino acid:[CP] ligases, catalyzing exactly the same reaction as adenylation domains in NRPS (Fig. 4). As such, aSerRS homologs might represent molecular fossils that link template based ribosomal protein synthesis and template-independent nonribosomal peptide synthesis with interesting implications considering coevolution of these fundamentally different systems for peptide synthesis.
Materials and Methods
Materials.
Bradyrhizobium japonicum USDA110 was obtained from USDA ARS Culture Collection. Agrobacterium tumefaciens C58C1, a derivative of sequenced strain C58, was from laboratory stocks. All proteins were His-tagged and purified by standard procedures. Bulk tRNA was isolated from A. tumefaciens and B. japonicum by phenol extraction and purified by chromatography on DEAE-cellulose. The details of protein and tRNA preparation are given in SI Text.
Crystallization and Structure Determination of Bll0957.
Crystals of purified Bll0957 were obtained in a hanging drop vapor diffusion experiment by equilibrating 1 μL of protein solution (21 mg/mL) and 1 μL of reservoir solution (0.17 M NH4OAc, 0.085 M NaOAc pH 4.6, 25.5% (w/v) PEG 4K and 15% (v/v) glycerol) at 18 °C. Bll0957:ATP and Bll0957:GlyAMS complexes were obtained by soaking Bll0957 crystals in the ligand solution. The crystal structure of Bll0957 was determined using the single-wavelength anomalous dispersion of zinc atoms. Data collection and refinement are described in SI Text, relevant statistics are given in Table S2. The coordinates of Bll0957 structures have been deposited in the Protein Data Bank (accession codes: 3MF2, AMP complex; 3MEY, ATP complex; 3MF1, GlyAMS complex).
Enzymatic Assays and Determination of Kinetic Parameters.
ATP-PPi exchange assay was performed at 30 °C in 50 mM TrisHCl pH 7.5, 150 mM KCl, 0.2 mg/mL BSA, 4 mM DTT, 20 mM MgCl2, 4 mM ATP, and 1 mM [32P] - PPi, and varying concentrations of amino acids. Reaction was quenched, products were separated on polyethylenimine-cellulose TLC plates and quantified by phosphorimaging (16).
Aminoacylation of CPs was monitored by assay similar to standard tRNA aminoacylation assay (16). Reaction was performed at 30 °C in 50 mM TrisHCl pH 7.5, 150 mM KCl, 0.4 mg/mL BSA, 10 mM MgCl2, 4 mM ATP, 200 μM [U - 14C]-Ala or [U - 14C]-Gly, 5 U/mL inorganic pyrophosphatase and varying concentrations of CPs. Aliquots were spotted on paper filters presoaked with trichloroacetic acid. The paper filters were washed, dried, and radioactive products were quantified by scintillation counting.
All determinations of kinetic parameters were repeated at least three times.
Fluorescent labeling and MS analysis of aminoacylated CPs is described in SI Text.
Supplementary Material
Table 1.
Kinetic parameters of aSerRS homologs for CP aminoacylation
| KM (μM) | kcat (s-1) | kcat/KM (s-1 M-1) | kcat/KM (rel.) | ||
| Bll0957 | vicinal CP | 1.19 ± 0.04 | 0.11 ± 0.02 | (91 ± 7)·103 | 1 |
| Bj ACP | - | - | 29 ± 3 | 3.1·10-4 | |
| Ec ACP | - | - | 37 ± 2 | 4.1·10-4 | |
| Bll6282 | vicinal CP | 3.9 ± 0.3 | 0.093 ± 0.006 | (23.91 ± 0.04)·103 | 1 |
| Bj ACP | - | - | - | - | |
| Ec ACP | - | - | - | - | |
| Atu2573 | vicinal CP | 4.7 ± 0.5 | 0.31 ± 0.01 | (68 ± 7)·103 | 1 |
| At ACP | - | - | 11.0 ± 0.2 | 1.6·10-4 | |
| Ec ACP | - | - | 16.5 ± 0.6 | 2.5·10-4 |
Enzymes were assayed under nonsaturating conditions with respect to amino acids (200 μM Gly for Bll0957 and Bll6282, 200 μM Ala for Atu2573). Abbreviations: At ACP, Bj ACP, or Ec ACP—acyl CP from A. tumefaciens, B. japonicum, or E. coli respectively; vicinal CP—cognate CP found in genomic vicinity of respective aSerRS homolog. Values are given as arithmetic mean ± standard error of mean (SEM).
Acknowledgments.
We are grateful to Deni Subasic for help in experiments with fusion proteins of mMbSerRS N-terminal domain and aSerRS homologs and to Dragana Ahel for valuable discussions. We are indebted to Eva Schaub for providing purified Sfp. The experimental data for Bll0957:AMP complex were collected during the EMBO Practical Course (X-ray Crystal Structure Determination of Macromolecules) held at the Synchrotron Soleil, Saint Aubin, France, September 14-20, 2008. This work was supported by grants from the Unity through Knowledge Fund (UKF, project 10/07), the Ministry of Science, Education and Sports of the Republic of Croatia (projects 119-0982913-1358 and 098-1191344-2943), Swiss National Science Foundation (SNSF) and the National Center of Excellence in Research Structural Biology program of the SNSF.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3MF2, 3MEY, and 3MF1).
See Commentary on page 14517.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007470107/-/DCSupplemental.
References
- 1.Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Park SG, Ewalt KL, Kim S. Functional expansion of aminoacyl-tRNA synthetases and their interacting factors: new perspectives on housekeepers. Trends Biochem Sci. 2005;30:569–574. doi: 10.1016/j.tibs.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Schimmel P, Ribas de Pouplana L. Footprints of aminoacyl-tRNA synthetases are everywhere. Trends Biochem Sci. 2000;25:205–207. doi: 10.1016/s0968-0004(00)01553-x. [DOI] [PubMed] [Google Scholar]
- 4.Geslain R, Ribas de Pouplana L. Regulation of RNA function by aminoacylation and editing? Trends Genet. 2004;20:604–610. doi: 10.1016/j.tig.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 5.Blaise M, et al. Glu-Q-tRNAAsp synthetase coded by the yadB gene, a new paralog of aminoacyl-tRNA synthetase that glutamylates tRNAAsp anticodon. Biochimie. 2005;87:847–861. doi: 10.1016/j.biochi.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Sareen D, Steffek M, Newton GL, Fahey RC. ATP-dependent L-cysteine:1D-myo-inosityl 2-amino-2-deoxy-alpha-D-glucopyranoside ligase, mycothiol biosynthesis enzyme MshC, is related to class I cysteinyl-tRNA synthetases. Biochemistry. 2002;41:6885–6890. doi: 10.1021/bi012212u. [DOI] [PubMed] [Google Scholar]
- 7.Roy H, Becker HD, Reinbolt J, Kern D. When contemporary aminoacyl-tRNA synthetases invent their cognate amino acid metabolism. Proc Natl Acad Sci USA. 2003;100:9837–9842. doi: 10.1073/pnas.1632156100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garg RP, Qian XL, Alemany LB, Moran S, Parry RJ. Investigations of valanimycin biosynthesis: elucidation of the role of seryl-tRNA. Proc Natl Acad Sci USA. 2008;105:6543–6547. doi: 10.1073/pnas.0708957105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng Y, Roy H, Patil PB, Ibba M, Chen S. Characterization of two seryl-tRNA synthetases in albomycin-producing Streptomyces sp. strain ATCC 700974. Antimicrob Agents Chemother. 2009;53:4619–4627. doi: 10.1128/AAC.00782-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bilokapic S, et al. Structure of the unusual seryl-tRNA synthetase reveals a distinct zinc-dependent mode of substrate recognition. EMBO J. 2006;25:2498–2509. doi: 10.1038/sj.emboj.7601129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bilokapic S, et al. Idiosyncratic helix-turn-helix motif in Methanosarcina barkeri seryl-tRNA synthetase has a critical architectural role. J Biol Chem. 2009;284:10706–10713. doi: 10.1074/jbc.M808501200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaric J, et al. Identification of amino acids in the N-terminal domain of atypical methanogenic-type seryl-tRNA synthetase critical for tRNA recognition. J Biol Chem. 2009;284:30643–30651. doi: 10.1074/jbc.M109.044099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finking R, Marahiel MA. Biosynthesis of nonribosomal peptides. Annu Rev Microbiol. 2004;58:453–488. doi: 10.1146/annurev.micro.58.030603.123615. [DOI] [PubMed] [Google Scholar]
- 14.Jakubowski H. Aminoacylation of coenzyme A and pantetheine by aminoacyl-tRNA synthetases: possible link between noncoded and coded peptide synthesis. Biochemistry. 1998;37:5147–5153. doi: 10.1021/bi972528v. [DOI] [PubMed] [Google Scholar]
- 15.Jakubowski H. Amino acid selectivity in the aminoacylation of coenzyme A and RNA minihelices by aminoacyl-tRNA synthetases. J Biol Chem. 2000;275:34845–34848. doi: 10.1074/jbc.C000577200. [DOI] [PubMed] [Google Scholar]
- 16.Francklyn CS, First EA, Perona JJ, Hou Y-M. Methods for kinetic and thermodynamic analysis of aminoacyl-tRNA synthetases. Methods. 2008;44:100–118. doi: 10.1016/j.ymeth.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woese CR, Olsen GJ, Ibba M, Söll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol Mol Biol R. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bilokapic S, Rokov Plavec J, Ban N, Weygand-Durasevic I. Structural flexibility of the methanogenic-type seryl-tRNA synthetase active site and its implication for specific substrate recognition. FEBS J. 2008;275:2831–2844. doi: 10.1111/j.1742-4658.2008.06423.x. [DOI] [PubMed] [Google Scholar]
- 19.Mercer AC, Burkart MD. The ubiquitous carrier protein—a window to metabolite biosynthesis. Nat Prod Rep. 2007;24:750–773. doi: 10.1039/b603921a. [DOI] [PubMed] [Google Scholar]
- 20.Hermanson GT. Bioconjugate Techniques. 2nd ed. London: Academic Press; 2008. pp. 183–184.pp. 408–410. [Google Scholar]
- 21.Gruic-Sovulj I, Jaric J, Dulic M, Cindric M, Weygand-Durasevic I. Shuffling of discrete tRNASer regions reveals differently utilized identity elements in yeast and methanogenic archaea. J Mol Biol. 2006;361:128–139. doi: 10.1016/j.jmb.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 22.May JJ, et al. Inhibition of the D-alanine:D-alanyl carrier protein ligase from Bacillus subtilis increases the bacterium’s susceptibility to antibiotics that target the cell wall. FEBS J. 2005;272:2993–3003. doi: 10.1111/j.1742-4658.2005.04700.x. [DOI] [PubMed] [Google Scholar]
- 23.Drake EJ, Nicolai DA, Gulick AM. Structure of the EntB multidomain nonribosomal peptide synthetase and functional analysis of its interaction with the EntE adenylation domain. Chem Biol. 2006;13:409–419. doi: 10.1016/j.chembiol.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 24.O’Donoghue P, Luthey-Schulten Z. On the evolution of structure in aminoacyl-tRNA synthetases. Microbiol Mol Biol R. 2003;67:550–573. doi: 10.1128/MMBR.67.4.550-573.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sissler M, et al. An aminoacyl-tRNA synthetase paralog with a catalytic role in histidine biosynthesis. Proc Natl Acad Sci USA. 1999;96:8985–8990. doi: 10.1073/pnas.96.16.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahel D, Slade D, Mocibob M, Söll D, Weygand-Durasevic I. Selective inhibition of divergent seryl-tRNA synthetases by serine analogues. FEBS Lett. 2005;579:4344–4348. doi: 10.1016/j.febslet.2005.06.073. [DOI] [PubMed] [Google Scholar]
- 27.Ribas de Pouplana L, Schimmel P. A view into the origin of life: aminoacyl-tRNA synthetases. Cell Mol Life Sci. 2000;57:865–870. doi: 10.1007/PL00000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng YQ. Deciphering the biosynthetic codes for the potent anti-SARS-CoV cyclodepsipeptide valinomycin in Streptomyces tsusimaensis ATCC 15141. ChemBioChem. 2006;7:471–477. doi: 10.1002/cbic.200500425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geiger O, López-Lara IM. Rhizobial acyl carrier proteins and their roles in the formation of bacterial cell-surface components that are required for the development of nitrogen-fixing root nodules on legume hosts. FEMS Microbiol Lett. 2002;208:153–162. doi: 10.1111/j.1574-6968.2002.tb11075.x. [DOI] [PubMed] [Google Scholar]
- 30.RajBhandary UL, Söll D. Aminoacyl-tRNAs, the bacterial cell envelope, and antibiotics. Proc Natl Acad Sci USA. 2008;105:5285–5286. doi: 10.1073/pnas.0801193105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ibba M, Söll D. Aminoacyl-tRNAs: setting the limits of the genetic code. Genes Dev. 2004;18:731–738. doi: 10.1101/gad.1187404. [DOI] [PubMed] [Google Scholar]
- 32.Francklyn CS, Minajigi A. tRNA as an active chemical scaffold for diverse chemical transformations. FEBS Lett. 2010;584:366–375. doi: 10.1016/j.febslet.2009.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weber T, Marahiel MA. Exploring the domain structure of modular nonribosomal peptide synthetases. Structure. 2001;9:R3–R9. doi: 10.1016/s0969-2126(00)00560-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.