Abstract
The innate immune response in Drosophila involves the inducible expression of antimicrobial peptide genes mediated by the Toll and IMD signaling pathways. Dorsal and DIF act downstream of Toll, whereas Relish acts downstream of IMD to regulate target gene expression. Dorsal, DIF, and Relish are NF-κB-related transcription factors and function as obligate dimers, but it is not clear how the various dimer combinations contribute to the innate immune response. We systematically examined the dimerization tendency of these proteins through the use of transgenic assays. The results show that all combinations of homo- and heterodimers are formed, but with varying degrees of efficiency. The formation of the DIF-Relish heterodimer is particularly interesting because it may mediate signaling for the seemingly independent Toll and IMD pathways. By incorporating a flexible peptide linker, we specifically tested the functions of the DIF^Relish (a ^ sign represents the peptide linker) linked heterodimer. Our results demonstrate that the linked heterodimer can activate target genes of both the Toll and IMD pathways. The DIF and Relish complex is detectable in whole animal extracts, suggesting that this heterodimer may function in vivo to increase the spectrum and level of antimicrobial peptide production in response to different infections.
Keywords: IMD, innate immunity, Toll
The self-defense response against microbial infection in Drosophila is similar to the innate immune response in mammals (1, 2). An important aspect of the innate immune response in Drosophila is the production of antimicrobial peptides (3). This appears to be a conserved self-defense mechanism, because mammalian neutrophils, macrophages, and intestinal cells also produce antimicrobial peptides (4).
The inducible expression of antimicrobial peptides in Drosophila is controlled by the Toll and IMD signal transduction pathways (1, 2). Gram-positive bacterial peptidoglycan or fungal glucan binds to upstream pattern recognition proteins and activates a protease cascade, which causes cleavage of the host protein Spätzle (5–6). Cleaved Spätzle acts as a ligand for the receptor Toll and stimulates the assembly of the receptor–adaptor complex (7–9). These signaling events target the transcription factors Dorsal and DIF, as well as the inhibitor Cactus, which are proteins related to those within the mammalian NF-κB/IκB complex (10–13). Toll signaling increases the nuclear localization of Dorsal and DIF, which in turn bind to κB motifs on the promoters of antimicrobial peptide genes (3, 13).
A number of peptidoglycan recognition proteins (PGRPs) act as receptors for Gram-negative bacterial peptidoglycan. The adaptor protein IMD interacts with the receptor and signals to downstream components including TAK1, TAB2, DIAP2, JNK, FADD, and other newly identified regulators (1, 2, 14, 15). These regulators converge signals onto the transcription factor Relish, the third Drosophila NF-κB-related protein that is homologous to NF-κB1 (p105) in mammals. Relish is cleaved during signal stimulation and the N-terminal fragment translocates to the nucleus (16). Once in the nucleus, Relish regulates a different subset of antimicrobial peptide genes (11, 17).
All NF-κB–related proteins contain the conserved Rel homology domain that is required for DNA binding and dimerization (13, 18). However, the relative extent of Dorsal, DIF, and Relish homo- and heterodimer formation and how these various dimers contribute to the observed immune response are not clear. In this study, we show that all homo- and heterodimer combinations are possible and that the DIF-Relish heterodimer can mediate signaling of both Toll and IMD pathways as a mechanism for activating the innate immune response.
Results
Dorsal, DIF, and Relish Dimer Combinations Are Detectable in a Transgenic Assay.
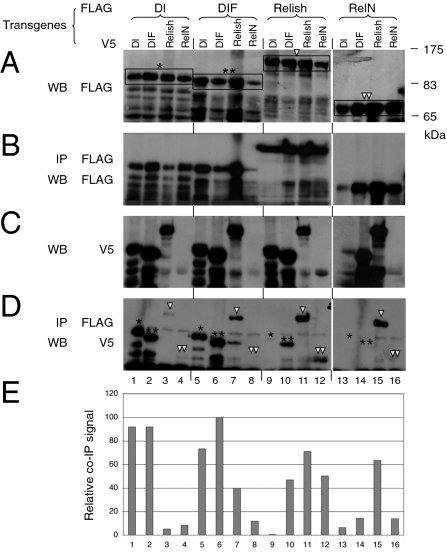
To evaluate the relative tendency of homodimer and heterodimer formation among the three NF-κB-related proteins, we systemically expressed these proteins with epitope tags in transgenic flies for use in a coimmunoprecipitation assay. The yolk protein 1 (YP1)-Gal4 driver was used to direct protein expression in adult female fat bodies, which are a major organ for antimicrobial peptide production. Western blots of whole fly extracts showed that the 3XFLAG- and V5-tagged Dorsal, DIF, and Relish were expressed at similar levels (Fig. 1 A and C). The major protein products matched the predicted sizes of the full-length proteins (indicated in brackets in Fig. 1A). Although we observed a consistently lower expression level of RelN (Fig. 1C, lanes 4, 8, and 12), previous studies have demonstrated that transgenic expression of RelN is sufficient for the activation of relevant target genes (17, 19).
Fig. 1.
Transgenic assay for dimerization of Drosophila NF-κB–related proteins. Transgenic flies all contained FLAG and V5 epitope-tagged constructs as indicated. (A) Whole fly extracts from the transgenic lines were analyzed by Western blot (WB) using an anti-FLAG antibody. The major protein bands of correct size were identified by the brackets and indicated with an asterisk for Dorsal (Dl), two asterisks for DIF, a triangle for Relish, and two triangles for RelN. These same notations are used in D to indicate the relevant protein bands in the immunoprecipitates. (B) The immunoprecipitated extracts were used for WB using the anti-FLAG antibody to assess the efficiency of immunoprecipitation (IP). (C) Whole extracts were analyzed by WB for protein expression using the V5 antibody. (D) Immunoprecipitated extracts were analyzed by Western blot using the V5 antibody to assess the efficiency of co-IP. (E) Quantification of D by measuring the intensity of the relevant bands indicated by the asterisks and triangles shown in D. The signals are normalized to that of the DIF-DIF homodimer (lane 6), which was set as 100%. The RelN blots shown in A, B, and C (lanes 13–16) were a longer exposure to show the expression levels.
We next assessed for the presence and relative levels of homo- and heterodimeric complexes in transgenic fly extracts by using a coimmunoprecipitation assay. In agreement with previous data showing that endogenous Dorsal can form homodimers (13), the V5-Dorsal protein clearly immunoprecipitated with 3XFLAG-Dorsal (Fig. 1D, lane 1). Using the highest signal of V5-DIF and 3XFLAG-DIF homodimer as a reference (100%) (Fig. 1E, lane 6), we estimated that the Dorsal and Relish homodimers formed with a similar efficiency, at approximately 90% and 70%, respectively, of the DIF homodimer (Fig. 1 D and E, lanes 1, 6, and 11).
The Dorsal-DIF heterodimer also formed efficiently, at approximately 80% of the DIF homodimer, whereas the DIF-Relish heterodimer formed at an intermediate efficiency of approximately 40%. In contrast, the Dorsal-Relish heterodimer formed much less efficiently at less than 7% (Fig. 1E). The reciprocal combination of the two tagged proteins yielded similar efficiencies of formation (e.g., Fig. 1 D and E, lanes 2 and 5 for Dorsal-DIF and DIF-Dorsal). These results demonstrate that all homodimer and heterodimer combinations of these proteins are possible but with varying degrees of efficiency.
RelN contains the N-terminal half and resembles the proteolytically cleaved and active form of Relish (17, 19). We observed that the tagged RelN in our coimmunoprecipitation assays had a strong tendency to dimerize with Relish but did not exhibit the same tendency toward Dorsal or DIF (lanes 13–16). Therefore, in these assays, RelN exhibited a different binding pattern than Relish for the interaction with DIF. However, we did not explore this difference further because overexpression of RelN may not be representative of the endogenous situation. Under physiologic conditions, full length Relish is produced inside the cell. The RelN homodimer should originate from the cleavage of the Relish homodimer, whereas the DIF-RelN heterodimer does not form de novo and should be derived from the preexisting DIF-Relish heterodimer.
Expression of Covalently Linked Homo- and Heterodimers.
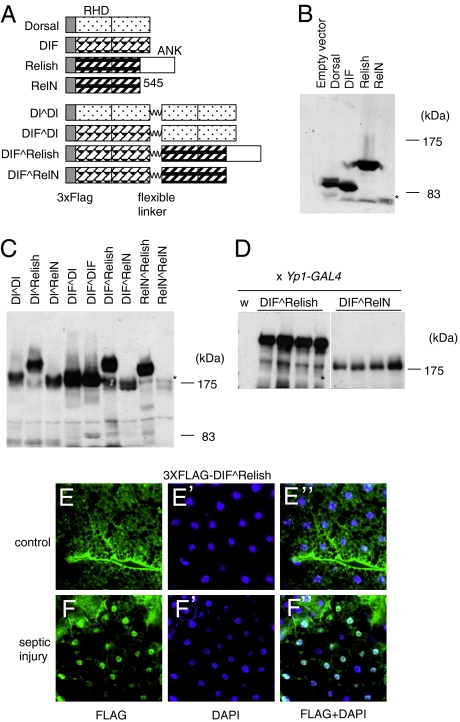
In consideration of all of the possible dimer combinations, the DIF-Relish heterodimer is the most interesting because it has a potential role in mediating cross-talk between the Toll and IMD pathways. To perform a more detailed analysis of the DIF-Relish heterodimer, we first generated constructs that would express covalently linked proteins containing a flexible peptide linker (Fig. 2A). The analysis of a linked protein should give a more definitive assessment of the heterodimer activity because the cotransfection of two nonlinked constructs will yield a mixture of homo- and heterodimers. The 20-aa peptide linker used for these constructs was derived from the gene-3-protein of the phage fd. The peptide has a linear length of approximately 70 Å, and it has been successfully used to link multimeric complexes for use in in vivo and in vitro assays (20). All combinations were constructed, with the exception of the Relish^Relish (a ^ sign represents the peptide linker), because it was possible that the first Relish protein could be cleaved and left with a nonlinked protein. The linked constructs all produced stable proteins of predicted size in S2 cells (Fig. 2 B and C). In addition, all of the proteins containing RelN were again expressed at lower levels (indicated by an asterisk in Fig. 2 B and C).
Fig. 2.
Construction and expression of linked dimers of NF-κB-related proteins. (A) An illustration of the various proteins and four representative linked constructs are included here. All constructs contain the 3XFLAG epitope at their N termini. RHD represents Rel Homology Domain. ANK is ankyrin repeats in the C terminus of Relish. RelN contains the N-terminal half of Relish and ends at amino acid 545, which is the natural protease cleavage site. The flexible linker (wavy line) is a 20-aa polypeptide from the gene-3-protein of phage fd. (B) A Western blot using the anti-FLAG antibody to show the expression of nonlinked proteins in transfected S2 cells. The asterisk indicates the RelN protein band, which had a lower expression level than the other three proteins. (C) A Western blot showing the expression of the linked proteins in transfected S2 cells. The asterisk indicates the RelN^RelN protein band, which had a lower expression level than the other proteins. (D) A Western blot showing DIF^Relish and DIF^RelN expression in four independent transgenic lines. The transgenic lines were crossed with the YP1-Gal4 driver line. The control extract from wild-type flies (W) showed no signal. (E and F) The subcellular localization of the DIF^Relish dimer in larval fat bodies. Mixed bacteria of S. aureus and E. coli were used for septic injury. The blue signal is DAPI staining of DNA and the green signal is the FLAG epitope. E–E’’ are fat body staining from unchallenged transgenic larvae, and F–F’’ are fat body staining from transgenic larvae 2 h after septic injury.
Transgenic flies were also generated that harbored the DIF^Relish and DIF^RelN constructs under the control of the UAS enhancer. Four independent lines showed stable protein expression as assayed by Western blot of whole fly extracts (Fig. 2D). The DIF^Relish protein had a higher steady state level than the DIF^RelN protein, which was consistent with the transfection results. We also observed the appearance of bands that resembled nonlinked NF-κB proteins in the transgenic fly or transfected cell extracts. It is possible that low-level cleavage of the linker or another mechanism, such as aberrant translation, may lead to the formation of nonlinked proteins. Nevertheless, the functional studies performed in this study suggest that low levels of these nonlinked proteins did not alter interpretation of the results.
We also examined the subcellular localization of the DIF^Relish heterodimer, whose expression was driven by the Cg-Gal4 in larval fat bodies (21). In unchallenged larvae, the anti-FLAG antibody revealed staining throughout the fat body cells (Fig. 2 E). However, in larvae subjected to septic injury, the staining was more concentrated in the nuclei (Fig. 2 F). Approximately 20% of the fat body nuclei of challenged larvae had markedly stronger staining, whereas less than 1% showed nuclear staining in the control fat bodies. These results support the hypothesis that DIF^Relish behaves similarly to other proteins of the NF-κB family.
DIF^RelN Linked Heterodimer Is a Potent Activator That Acts Through a Canonical κB Site.
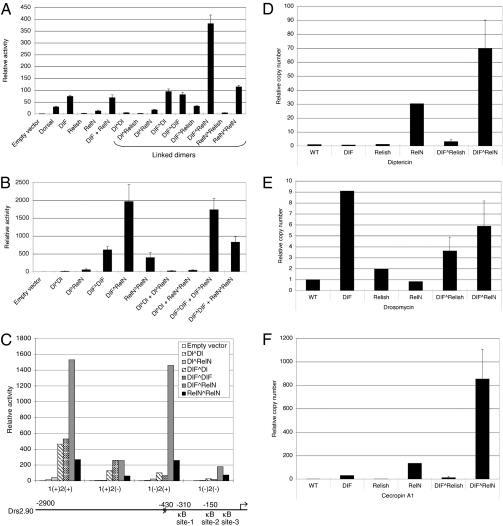
The activities of the linked dimer constructs were assayed by transient transfection in S2 cells without ligand stimulation. Transfection of wild-type Dorsal, DIF, and Relish constructs gave similar results as previously shown. DIF expression induced high expression of the Drosomycin promoter-luciferase reporter (75-fold), whereas Relish expression only caused a modest increase (fivefold) (Fig. 3A). Among all of the linked dimer constructs, DIF^RelN induced a particularly strong activation of the Drosomycin reporter (350-fold). However, DIF^Relish exhibited very low activity, which is consistent with the hypothesis that full-length Relish contains a C-terminal inhibitory domain and is normally retained in the cytoplasm (Fig. 2 E and F). In addition, the very high DIF^RelN activity that was observed was not recapitulated by cotransfection of DIF and RelN as separate plasmids. We hypothesize that only a small proportion of heterodimers are formed when DIF and RelN are coexpressed from individual plasmids (Fig. 1) and therefore the low level of heterodimer that is formed is not sufficient for reproducing the DIF^RelN activity.
Fig. 3.
Activation of Toll and IMD pathway target genes by the linked dimers. (A) Luciferase assays were performed using extracts of S2 cells transfected with the plasmids as indicated. The luciferase activities were normalized using β-galactosidase activity of a cotransfected LacZ plasmid. The normalized activity of the UAS empty vector was then set at one and the activities of other constructs were calculated and plotted as relative activity. Error bars represent SD. The Drosomycin-luciferase reporter used in these assays contained 2.9 kb of upstream sequence (C Lower). (B) Similar luciferase assays for the cotransfection of the various linked dimers as indicated, using a 0.43-kb Drosomycin reporter, which has the κB site-3 mutated and site-1 and site-2 intact [1(+)2(+)]. The activity of the empty vector was set to one and used to calculate the relative activity of the other samples. (C) Luciferase assays using the Drosomycin-luciferase reporter with the κB site mutated and the various linked dimers. All of the luciferase reporters contained the same 0.43 kb of upstream sequence and had site-3 mutated. For example, the 1(+)2(−) construct contained a wild-type site-1 and mutated site-2 and 3. The plasmids encoding the linked dimers for transfection are indicated (Inset). The activity of the 1(+)2(+) template with the empty UAS vector was set as 1 and the relative activities of all other samples were plotted against this value. (D to F) The RT-PCR results of three endogenous antimicrobial peptide genes: Diptericin, Drosomycin, and CecropinA1. The mRNA isolated from control flies (WT) and transgenic lines as indicated after crossing with YP1-Gal4 were used as templates for quantitative RT-PCR. The RT-PCR result of each gene in a sample was normalized with that of the rp49. The level of each antimicrobial peptide gene in WT flies was then set to 1 and the other samples were plotted relative to this level as shown.
We next performed a series of experiments using the linked dimers to test whether the linked DIF^RelN was able to interact with other NF-κB–related proteins. Cotransfection of DIF^RelN and DIF^DIF resulted in similar activity as DIF^RelN alone, suggesting that the functions of DIF^RelN are not augmented by the presence of DIF^DIF (Fig. 3B). Moreover, cotransfection of DIF^DIF and RelN^RelN had only an additive effect, which is consistent with the idea that these two dimers occupy two independent binding sites on the Drosomycin promoter (22). Overall, these results suggest that DIF^RelN functions as a self-contained dimer and that overexpression of other NF-κB proteins does not enhance its function.
Previous results have shown that the Drosomycin proximal promoter contains three κB sites, with site-1 and site-2 being essential and site-3 not being essential (Fig. 3C) (22). Therefore, we assessed the activities of the DIF^RelN linked dimer on the wild-type and mutated promoter. When site-1 was mutated [template 1(−)2(+)], the DIF^RelN-stimulated activity remained the same (Fig. 3C). In contrast, when site-2 was mutated [template 1(+)2(−)], the DIF^RelN-stimulated activity was reduced by 80%. Therefore, the DIF^RelN protein activates the Drosomycin promoter through κB site-2. In addition, the Dorsal^DIF and DIF^DIF activity depended predominantly on site 1. These results are consistent with previous experiments using nonlinked constructs (22). Taken together, our data have shown that the linked dimers act through canonical κB sites, which again suggests that they have similar activities as other NF-κB protein dimers.
We next examined whether the linked heterodimers could activate antimicrobial peptide genes in whole animals. Transgenic flies expressing DIF^Relish, DIF^RelN, or nonlinked proteins were assayed for antimicrobial peptide gene expression using quantitative RT-PCR. Diptericin is the most widely used readout for the IMD pathway. Overexpression of RelN, but not DIF, increased Diptericin expression by 30-fold, recapitulating the regulatory specificity (Fig. 3D). In comparison, overexpression of DIF^RelN increased Diptericin expression even more, to approximately 70-fold. Drosomycin expression is preferentially regulated by the Toll pathway. Overexpression of DIF, but not RelN, increased Drosomycin expression by ninefold. DIF^RelN also caused an increase (sixfold) in Drosomycin expression (Fig. 3E). Although the induction was weaker than that observed in the transient transfection assay, it nonetheless showed that the heterodimer can activate Drosomycin in vivo. CecropinA1 can be regulated by both the Toll and IMD pathways. DIF^RelN again showed a marked stimulation of CecropinA1 (Fig. 3F). Taken together, we have demonstrated that the linked DIF^RelN heterodimer expressed in transgenic flies has stimulatory activity toward antimicrobial targets of both the Toll and IMD pathways. DIF^Relish was not active in these assays, which is consistent with the observation that it was located in the cytoplasm in the absence of microbial challenge in our overexpression assays.
DIF^Relish Can Substitute DIF or Relish in Whole Animals.
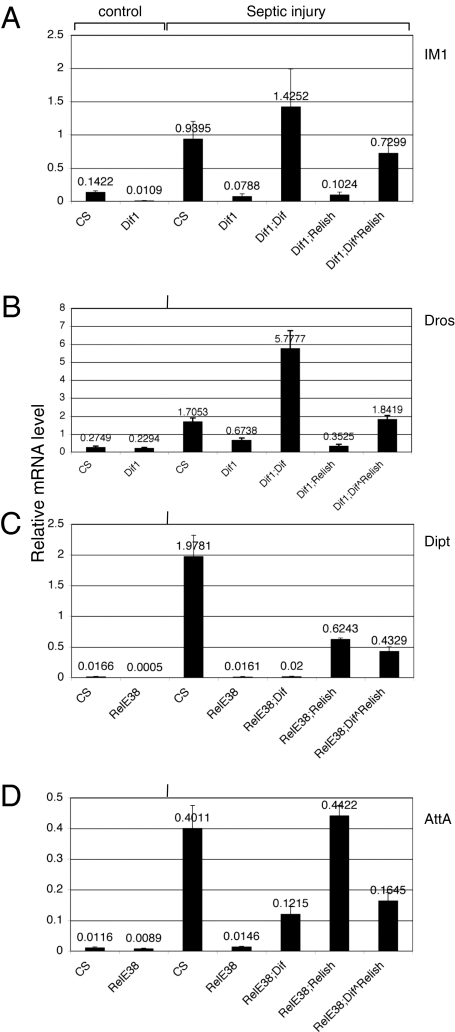
The DIF-Relish heterodimer should be the naturally existing protein that is cleaved and gives rise to DIF-RelN after signal stimulation. To demonstrate that the DIF^Relish linked heterodimer is functional in a physiological setting, we performed a genetic rescue experiment. The Dif1 mutant contains a point mutation in the gene and alters the DNA binding domain, which causes a marked reduction in the expression of the Toll pathway target genes Drosomycin and IM1 (23). The RelishE38 mutant contains a small deletion, which causes a defect in the expression of the IMD pathway target genes Diptericin and Attacin (17). Therefore, we established fly stocks that contained Dif1 or RelishE38 and expressed the DIF^Relish. Adult flies were subject to septic injury by injecting the Gram-positive bacteria Staphylococcus aureus in the Dif1 mutant and the Gram-negative bacteria Escherichia coli in the RelishE38 mutant. In wild-type flies, septic injury induced the expression of all four target genes, whereas the mutants showed defects in the expression of the target genes in the respective pathways (Fig. 4). Furthermore, the expression of IM1 and Drosomycin in the Dif1 mutant was rescued by the UAS-DIF but not by UAS-Relish. Conversely, Diptericin regulation in the RelishE38 mutant was rescued by UAS-Relish but not by UAS-DIF. AttacinA was strongly rescued by UAS-Relish and moderately rescued by UAS-DIF. These rescue results recapitulate the preferential regulation of the target genes within the two pathways.
Fig. 4.
The linked dimer of DIF^Relish can rescue antimicrobial gene expression. (A and B) S. aureus was used for septic injury of wild-type CantonS (CS), Dif1, and the corresponding rescue lines. The rescue lines for Dif1 were Dif1;YP1Gal4 crossed with Dif1;UAS-DIF (labeled as Dif1; DIF), Dif1;UAS-Relish (Dif1;Relish), and Dif1;UAS-DIF^Relish (Dif1;DIF^Relish). (C and D) E. coli was used for septic injury of the RelishE38 mutant and rescued flies. The rescue lines were similar crosses between YP1Gal4;RelishE38 and UAS-DIF;RelishE38 (labeled as RelE38;DIF), UAS-Relish;RelishE38 (RelE38;Relish), and UAS-DIF^Relish; RelishE38 (RelE38;DIF^Relish). Total RNA was isolated from adult flies 8 h after septic injury. The RNA samples were analyzed by quantitative RT-PCR. Antimicrobial peptide gene expression was analyzed in these rescue lines using IM1-, Drosomycin-, Diptericin-, and AttacinA-specific primers. The PCR cycle number for each gene was compared with that of rp49 in each sample (Materials and Methods) and the relative expression level plotted as shown. A value of one had the same mRNA copy number as rp49. The results shown are an average of three independent experiments and the error bars represent the SD.
Importantly, the UAS-DIF^Relish transgene rescued IM1 and Drosomycin expression in the Dif1 mutant background to levels that were comparable to the response seen in wild-type flies. The rescue by UAS-DIF induced a much higher expression than wild-type, which is consistent with previous observations showing that DIF is a strong transcriptional activator when overexpressed (19). Meanwhile, the rescue of Diptericin expression in the RelishE38 mutant by UAS-DIF^Relish was similar to that by UAS-Relish, although both rescued expression levels were lower than the response observed in wild-type flies. AttacinA expression in the RelishE38 mutant was efficiently rescued by UAS-Relish, whereas the UAS-DIF^Relish rescue reached approximately 34% of that expression level. These results together demonstrate that the linked DIF^Relish heterodimer is biologically active and can regulate target genes that respond to both the Toll and IMD pathways.
Detection of DIF-Relish Heterodimers in Whole Animal Extracts.
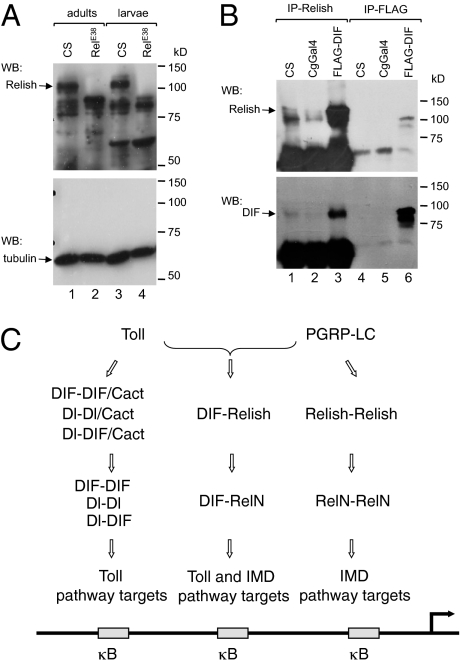
The results presented above suggest that the DIF-Relish heterodimer has an important function in vivo. To show that this heterodimer can exist at physiological levels, we performed a coimmunoprecipitation experiment on the endogenous proteins. We raised an antibody against a Relish peptide sequence (Materials and Methods). Western blot of adult and larval crude extracts using this antibody showed that the endogenous protein was detectable (Fig. 5A). Importantly, the full-length protein band (Fig. 5A, Upper, indicated by an arrow) disappeared in the RelishE38 deletion mutant extracts, indicating that the antibody can recognize endogenous Relish.
Fig. 5.
DIF-Relish complex formation and a model for NF-κB-related protein dimers. (A) Western blot (WB) analysis of endogenous Relish in adult and larval extracts. The anti-Relish antibody detected a major band with the predicted size of full length Relish (arrow). In the null RelishE38 mutant, this major band disappeared, whereas most other bands remained. Therefore, the other bands are most likely a result of cross reactivity with the serum. (Lower) is a WB using an anti-tubulin antibody and serves as the loading control. (B) An immunoprecipitation (IP) using anti-Relish or anti-FLAG antibodies, followed by a WB using anti-Relish (Upper) or anti-DIF (Lower) antibodies. The Relish precipitation showed a much stronger Relish signal when DIF was overexpressed (Upper, lane 3). DIF was detectable in the anti-Relish precipitated wild-type extracts and the signal was stronger in the FLAG-DIF over-expression sample (arrow, Lower). DIF was clearly detected in the anti-FLAG precipitated extract from the over-expression sample (Lower, lane 6). (C) A model figure illustrating the DIF-Relish heterodimer in the Toll and IMD pathways. Dorsal and DIF homodimers and heterodimers regulate target genes of the Toll pathway. Relish homodimers regulate target genes of the IMD pathway. DIF-Relish heterodimers regulate target genes of both Toll and IMD pathways. Most antimicrobial peptide gene promoters have multiple κB sites and therefore can interact with multiple different dimers at the same time.
The larval extracts were used for immunoprecipitation using the anti-Relish antibodies. The immunoprecipitated extracts were then analyzed by Western blot using anti-Relish and anti-DIF antibodies (Fig. 5B). Endogenous DIF was detected in the extracts that were immunoprecipitated with the Relish antibody and was consistent with the protein expressed by transgenic FLAG-DIF (Fig. 5B Lower, lanes 1, 2, and 6). Moreover, the amount of DIF protein that was coprecipitated with Relish was substantially higher in the FLAG-DIF transgenic fly extract (Fig. 5B, lane 3). Conversely, the FLAG-DIF transgenic flies had a substantially higher level of Relish protein in the immunoprecipitate (Fig. 5B Upper, lane 3), again suggesting an interaction between DIF and Relish. Together, these results demonstrate that endogenous DIF and Relish can form a detectable complex in normal larval extracts.
Discussion
In this study, we have shown that DIF-Relish heterodimers can form in whole animals. Both DIF and Relish have been previously detected in the fat body and hemocytes. We cannot completely rule out the possibility that the coimmunoprecipitation of the two proteins could be an artifact that occurs when the cells are lysed. However, it is more difficult thermodynamically to have two endogenous proteins come together after cell lysis if they are not already bound because there is a dilution effect from the added buffer used for tissue homogenization. Therefore, the existence of heterodimers in the cells is the most logical interpretation based on the coimmunoprecipitation of two endogenous proteins. Because the linked heterodimer rescued the target gene expression when either Gram-positive or Gram-negative bacteria were used, the data suggest that this dimeric complex can mediate signals from both the Toll and IMD pathways.
An interesting question remains is whether the DIF-Relish heterodimer interacts with the cytoplasmic inhibitor Cactus. Cactus normally binds to Dorsal and DIF for cytoplasmic retention of these proteins (13). Moreover, Relish contains ankyrin repeats in the C terminus that serve as a self-inhibitory domain. The crystal structure of mammalian NF-κB/IκB indicates that the inhibitor binds to the NF-κB dimer in a one-to-one ratio (18). Therefore, it is not clear how Cactus positions on the DIF-Relish complex that already contains C-terminal ankyrin repeats within Relish. However, in this study we observed the formation of a Relish homodimer, which should contain two inhibitory domains. Therefore, it is possible to have more than one inhibitor in the complex. The mammalian NF-κB complex contains RelB/NF-κB2, in which NF-κB2 is the full-length p110 protein and RelB is similar to DIF. Therefore, this may resemble the DIF-Relish complex (24).
The mechanism by which two different signaling pathways regulate the DIF-Relish heterodimer remains to be investigated. As mentioned above, it is not yet clear whether Cactus can form a complex with the DIF-Relish heterodimer, and if so, whether Toll signaling can cause similar Cactus phosphorylation and degradation while in a complex with this heterodimer. Interestingly, if the Toll pathway can regulate this heterodimer, then the Relish in the DIF-Relish heterodimer should still contain the C-terminal inhibitory domain, but it is not clear how the complex is translocated into the nucleus. One possibility is that this heterodimer has to be activated by both pathways simultaneously, thereby eliminating all of the inhibitors. In nature, injury often leads to infection by multiple microbes. Therefore, the activation of both IMD and Toll in the regulation of this heterodimer may be a common occurrence in animals. In conclusion, our demonstration that the DIF-Relish heterodimer is formed efficiently and has gene-activating potential provides an additional regulatory mechanism for the innate immune response.
Materials and Methods
Molecular Cloning.
The peptide linker sequence used was ASGAGGSEGGGSEGGTSGAT and was named L20 (20). The oligonucleotide sequence used in the creation of this peptide was as follows: GCCTCGGGCGCTGGAGGTTCCGAGGGAGGTGGCTCGGAAGGTGGAACCTCGGGAGCTACA. The annealed, double-stranded oligonucleotide contained HindIII and XmaI sites at each end. The wild-type and linked NF-κB–related protein constructs were first made in the pBluescript vector. The order of relevant sequence and restriction sites for the wild-type proteins are as follows: KpnI–3XFLAG–XmaI–NF-κB with a stop codon–XbaI. For the linked proteins, the following sequences and restriction sites were used: KpnI–3XFLAG–XmaI–NF-κB without a stop codon: HindIII–L20 linker–XmaI–NF-κB with a stop codon–XbaI. The fragments between the KpnI and XbaI sites were then cloned into the pUAST vector. A modified pUAST vector that lacks 3 kb of sequence between the EcoRV and StuI sites, which contains the w+ gene, was also used for the cloning of the large DNA fragments that encoded the linked dimers when cloning into the original pUAST vector was not successful. These modified pUAST constructs were used for the transfection assays only.
Fly Stocks, Transgenics, and Septic Injury.
Flies were maintained on cornmeal-yeast-molasses-agar media at 23 °C. CantonS or w1118 were used as controls. The RelishE38, Dif1, Cg-Gal4, and YP1-Gal4 constructs have been previously described (17, 21, 25). The transgenic constructs were injected into w1118 embryos. The genotypes for the rescue crosses were as follows: Dif1;YP1Gal4, Dif1;UASDif, Dif1;UASRelish, Dif1;UASDif^Relish, YP1Gal4;RelishE38, UASDif; RelishE38, UASRelish;RelishE38, and UASDIF^Relish;RelishE38.
Septic injury was carried out by puncturing the animals with a sharp tungsten needle that had been dipped in bacteria. E. coli was cultured in 2xYT medium at 37 °C overnight. S. aureus was cultured in 2xYT medium supplemented with 10 μg/mL chloramphenicol at 37 °C overnight. The E. coli culture was concentrated by 10-fold before use and the S. aureus culture was used directly. Adult flies were punctured with a needle that had been dipped in E. coli (for the RelishE38 mutants) or S. aureus (for the Dif1 mutants) and placed in a normal fly food vial at 29 °C for 8 h. For the larval fat body staining, mixed bacteria were used. Injured larvae were placed on a clean Whatman filter paper that was kept humid inside a Petri dish for 2 h before dissection of the fat body for antibody staining.
Immunohistochemistry, Immunoprecipitation, and Western Blot.
Extract preparation, immunoprecipitation, and Western blots were performed as previously described (26). The quantification of the Western blot signals was carried out by scanning the X-ray film as shown in Fig. 1D. The relevant bands were quantified by the Image J program and the signal was plotted relative to the darkest band as shown in Fig. 1E. For the coimmunoprecipitation experiments, a Relish antibody was raised in rabbits against amino acids 202–301 using the genomic antibody technology and affinity purified (Strategic Diagnostics Inc.). One microliter of the antiserum was used for immunoprecipitation and a 1:5,000 dilution was used for the Western blot. The DIF antibody has been previously described (23). For immunofluorescent staining of the fat body, third instar larvae were pulled open directly in fixation medium containing 1×PBS and 4% formaldehyde (Mallinckrodt Chemicals). The tissues were fixed in this medium for 30 min. Subsequent rinses, washes, and incubations with primary and secondary antibodies were done in a solution containing 1×PBS, 0.5% BSA, and 0.1% Triton X-100. The anti-FLAG M2 (1:200) (Sigma) was used as the primary antibody and a goat anti-mouse IgG conjugated to Alexa 488 (1:1,500 dilution) was used as the secondary antibody (Molecular Probes).
Transfection and Gene Expression Assays.
Cell maintenance, transient transfection, luciferase assays, and Western blots were performed as previously described (22). RT-PCR was carried out with the iScript cDNA Synthesis Kit (BIO-RAD) for cDNA synthesis and the iQ SYBR Green Supermix (BIO-RAD) and MyiQ Single Color Real-Time PCR Detection System (BIO-RAD) for real-time PCR. The cycle number of each primer set was compared with the cycle number of rp49 within each sample and the expression level was calculated as 1/2(cycle#x-cycle#rp49). The following gene-specific primers were used:
Drosomycin, 5′-TACTTGTTCGCCCTCTTCG and 5′-GTATCTTCCGGACAGGCAGT;
Diptericin, 5′-CCGCAGTACCCACTCAATCT and 5′-ACTGCAAAGCCAAAACCATC;
CecropinA1, 5′-CATCTTCGTTTTCGTCGCTC and 5′-CGACATTGGCGGCTTGTTGA;
IM1, 5′-CTCAGTCGTCACCGTTTTTG and 5′-CATTGCACACCCTGCAATC;
AttacinA, 5′-AGGTTCCTTAACCTCCAATC and 5′-CATGACCAGCATTGTTGTAG;
rp49, 5′-AAGCTAGCCCAACCTGCTTC and 5′-GTGCGCTTCTTCACGATCT.
Acknowledgments
We thank Neal Silverman (University of Massachusetts Medical School), Steve Wasserman (University of California San Diego), and Young-Joon Kim (Yonsei University) for allowing us to test their Relish antibodies and Dominique Ferrandon (Institut de Biologie Moleculaire et Cellulaire, Strasbourg) for the DIF antibody. We thank Scot Wolfe and Dan Bolon for help with the peptide linker design. This work was supported by a grant from the National Institutes of Health (GM53269). E.-Y.Y. was supported by the Korea Research Foundation Grant funded by the Korean Government (MOEHRD) (KRF-2007-611-C00001). Y.T.I. is a member of the University of Massachusetts Diabetes Endocrinology Research Center, and core resources supported by the Diabetes Endocrinology Research Center were also used (DK32520).
Footnotes
The authors declare no conflict of interest.
References
- 1.Aggarwal K, Silverman N. Positive and negative regulation of the Drosophila immune response. BMB Rep. 2008;41:267–277. doi: 10.5483/bmbrep.2008.41.4.267. [DOI] [PubMed] [Google Scholar]
- 2.Vallet-Gely I, Lemaitre B, Boccard F. Bacterial strategies to overcome insect defences. Nat Rev Microbiol. 2008;6:302–313. doi: 10.1038/nrmicro1870. [DOI] [PubMed] [Google Scholar]
- 3.Uvell H, Engström Y. A multilayered defense against infection: Combinatorial control of insect immune genes. Trends Genet. 2007;23:342–349. doi: 10.1016/j.tig.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Pasparakis M. IKK/NF-kappaB signaling in intestinal epithelial cells controls immune homeostasis in the gut. Mucosal Immunol. 2008;1(Suppl 1):S54–S57. doi: 10.1038/mi.2008.53. [DOI] [PubMed] [Google Scholar]
- 5.Buchon N, et al. A single modular serine protease integrates signals from pattern-recognition receptors upstream of the Drosophila Toll pathway. Proc Natl Acad Sci USA. 2009;106:12442–12447. doi: 10.1073/pnas.0901924106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El Chamy L, Leclerc V, Caldelari I, Reichhart JM. Sensing of ‘danger signals’ and pathogen-associated molecular patterns defines binary signaling pathways ‘upstream’ of Toll. Nat Immunol. 2008;9:1165–1170. doi: 10.1038/ni.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gangloff M, et al. Structural insight into the mechanism of activation of the Toll receptor by the dimeric ligand Spätzle. J Biol Chem. 2008;283:14629–14635. doi: 10.1074/jbc.M800112200. [DOI] [PubMed] [Google Scholar]
- 8.Towb P, Sun H, Wasserman SA. Tube Is an IRAK-4 homolog in a Toll pathway adapted for development and immunity. J Innate Immun. 2009;1:309–321. doi: 10.1159/000200773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moncrieffe MC, Grossmann JG, Gay NJ. Assembly of oligomeric death domain complexes during Toll receptor signaling. J Biol Chem. 2008;283:33447–33454. doi: 10.1074/jbc.M805427200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pal S, Wu J, Wu LP. Microarray analyses reveal distinct roles for Rel proteins in the Drosophila immune response. Dev Comp Immunol. 2008;32:50–60. doi: 10.1016/j.dci.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Busse MS, Arnold CP, Towb P, Katrivesis J, Wasserman SA. A kappaB sequence code for pathway-specific innate immune responses. EMBO J. 2007;26:3826–3835. doi: 10.1038/sj.emboj.7601798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ayyar S, et al. NF-kappaB/Rel-mediated regulation of the neural fate in Drosophila. PLoS ONE. 2007;2:e1178. doi: 10.1371/journal.pone.0001178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minakhina S, Steward R. Nuclear factor-kappa B pathways in Drosophila. Oncogene. 2006;25:6749–6757. doi: 10.1038/sj.onc.1209940. [DOI] [PubMed] [Google Scholar]
- 14.Pal S, Wu LP. Pattern recognition receptors in the fly: Lessons we can learn from the Drosophila melanogaster immune system. Fly (Austin) 2009;3:121–129. doi: 10.4161/fly.8827. [DOI] [PubMed] [Google Scholar]
- 15.Charroux B, Rival T, Narbonne-Reveau K, Royet J. Bacterial detection by Drosophila peptidoglycan recognition proteins. Microbes Infect. 2009;11:631–636. doi: 10.1016/j.micinf.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Ertürk-Hasdemir D, et al. Two roles for the Drosophila IKK complex in the activation of Relish and the induction of antimicrobial peptide genes. Proc Natl Acad Sci USA. 2009;106:9779–9784. doi: 10.1073/pnas.0812022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiklund ML, Steinert S, Junell A, Hultmark D, Stöven S. The N-terminal half of the Drosophila Rel/NF-kappaB factor Relish, REL-68, constitutively activates transcription of specific Relish target genes. Dev Comp Immunol. 2009;33:690–696. doi: 10.1016/j.dci.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann A, Natoli G, Ghosh G. Transcriptional regulation via the NF-kappaB signaling module. Oncogene. 2006;25:6706–6716. doi: 10.1038/sj.onc.1209933. [DOI] [PubMed] [Google Scholar]
- 19.Yagi Y, Ip YT. Helicase89B is a Mot1p/BTAF1 homologue that mediates an antimicrobial response in Drosophila. EMBO Rep. 2005;6:1088–1094. doi: 10.1038/sj.embor.7400542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin A, Baker TA, Sauer RT. Rebuilt AAA + motors reveal operating principles for ATP-fuelled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- 21.Bettencourt R, Asha H, Dearolf C, Ip YT. Hemolymph-dependent and -independent responses in Drosophila immune tissue. J Cell Biochem. 2004;92:849–863. doi: 10.1002/jcb.20123. [DOI] [PubMed] [Google Scholar]
- 22.Tanji T, Hu X, Weber AN, Ip YT. Toll and IMD pathways synergistically activate an innate immune response in Drosophila melanogaster. Mol Cell Biol. 2007;27:4578–4588. doi: 10.1128/MCB.01814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rutschmann S, et al. The Rel protein DIF mediates the antifungal but not the antibacterial host defense in Drosophila. Immunity. 2000;12:569–580. doi: 10.1016/s1074-7613(00)80208-3. [DOI] [PubMed] [Google Scholar]
- 24.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 25.Goto A, et al. Akirins are highly conserved nuclear proteins required for NF-kappaB-dependent gene expression in drosophila and mice. Nat Immunol. 2008;9:97–104. doi: 10.1038/ni1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu X, Yagi Y, Tanji T, Zhou S, Ip YT. Multimerization and interaction of Toll and Spätzle in Drosophila. Proc Natl Acad Sci USA. 2004;101:9369–9374. doi: 10.1073/pnas.0307062101. [DOI] [PMC free article] [PubMed] [Google Scholar]