Abstract
Neuronal ceroid lipofuscinoses (NCLs) represent the most common group of inherited progressive encephalopathies in children. They are characterized by progressive loss of vision, mental and motor deterioration, epileptic seizures, and premature death. Rare adult forms of NCL with late onset are known as Kufs’ disease. Loci underlying these adult forms remain unknown due to the small number of patients and genetic heterogeneity. Here we confirm that a late-onset form of NCL recessively segregates in US and French pedigrees of American Staffordshire Terrier (AST) dogs. Through combined association, linkage, and haplotype analyses, we mapped the disease locus to a single region of canine chromosome 9. We eventually identified a worldwide breed-specific variant in exon 2 of the Arylsulfatase G (ARSG) gene, which causes a p.R99H substitution in the vicinity of the catalytic domain of the enzyme. In transfected cells or leukocytes from affected dogs, the missense change leads to a 75% decrease in sulfatase activity, providing a functional confirmation that the variant might be the NCL-causing mutation. Our results uncover a protein involved in neuronal homeostasis, identify a family of candidate genes to be screened in patients with Kufs' disease, and suggest that a deficiency in sulfatase is part of the NCL pathogenesis.
Keywords: animal model, ataxia, dog, neurodegenerative, lysosome
In humans, neuronal ceroid lipofuscinoses (NCLs) are neurodegenerative disorders characterized by various neurologic alterations and intracellular accumulation of autofluorescent storage material. NCLs represent the most common group of progressive encephalopathies in children (1). Some 10% of NCLs are sporadic or inherited adult-onset forms known as Kufs’ disease (2 –4). All NCLs, except rare autosomal dominant adult forms, segregate as autosomal recessive morbidity traits and are subclassified into 10 genetic forms, designated CLN1–CLN10, 8 of which (CLN1–3, CLN5–8, and CLN10) have been characterized molecularly (1). Genes underlying Kufs’ disease (CLN4) and CLN9 remain to be identified.
NCLs compose a subgroup of lysosomal disorders in which altered biochemical pathways affect primarily neurons. Because these postmitotic neurons are metabolically very active, individuals with an NCL cannot dilute aggregates during cell division. These aggregates induce cellular damage or oxidative stress, eventually leading to early neuronal apoptosis (5). Although most CLN proteins have been identified, their specific contribution to NCL pathogenesis remains largely unknown. Indeed, CLN2 is a soluble lysosomal enzyme, but its various physiological substrates are not accumulated in the storage material (reviewed in ref. 1). More likely, a large panel of lysosomal and nonlysosomal CLN proteins are involved in cellular mechanisms, including compartments outside the lysosome (5). For instance, intracellular autofluorescent storage material comparable to that found in NCLs has been reported in lymphoblasts treated with pharmacological inhibitors of microtubule assembly (6). To date, the three main cellular mechanisms likely impaired in NCLs are intracellular and membrane trafficking, autophagy, and calcium storage. First, cytoskeleton-associated proteins display abnormalities in CLN1- and CLN5-deficient neurons that are associated with changes in the growth cone assembly (7), and CLN1 modulates the early stages of endocytic vesicule formation (8). Second, reduced autophagy through inhibition of fusion between autophagosomes and lysosomes is observed in most NCLs (5). This results in the accumulation of polyubiquitinated proteins and damaged mitochondria (potent generators of reactive oxygen species), and also in the activation of the endoplasmic reticulum stress, whose pathways converge to the caspase 9-induced apoptosis signal (5, 9). Third, lysosomal dysfunction may promote calcium-mediated cell death through membrane, mitochondrial, or endoplasmic dysfunction. Elevated calcium concentrations are cytotoxic. Specialized proteins from these compartments are required to maintain low calcium levels. An example of altered calcium cycling in CLN3-deficient neurons has been reported (10). Finally, these cellular mechanisms are presumably interconnected, given that, for example, CLN5 has been found to interact with CLN1, CLN2, CLN3, CLN6, and CLN8 (11).
Several mammalian models have been described, most of which recapitulate early- or juvenile-onset NCLs (www.caninegeneticdiseases.net/CL_site/mainCL.htm and refs. 1, 12, and 13). Adult American Staffordshire terriers (ASTs) may suffer from ataxia, a condition defined by uncoordinated and inaccurate movements and awkward positions (14, 15). Histopathological brain examination in affected ASTs has revealed severe cerebellar cortical abiotrophy and remodeling with loss of Purkinje cells (15, 16). Segregation analysis of ataxia in ASTs supports an autosomal recessive mode of inheritance with an alarming incidence of the deleterious allele, estimated at ~40% (15). Another study of five unrelated Swiss cases revealed the presence of autofluorescent cytoplasmic storage material in Purkinje cells, suggesting that the ataxia might result from NCLs (16). Here we confirm that ASTs represent a model of a late-onset form of NCL that led us to identify tentatively arylsulfatase G, a sulfatase ensuring long-term neuronal survival, as a candidate protein for CLN4.
Results
Affected Ataxic ASTs Suffer from a Late-Onset NCL.
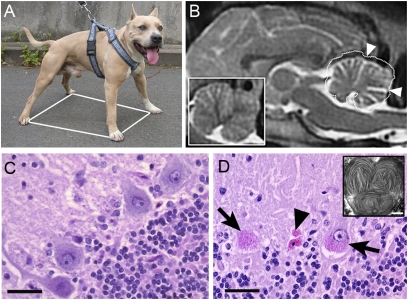
We analyzed a total of 138 French and US ASTs that exhibited locomotor disabilities. All of the dogs displayed static and dynamic ataxia, a condition detected early by owners (Fig. 1A). Ataxia initially involved loss of balance and stumbling when turning corners, walking uphill or downhill, or negotiating stairs (Movie S1). Neither owners nor veterinarians reported visual impairment in any of the affected dogs. A majority of the dogs (70%) first displayed locomotor ataxia between 3 and 5 y of age (Fig. S1). All 58 of the 138 dogs diagnosed by MRI exhibited significant cerebellar atrophy (Fig. 1B and ref. 17). At necropsy, the affected dogs displayed cerebellar atrophy. The most prominent histological finding recorded in each of these dogs was a marked loss of Purkinje cells (Fig. 1C and D). PAS reagent–, Luxol fast blue–, and Sudan black–positive material was observed in the cytoplasm of surviving or remnants of Purkinje cells (Fig. 1D), macrophages and neurons in the occipital cortex, the hypoglossal nuclei, the posterior nuclei of the thalamus (Fig. S2), and pyramidal cells from the hippocampus and Ammon's horn. This material was highly suggestive of pathological ceroid lipofuscin accumulation. Ultrastructural examination of Purkinje cells revealed abnormal lysosomes filled with inclusions of material of medium electron density displaying curved, straight, or concentric profiles with alternate clear and dense bands (Fig. 1D, Inset). The white matter, lymph nodes, and spleen appeared histologically normal in the necropsied dogs. Unlike in other NCL dog models, degenerative lesions or lipofuscinosis were not observed in the retina.
Fig. 1.
Clinical and histopathological features of the disease. (A) The wide-based stance (white polygon) of a 5-y-old affected AST illustrates loss of motor coordination. (B) Sagittal 2-T weighted MRI of the brain from a 6-y-old AST through the cerebellum. (Inset) A similar image at the same scale of the cerebellum from an age-matched healthy AST, the outline of which has been projected on the cerebellum of the main image (white dotted line). A reduction of gray matter is demonstrated by the enlarged sulci (arrowheads). (C and D) Transverse sections of the cerebellum from healthy dogs (C) and affected dogs (D), stained with PAS reagent and counterstained in Mayer's hematoxylin solution. (C) In a normal cerebellum, the large PAS-negative Purkinje neurons lie between the molecular (top left) and granular (bottom right) layers. (D) In the cerebellum from an affected dog, massive Purkinje cell loss results in a blurry line. The remaining Purkinje neurons (arrows) or neuron remnants (arrowheads) accumulate perinuclear PAS-positive granular material. (Inset) Affected Purkinje neurons imaged on transmission electron microscopy show accumulated lysosomal material composed of concentric straight or curved profiles with alternating clear and dense bands. (Scale bars: 50 μm for the sections; 250 nm for the inset.)
Based on these clinical and histopathological features, we confirmed that the ataxia frequently seen in French and US AST populations is highly reminiscent of an NCL. Our earlier conclusion based on a panel of US ASTs suggesting that NCL segregates as an autosomal recessive trait (15, 18) was confirmed once we included 73 French ASTs for which pedigree information was available (Table S1).
Mapping the NCL Locus and Identification of an ARSG Missense Mutation.
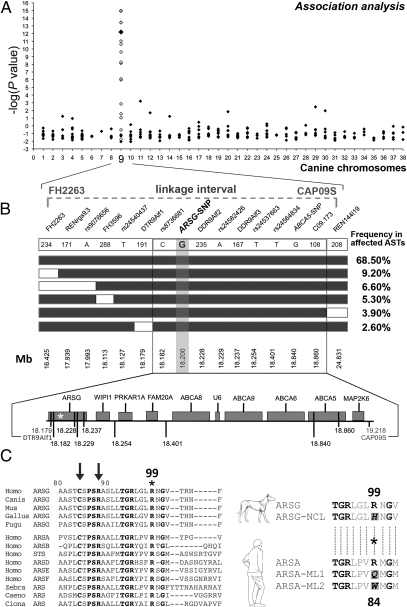
To map the NCL locus, we genotyped 77 unrelated French ASTs with 247 microsatellites from the MSS2 screening set (19). The cohort included 39 affected dogs (22 males and 17 females) and 38 nonaffected dogs age >3 y (22 males and 16 females). A single marker strongly associated with NCL (C09.173; χ2 test, Bonferroni-corrected P = 4.1 × 10−13) (Fig. 2A) was identified on canine chromosome 9 (CFA09). This association was confirmed by genotyping the cohort with 14 additional polymorphic microsatellites or SNPs (Table S2 and Fig. 2A). The highest P value was obtained for ABCA5-SNP (χ2 test, Bonferroni-corrected P = 1.1 × 10−15). In parallel, we genotyped 48 related US ASTs with a genome-wide panel of 315 microsatellites. The NCL locus mapped to the same region on CFA09. A peak in linkage yielding a maximum LOD score of 9.4 was detected for a 2.8-Mb segment between FH2263 and CAP09S (18) (Fig. 2B). To identify additional informative markers, we genotyped 12 affected and 10 healthy French ASTs using the Illumina Infinium CanineSNP170 array. We chose six polymorphic SNPs to genotype the cohort of 77 French ASTs. A haplotype spanning a 678-kb region between rs8736681 and C09.173 was common to 100% of the affected dogs and 58.5% of the healthy dogs (χ2 = 40.7; P < 0.001) (Fig. 2B). This haplotype includes four Illumina SNPs, the ABCA5-SNP, C09.173 from the MSS2, and two microsatellites identified in our laboratory (Table S2). By combining the haplotype and linkage data, we defined a 1,039-kb critical region delimited by DTR9Alf1 and CAP09S. This region is syntenic to human chromosome 17 and to mouse chromosome 11, on which neither NCL nor cerebellar ataxia loci have been mapped (www.ncbi.nlm.nih.gov/sites/entrez?db=OMIM and www.informatics.jax.org).
Fig. 2.
Mapping, fine-mapping, and identification of a candidate gene for NCL in ASTs. (A) A single locus with strong genome-wide significance was identified on CFA09 (larger black diamonds) and confirmed (white diamonds); see Table S2. The −log(P values) are reported on the y-axis. The 2.8-Mb candidate region defined by our linkage analysis (18) is shown. (B) Fine-mapping using haplotype analysis for NCL. Haplotypes identified in affected ASTs are shown as boxes (NCL allele, black boxes; alternative allele, white boxes). Genotyped microsatellites and SNPs are indicated in their 5′-3′ position, and their position on CFA09 is shown. The Haplotypes with a frequency <2% are omitted. In the bottom part, candidate genes within the critical region are shown. The relative positions of markers are indicated by vertical black lines, and the nonsynonymous associated ARSG-SNP is represented by an asterisk. (C) (Left) N-terminal sequence of ARSG or predicted ARS from metazoans is aligned with the N-terminal amino acids of human ARS. Conserved amino acids are in bold type. Arrows point to two of the 10 residues involved in the catalytic site of arylsulfatases (22), with positions referring to the canine ARSG. The R99 mutant in affected dogs is highlighted by an asterisk. (Right) The sequence flanking the R99 of the canine WT ARSG protein (ARSG) is aligned with the corresponding sequence of the mutated protein (ARSG-NCL) and best aligned with the R84 of the human WT ARSA (ARSA), replaced by a glutamine (Q) or a tryptophane (W) in some patients affected by metachromatic leukodystrophy [ARSA-ML1; (25) and ARSA-ML2, (24)].
The 1,039-kb critical region spans nine protein-coding genes, a U6 noncoding small nuclear RNA gene (Fig. 2B), and two nonannotated genes. To select candidate genes, we compiled functional and expression data on the described or putative function of the 12 genes, their expression pattern in human and mouse tissues, and their expression pattern in dog tissues assessed by RT-PCR (Fig. S3 and Table S3). Three genes were candidates: the ATP-binding cassette, subfamily A, member 5 (ABCA5) gene, because the C09.173 marker is located within ABCA5 intronic sequence, and the Arylsulfatase G (ARSG) and the Mitogen-activated protein kinase 6 (MAP2K6) genes, because they are expressed in the central nervous system and may play a role in neuronal homeostasis. Sequence analysis of the coding and intron–exon boundaries of the three genes in four healthy dogs and four affected dogs revealed two differences between the healthy and affected dogs: (i) a synonymous ABCA5-SNP associated with NCL (Fig. 2A and Table S3) and (ii) a nonsynonymous c.296G > A substitution in exon 2 of the ARSG gene (ARSG-SNP). This nonsynonymous c.296G > A substitution was detected in two copies in all 138 affected ASTs, but not in 47% (86/181) of healthy ASTs, and was detected in a single copy in 50% (91/181) of healthy ASTs from the French and US panels (Table 1). The presence in 50% of healthy carriers was compatible with the high prevalence of the disease allele inferred in the AST breed (15). Moreover, the c.296G>A substitution was absent in 525 healthy dogs from 54 other breeds (Table S4). Remarkably, four ASTs from the healthy group were genotyped A/A and shared the 678-kb spanning haplotype specific to NCL. Three of these dogs were 9 y old at the time of the study (June 2010). Because their owners did not give consent, these dogs could not be evaluated by MRI, but their clinical status is currently being monitored. The fourth dog died at age 13 y from a nonneurologic disease. However, histopathological examination of this dog's brain revealed mild accumulation of ceroid lipofuscin in Purkinje neurons, the level and staining pattern of which were clearly different from those routinely observed in aging dogs. Thus, this accumulation was considered a subclinical consequence of the low expressivity of the mutation in this dog. In conclusion, multibreed segregation analyses of the c.296G>A substitution in the ARSG gene confirmed that the mutation is specifically associated with NCL in ASTs.
Table 1.
Genotype frequencies for the ARSG-SNP in healthy and affected ASTs
| Clinical status of dogs | Healthy | Affected | ||
| Genotype at the ARSG-SNP locus | G/G | G/A | A/A | A/A |
| French ASTs | 53.5% (n = 38) | 45.1% (n = 32) | 1.4% (n = 1) | 100% (n = 66) |
| US ASTs | 43.6% (n = 48) | 53.6% (n = 59) | 2.7% (n = 3) | 100% (n = 72) |
The number of dogs is indicated in brackets. The high frequency of heterozygotes (50%) is compatible with the frequency of the deleterious allele that has been estimated to ~40% in US pedigrees (15).
Reduced Arylsulfatase Activity of the Mutated ARSG.
The c.296G>A substitution results in the p.R99H substitution in the canine protein. ARSG belongs to a large family of sulfatases conserved in metazoans (20). To evaluate the functional impact of the p.R99H substitution, we assessed the selective pressure exerted on the R99 residue by sequence alignments. The canine R99 is conserved with all of the functionally characterized human ARSs (Fig. 2C), a maximum conservation level previously reported for the 10 residues of the catalytic domain (20, 21). Moreover, the R99 of canine ARSG best aligns with R84 of human ARSA (Fig. 2C). The crystal structure of ARSA indicates that R84 might be essential for its enzymatic activity (22). Indeed, p.R84Q or p.R84W substitution in human ARSA induces decreased enzymatic activity, eventually leading to metachromatic leukodystrophy (Fig. 2C) (23 –25).
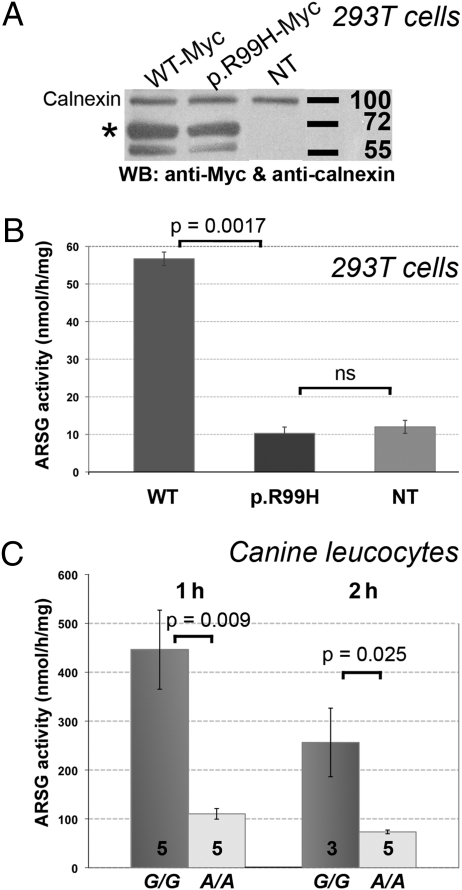
To test whether R99 plays a role in ARSG activity that would be impaired in affected dogs, we overexpressed the WT and p.R99H variants in HEK293T and compared the resulting ARSG-specific activity (Fig. 3 A and B). To prevent redundant activity due to other ARSs (26 –28), we applied specific inhibiting conditions. ARSC, ARSE, and ARSF were inhibited at pH 4.6, and ARSB was inhibited in the presence of 10% NaCl. In addition, ARSA and ARSE activity was blocked by warfarin at 187.5 mM (Fig. S4). In HEK293T cells transfected with the p.R99H variant–expressing vector, the ARSG activity reached only 18.1% of the ARSG activity measured in HEK293T cells transfected with the WT-expressing vector (10.28 ± 1.66 and 56.7 ± 1.78, respectively; n = 7; Mann–Whitney–Wilcoxon P = 0.0017). ARSG activity did not differ significantly in HEK293T cells transfected with the p.R99H variant–expressing vector and with the control vector (12 ± 1.73; n = 7; Mann–Whitney–Wilcoxon P = 0.403). These findings indicate that the p.R99H variant is unable to restore WT ARSG activity in overexpressing cells.
Fig. 3.
The highly conserved R99 is critical for ARSG activity. (A) In HEK293T cells, similar overexpression levels of C-ter Myc-tagged WT (WT-Myc) or variant (p.R99H-Myc) human ARSG are validated in a Western blot assay (WB) by their specific immunoreactivity with an anti-Myc antibody (asterisk). Proteins are detected at the expected molecular weight (63 kDa). NT, nontransfected cells. Adjusted levels of total proteins are confirmed by levels of the housekeeping calnexin. (B) These HEK293T cells were used to measure ARSG activity, expressed in nmol/h/mg of protein, using a method adapted from Frese et al. (32). Vertical bars represent the SEM for seven experiments. (C) Total leukocyte arylsulfatase activity was compared between homozygous (G/G) healthy controls, taken as the 100% reference activity level, and homozygous (A/A) affected ASTs. The number of sampled dogs is identified within each histogram, and vertical bars represent SEM.
To confirm the loss of activity of the p.R99H variant under physiological conditions, we compared the ARSG activity in healthy and affected ASTs. Because the canine ARSG gene is ubiquitously expressed in dogs (Table S3), we evaluated the arylsulfatase activity in leukocytes (Fig. 3C). After 1 h of incubation with the substrate, the ARSG activity of leukocytes was significantly lower in affected dogs compared with healthy dogs [110.2 ± 10.82 (n = 5) vs. 446.2 ± 80.89 (n = 5); Mann–Whitney–Wilcoxon P = 0.009]. This reduced activity remained significantly lower in affected dogs after 2 h of incubation [73.2 ± 3.79 (n = 5) vs. 256.3 ± 70.15 (n = 3); Mann–Whitney–Wilcoxon P = 0.025].
Taken together, these data support the assumption that the p.R99H substitution abolishes most of ARSG activity and that it is the pathogenic mutation. The strong association between ARSG-SNP and NCL was used to develop a predictive DNA test currently exploited by AST breeders worldwide.
Discussion
AST Canine Model Identifies ARSG as a Candidate Gene for Human Late-Onset NCLs.
In humans, CLN4 includes a rare adult-onset form of NCL known as Kufs’ disease. Two groups of phenotypes have been recognized, with some cases demonstrating characteristics of both (2, 29). Because neither the gene mutation nor the pathological mechanism has been identified in inherited forms of CLN4, the diagnosis is currently based on clinical, histopathological, and ultrastructural data (4). Animal models have seldom been described, and none of these models fully recapitulates human symptoms. Historically, the closest and most-studied model has been the Tibetan Terrier, but nevertheless, the molecular etiology of this inherited canine disorder remains obscure (30, 31). We confirmed that AST dogs suffering from an inherited form of locomotor ataxia are affected by a form of NCL that shares various features with Kufs’ disease, including late onset and slow progress, absence of visual impairment, marked cerebellar atrophy, and accumulation of PAS-positive lipopigment in Purkinje cells and thalamic neurons.
Using combined association, linkage, and haplotype analyses, we identified a variant in the ARSG gene that specifically segregates with the disease. Because the complete 1,039-kb candidate region has not yet been fully sequenced, we cannot ascertain whether the c.296G>A substitution found in ARSG is the only important variant in linkage disequilibrium with NCL. However, p.R99H impairs ARSG activity when overexpressed in HEK293T cells or when naturally expressed in leukocytes of affected dogs, suggesting a strong genotype–phenotype correlation. Further reports of ARSG mutations in Kufs’ patients or analysis of ARSG-mutagenized mice would help to validate this causative hypothesis. Meanwhile, we suggest that screening of ARS enzymatic activity might be useful in the early diagnosis of patients affected by Kufs’ disease.
Linking ARSG Deficiency to NCL Pathogenic Mechanisms.
NCLs represent a subgroup of lysosomal storage disorders characterized by the accumulation of autofluorescent ceroid or lipofuscin lipopigments in neural and peripheral tissues (1, 3). ARSG acts as a monomeric lysosomal enzyme (32). How a deficiency in ARSG leads to NCL can be explained by a multistep mechanism. In the first step, neurons lacking ARSG activity might accumulate undegraded substrates in their lysosomes. In the second step, lysosomal storage may induce impaired autophagy, cellular trafficking, or calcium dynamics, which are common secondary changes seen in NCLs (5, 8). Neurons are metabolically very active and sensitive to the effects of lysosomal storage; indeed, increasing autophagy efficiency in cultured Pukinje neurons protects these neurons from death (33).
The specific composition of the storage material in neurons of affected ASTs is unknown. This composition might not be informative, however, because the storage material is not specific to a NCL form, and because it does not provide precise clues regarding the biochemical mechanisms altered by the pathogenic mutation. Indeed, although NCLs are genetically heterogeneous, the storage material is composed essentially of subunit c of mitochondrial ATP synthase or of sphingolipid activator proteins A and D (1). Nonspecific secondary lipid storage materials also are frequently associated (34). In contrast, characterization of ARSG endogenous substrate(s) may be relevant because it may identify essential molecular pathways for neuronal homeostasis. ARSG belongs to a large family of 17 sulfatases known to catalyze the hydrolysis of sulfate esters and sulfamates in a wide variety of substrates, including steroids, carbohydrates, proteoglycans, and glycolipids (20). The 11 known arylsulfatases form a subgroup defined by the ability of most of its members to hydrolyze in vitro small aryl substrates, such as p-nitrocatechol sulfate (pNCS); however, only a few endogenous substrates have been identified as sulfatides for ARSA or chondroitin and heparan sulfates for ARSB (35, 36). The identification of ARSG substrates might benefit the canine NCL model.
Defective mutations characterized in the six lysosomal proteins of the sulfatase family have been found to induce non-NCL lysosomal storage diseases (20, 32). Thus, the AST canine model represents a relevant example of a lysosomal sulfatase deficiency leading to an NCL and offers a unique opportunity to establish a functional link, as-yet unelucidated, between lysosomal sulfatase activity and CLN proteins. The fact that 4 of 181 ASTs from our clinically healthy group were homozygous for the affected haplotype and the ARSG variant demonstrates that NCL penetrance is variable. In addition, clinical and pathological differences observed among affected dogs show varying expressivity. This phenotypic diversity in dogs sharing the same disease-causing mutation may reflect environmental or genetic interactions. In a Drosophila model of CLN10, modifiers have been identified that are involved in endocytosis and metabolism/oxidation of lipids (37). These data emphasize that lysosomal function is supported by a still-growing complex network. Collection of more fully phenotyped affected dogs is ongoing and should help to identify modifiers and new candidates for NCL pathogenesis in humans.
Methods
See SI Methods for more details.
Dogs.
A total of 138 affected ASTs were included. Of these 138 dogs, 116 were diagnosed by a European or American board-certified veterinary neurologist. For each of these dogs, a clinical history was compiled and complete clinical and neurologic evaluations were performed. The remaining 22 affected ASTs were diagnosed by their regular veterinarian. The 181 healthy control dogs were ≥3-y-old French (n = 71) and US (n = 110) ASTs that failed to exhibit any of the symptoms seen in affected dogs. Thirty-eight of the healthy French dogs were chosen to compose the control group for the genome scan.
Histology and Electronic Microscopy.
Whole brains including the cerebellum were removed from 23 affected ASTs aged 4–8 y and euthanized for medical reasons. Lymph node, eye and spleen samples were collected. Routine histological examinations were performed. Selected areas of the cerebellar cortex from a 4-y-old affected female were used for observations by transmission electron microscopy.
MSS2 and SNP Marker Genotyping.
DNA was extracted from blood. Microsatellite markers from the MSS 2 (19) were individually amplified for each dog using a fluorescent primer in PCR. Labeled products were size-separated. The CanineHD BeadChip Panel (Illumina) featuring validated SNPs derived from the CanFam2.0 assembly (38) was used to obtain genotype calls. Microarray work was performed following the manufacturer's recommendations by the Centre National du Génotypage, Evry, France. Select SNP genotyping of our complete cohort of ASTs was determined using a pyrosequencing method adapted from Ahmadian et al. (39) on a Biotage PSQTM 96 pyrosequencer. The PCR primers are discussed in SI Methods and listed in Table S3.
Mapping Strategy and Association Analysis.
Our genome-wide mapping strategy relied on a case-control association analysis. In the first step, dogs were tested for both phenotype and genotype at each MSS2 locus using the STRAT software (40). The P values were corrected according to Bonferroni procedures. In the second step, fine-mapping of the CFA09 critical region was performed using the PLINK software package (41). Individuals were tested for both phenotype and a genotype at each locus. We assumed that both genotype and phenotype were binary, denoting the alleles by A and non-A. The A allele was defined as the disease-associated allele (i.e., the most frequent allele in the affected dog cohort). Multiallelic loci were accommodated by focusing on the A allele and grouping the remaining as non-A alleles. The P values were corrected according to Bonferroni procedures. NCL-associated haplotypes were identified using HAPLOVIEW software (42).
RT-PCR.
Organ samples were collected from a 7-y-old affected AST and a 4-y-old Labrador Retriever suffering from an incurable nonneurologic disease. Samples were frozen and stored at −80 °C. RT-PCR is described in more detail in SI Methods.
Expression of the p.R99H Protein.
A Myc-FLAG-ARSG expression vector was purchased from Origene (human ARSG, NM_014960). A single amino acid substitution was obtained using the QuikChange II Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer's protocol. PCR and sequencing primers are provided in SI Methods. Transfection of the expression vectors into HEK293T cells and Western blot analysis were performed following a protocol reported in detail in SI Methods.
Enzymatic Assays.
Arylsulfatase activity was assessed in transfected HEK293T cells and leukocytes from affected and healthy ASTs using a method adapted from Frese et al. (32) and detailed in SI Methods.
Statistical Analysis.
Correction for multiple testing was done according to the Bonferroni method. The χ2 test and the nonparametric Mann–Whitney–Wilcoxon test for small distributions were performed with StatView F-4.1 (SAS Institute). All error bars represent SEM.
GenBank Accession Codes.
Reference and submitted sequences were Homo sapiens ARSG cDNA, NM_014960; Canis familiaris ABCA5 exon 36 containing the ABCA5-SNP, FM211813; Canis familiaris ARSG exon 2 containing the ARSG-SNP, FM246885.
Supplementary Material
Acknowledgments
We thank J. L. Kessler, V. Gkouni, and A. Uriarte for technical assistance; L. Dandolo for fruitful discussions; and G. Egidy-Maskos and M. Chodkiewicz for corrections to the manuscript. We gratefully acknowledge the contributions of the participating dog owners and veterinarians, the Club Français des Amateurs de Bull terrier, d’American Staffordshire terrier et de Staffordshire Bull terrier, and the Staffordshire Terrier Club of America. This work was funded by grants from the European Commission (FP6 EuroTransBio “Biomarks” and FP7 LUPA GA-201370), the “Centronuclear myopathy” Project (www.labradorcnm.com), the American Kennel Club-Canine Health Foundation, the Centre National de la Recherche Scientifique, and Royal Canin.
Footnotes
Conflict of interest statement: INRA and ENVA have applied for a patent covering the use of the canine ARSG SNP for the diagnosis of NCL or for selective breeding of dogs. M.A. and S. Blot are listed as inventors in this application.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession nos. FM246885, FM211814, FM211419, FM211812, and FM211813).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0914206107/-/DCSupplemental.
References
- 1.Jalanko A, Braulke T. Neuronal ceroid lipofuscinoses. Biochim Biophys Acta. 2009;1793:697–709. doi: 10.1016/j.bbamcr.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 2.Berkovic SF, Carpenter S, Andermann F, Andermann E, Wolfe LS. Kufs’’ disease: A critical reappraisal. Brain. 1988;111:27–62. doi: 10.1093/brain/111.1.27. [DOI] [PubMed] [Google Scholar]
- 3.Haltia M. The neuronal ceroid-lipofuscinoses: From past to present. Biochim Biophys Acta. 2006;1762:850–856. doi: 10.1016/j.bbadis.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 4.Lewandowska E, et al. Kufs’’ disease: Diagnostic difficulties in the examination of extracerebral biopsies. Folia Neuropathol. 2009;47:259–267. [PubMed] [Google Scholar]
- 5.Bellettato CM, Scarpa M. Pathophysiology of neuropathic lysosomal storage disorders. J Inherit Metab Dis. 2010;33:347–362. doi: 10.1007/s10545-010-9075-9. [DOI] [PubMed] [Google Scholar]
- 6.Seehafer SS, Pearce DA. Spectral properties and mechanisms that underlie autofluorescent accumulations in Batten disease. Biochem Biophys Res Commun. 2009;382:247–251. doi: 10.1016/j.bbrc.2009.02.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.von Schantz C, et al. Brain gene expression profiles of Cln1- and Cln5- deficient mice unravels common molecular pathways underlying neuronal degeneration in NCL diseases. BMC Genomics. 2008;9:146–160. doi: 10.1186/1471-2164-9-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saja S, Buff H, Smith AC, Williams TS, Korey CA. Identifying cellular pathways modulated by Drosophila palmitoyl-protein thioesterase 1 function. Neurobiol Dis. 2010 doi: 10.1016/j.nbd.2010.02.010. 10.1016/j.nbd.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao L, Ackerman SL. Endoplasmic reticulum stress in health and disease. Curr Opin Cell Biol. 2006;18:444–452. doi: 10.1016/j.ceb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 10.Luiro K, et al. Batten disease (JNCL) is linked to disturbances in mitochondrial, cytoskeletal, and synaptic compartments. J Neurosci Res. 2006;84:1124–1138. doi: 10.1002/jnr.21015. [DOI] [PubMed] [Google Scholar]
- 11.Lyly A, et al. Novel interactions of CLN5 support molecular networking between neuronal ceroid lipofuscinosis proteins. BMC Cell Biol. 2009;10:83–94. doi: 10.1186/1471-2121-10-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fiske RA, Storts RW. Neuronal ceroid-lipofuscinosis in Nubian goats. Vet Pathol. 1988;25:171–173. doi: 10.1177/030098588802500213. [DOI] [PubMed] [Google Scholar]
- 13.Kuwamura M, et al. Neuronal ceroid-lipofuscinosis in a Japanese domestic shorthair cat. J Vet Med Sci. 2009;71:665–667. doi: 10.1292/jvms.71.665. [DOI] [PubMed] [Google Scholar]
- 14.Hanzlícek D, et al. Cerebellar cortical abiotrophy in American Staffordshire terriers: clinical and pathological description of 3 cases. Schweiz Arch Tierheilkd. 2003;145:369–375. doi: 10.1024/0036-7281.145.8.369. (in German) [DOI] [PubMed] [Google Scholar]
- 15.Olby N, et al. Cerebellar cortical degeneration in adult American Staffordshire Terriers. J Vet Intern Med. 2004;18:201–208. doi: 10.1892/0891-6640(2004)18<201:ccdiaa>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 16.Sisó S, Navarro C, Hanzlícek D, Vandevelde M. Adult-onset thalamocerebellar degeneration in dogs associated to neuronal storage of ceroid lipopigment. Acta Neuropathol. 2004;108:386–392. doi: 10.1007/s00401-004-0902-7. [DOI] [PubMed] [Google Scholar]
- 17.Thames RA, et al. Development of a morphometric magnetic resonance image parameter suitable for distinguishing between normal dogs and dogs with cerebellar atrophy. Vet Radiol Ultrasound. 2010;51:246–253. doi: 10.1111/j.1740-8261.2009.01655.x. [DOI] [PubMed] [Google Scholar]
- 18.Olby N, et al. Linkage analysis in American Staffordshire Terriers with hereditary cerebellar cortical degeneration. J Vet Intern Med. 2008;22:723–724. [Google Scholar]
- 19.Clark LA, et al. Chromosome-specific microsatellite multiplex sets for linkage studies in the domestic dog. Genomics. 2004;84:550–554. doi: 10.1016/j.ygeno.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 20.Sardiello M, Annunziata I, Roma G, Ballabio A. Sulfatases and sulfatase modifying factors: An exclusive and promiscuous relationship. Hum Mol Genet. 2005;14:3203–3217. doi: 10.1093/hmg/ddi351. [DOI] [PubMed] [Google Scholar]
- 21.Ferrante P, Messali S, Meroni G, Ballabio A. Molecular and biochemical characterisation of a novel sulphatase gene, Arylsulfatase G (ARSG) Eur J Hum Genet. 2002;10:813–818. doi: 10.1038/sj.ejhg.5200887. [DOI] [PubMed] [Google Scholar]
- 22.Ghosh D. Three-dimensional structures of sulfatases. Methods Enzymol. 2005;400:273–293. doi: 10.1016/S0076-6879(05)00016-9. [DOI] [PubMed] [Google Scholar]
- 23.Biffi A, et al. Metachromatic leukodystrophy: Mutation analysis provides further evidence of genotype–phenotype correlation. Clin Genet. 2008;74:349–357. doi: 10.1111/j.1399-0004.2008.01058.x. [DOI] [PubMed] [Google Scholar]
- 24.Gort L, Coll MJ, Chabás A. Identification of 12 novel mutations and two new polymorphisms in the arylsulfatase A gene: Haplotype and genotype–phenotype correlation studies in Spanish metachromatic leukodystrophy patients. Hum Mutat. 1999;14:240–248. doi: 10.1002/(SICI)1098-1004(1999)14:3<240::AID-HUMU7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 25.Kappler J, von Figura K, Gieselmann V. Late-onset metachromatic leukodystrophy: Molecular pathology in two siblings. Ann Neurol. 1992;31:256–261. doi: 10.1002/ana.410310305. [DOI] [PubMed] [Google Scholar]
- 26.Bostick WD, Dinsmore SR, Mrochek JE, Waalkes TP. Separation and analysis of arylsulfatase isoenzymes in body fluids of man. Clin Chem. 1978;24:1305–1316. [PubMed] [Google Scholar]
- 27.Franco B, et al. A cluster of sulfatase genes on Xp22.3: Mutations in chondrodysplasia punctata (CDPX) and implications for warfarin embryopathy. Cell. 1995;81:15–25. doi: 10.1016/0092-8674(95)90367-4. [DOI] [PubMed] [Google Scholar]
- 28.Puca AA, et al. Identification by shotgun sequencing, genomic organization, and functional analysis of a fourth arylsulfatase gene (ARSF) from the Xp22.3 region. Genomics. 1997;42:192–199. doi: 10.1006/geno.1997.4716. [DOI] [PubMed] [Google Scholar]
- 29.Josephson SA, Schmidt RE, Millsap P, McManus DQ, Morris JC. Autosomal dominant Kufs’’ disease: A cause of early-onset dementia. J Neurol Sci. 2001;188:51–60. doi: 10.1016/s0022-510x(01)00546-9. [DOI] [PubMed] [Google Scholar]
- 30.Katz ML, Sanders DN, Mooney BP, Johnson GS. Accumulation of glial fibrillary acidic protein and histone H4 in brain storage bodies of Tibetan terriers with hereditary neuronal ceroid lipofuscinosis. J Inherit Metab Dis. 2007;30:952–963. doi: 10.1007/s10545-007-0683-y. [DOI] [PubMed] [Google Scholar]
- 31.Riis RC, Cummings JF, Loew ER, de Lahunta A. Tibetan terrier model of canine ceroid lipofuscinosis. Am J Med Genet. 1992;42:615–621. doi: 10.1002/ajmg.1320420437. [DOI] [PubMed] [Google Scholar]
- 32.Frese MA, Schulz S, Dierks T. Arylsulfatase G, a novel lysosomal sulfatase. J Biol Chem. 2008;283:11388–11395. doi: 10.1074/jbc.M709917200. [DOI] [PubMed] [Google Scholar]
- 33.Bains M, Florez-McClure ML, Heidenreich KA. Insulin-like growth factor-I prevents the accumulation of autophagic vesicles and cell death in Purkinje neurons by increasing the rate of autophagosome-to-lysosome fusion and degradation. J Biol Chem. 2009;284:20398–20407. doi: 10.1074/jbc.M109.011791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walkley SU, Vanier MT. Secondary lipid accumulation in lysosomal disease. Biochim Biophys Acta. 2009;1793:726–736. doi: 10.1016/j.bbamcr.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pungor E, Jr, et al. Development of a functional bioassay for arylsulfatase B using the natural substrates of the enzyme. Anal Biochem. 2009;395:144–150. doi: 10.1016/j.ab.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 36.Schenk M, et al. Interaction of arylsulfatase-A (ASA) with its natural sulfoglycolipid substrates: A computational and site-directed mutagenesis study. Glycoconj J. 2009;26:1029–1045. doi: 10.1007/s10719-008-9222-9. [DOI] [PubMed] [Google Scholar]
- 37.Kuronen M, Talvitie M, Lehesjoki AE, Myllykangas L. Genetic modifiers of degeneration in the cathepsin D–deficient Drosophila model for neuronal ceroid lipofuscinosis. Neurobiol Dis. 2009;36:488–493. doi: 10.1016/j.nbd.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 38.Lindblad-Toh K, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 39.Ahmadian A, et al. Single-nucleotide polymorphism analysis by pyrosequencing. Anal Biochem. 2000;280:103–110. doi: 10.1006/abio.2000.4493. [DOI] [PubMed] [Google Scholar]
- 40.Pritchard JK, Stephens M, Rosenberg NA, Donnelly P. Association mapping in structured populations. Am J Hum Genet. 2000;67:170–181. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.