Abstract
Among the arsenal of plant-derived compounds activated upon attack by herbivores and pathogens are small peptides that initiate and amplify defense responses. However, only a handful of plant signaling peptides have been reported. Here, we have isolated a 12-aa peptide from soybean (Glycine max) leaves that causes a pH increase of soybean suspension-cultured cell media within 10 min at low nanomolar concentrations, a response that is typical of other endogenous peptide elicitors and pathogen-derived elicitors. The amino acid sequence was determined and was found to be derived from a member of the subtilisin-like protease (subtilase) family. The sequence of the peptide was located within a region of the protein that is unique to subtilases in legume plants and not found within any other plant subtilases thus far identified. We have named this peptide signal Glycine max Subtilase Peptide (GmSubPep). The gene (Glyma18g48580) was expressed in all actively growing tissues of the soybean plant. Although transcription of Glyma18g48580 was not induced by wounding, methyl jasmonate, methyl salicylate, or ethephon, synthetic GmSubPep peptide, when supplied to soybean cultures, induced the expression of known defense-related genes, such as Cyp93A1, Chib-1b, PDR12, and achs. GmSubPep is a unique plant defense peptide signal, cryptically embedded within a plant protein with an independent metabolic role, providing insights into plant defense mechanisms.
Keywords: damaged-self recognition, plant defense, signaling peptide, subtilase
The modification of survival mechanisms during coevolution of plant hosts with their biotic attackers resulted in the present-day complexity of plant–pathogen and plant–insect interactions. Whereas the invading species has developed methods of adhesion, penetration, and feeding, the plant has evolved mechanisms for perception of attack and activation of defense responses, based on surveillance of its own tissue. Damaged-self recognition occurs when signaling molecules are released from damaged cells and perceived by plant receptors to elicit a defense response (1). These elicitors can be of a heterogeneous nature, such as cutin monomers or cell wall fragments of various sizes (2–4), which exist in all plant species, or they can be a more specific, fine-tuned signal, such as endogenous peptide signals, which may be limited to a single phylogenetic family, coevolving with a specific predator (5–7). Collectively, these diverse compounds are termed endogenous elicitors or damage-associated molecular patterns (DAMPs).
Plants have also evolved receptor-mediated recognition systems for fungal and microbial biochemicals from the sites of infection, termed microbe-associated molecular patterns (MAMPs). These signals include bacterial peptide fragments, such as flg22 and elf18 (8, 9), fungal peptide elicitors, such as Pep13, AVR9, and elicitins (10–12), chitin fragments from fungal cell walls (13), and the heptaglucoside elicitor from the oomycete Phytophtora megasperma (3). MAMPs and DAMPs are perceived on the plant cell surface by receptors that transduce a signal intracellularly, initiating a defense pathway. For example, upon wounding of tomato plants, the plant peptide signal systemin is released from its precursor and, through receptor-mediated events, initiates the jasmonate signaling pathway, producing protease inhibitors and other defense compounds that protect the plant from further attack (14).
Relatively few endogenous peptide defense signals have been isolated thus far. These include a family of glycopeptides from the Solanaceae that are functionally related to systemin, hydroxyproline-rich glycopeptide systemins (HypSys) (15), and a family of signaling peptides from Arabidopsis (AtPeps) that amplify the innate immune responses through the jasmonate/ethylene and salicylate signaling pathways (16, 17). Another plant peptide signal generated from a plant has been found that is derived from an intracellular protein. When cowpea (Vigna unguiculata) leaves were consumed by armyworm larvae, a proteolyzed fragment of the cowpea chloroplastic ATP synthase was produced in the insect gut that was found to elicit defense responses when deposited on the leaves in the oral secretions (18). The peptide was termed inceptin and is an example of an indirect signal generated by the insect that initiates a specific plant defense response.
The complexity of the host–herbivore/pathogen relationship is becoming a common theme in plant biology. In recent work with inceptin, the peptide was active in inducing defense responses in cowpea and not in another member of the Fabaceae family, Glycine max (19). This is similar to systemin, which was found only in one clade of the Solanaceae. In a continuing search for plant elicitors of defense responses, we have isolated a 12-aa peptide from Glycine max that induces the expression of defense genes. The peptide is processed from a unique region of an extracellular subtilisin-like protease (subtilase), providing insight into the mechanism by which host plant–derived, damage-associated signals mediate immune responses.
Results and Discussion
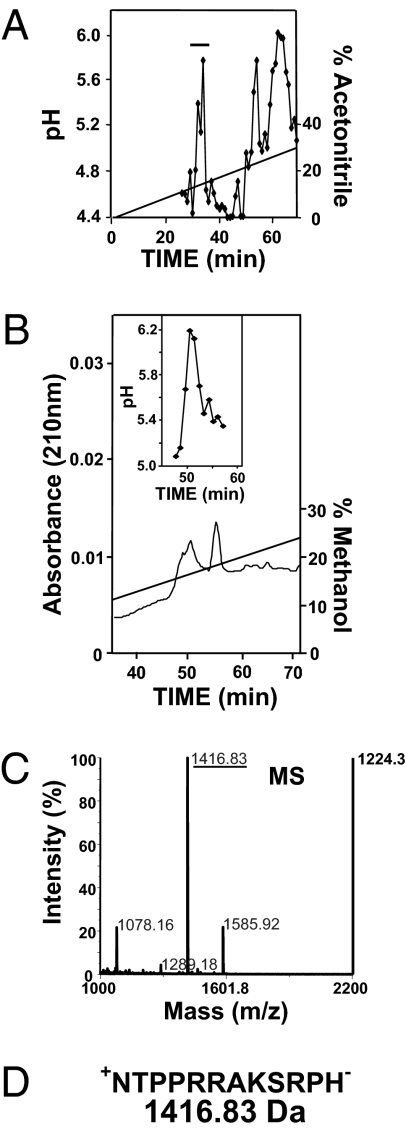
In our investigations of defense peptides, a bioassay has been used that takes advantage of a dramatic increase in pH of the media of suspension cells when a bioactive peptide binds to its receptor (20–22). A crude peptide fraction obtained from soybean leaves displayed the ability to alkalinize soybean suspension cell media when separated on a C18 reversed-phase column (Fig. 1A). Along with large, late-eluting peaks corresponding in retention time to the previously reported rapid alkalinization factor (22), a more hydrophilic peak was detected at 32–34 min. Through a series of HPLC steps and in a manner previously used for the purification of other bioactive peptides (21, 22) (Materials and Methods and SI Materials and Methods), a fraction containing very little UV-absorbing material was shown to induce a strong (over one pH unit) alkalinization response in soybean suspension cell media (Fig. 1B). The active fraction was analyzed by MALDI-MS, and a major mass peak was detected at 1,416.83 Da (Fig. 1C). MALDI-MS/MS fragmentation data coupled with a BLAST search using the soybean genome database (Phytozome; http://www.phytozome.net/) revealed a potential peptide sequence consisting of 12 amino acids: NTPPRRAKSRPH. The peptide was synthesized and confirmed to have the same MS/MS fragmentation pattern as the native peptide (Fig. S1).
Fig. 1.
HPLC purification of GmSubPep from soybean leaf extracts. (A) A crude leaf extract (see Materials and Methods) was applied to a reversed-phase C18 semipreparative HPLC column in 0.1% trifluoroacetic acid/H2O and eluted with an acetonitrile gradient. Fractions (2 mL) were collected, and 10-μL aliquots were assayed for alkalinizing activity. The bioactive fraction (32–34) designated by the bar was pooled for further purification. (B) After several HPLC purification steps, the active fraction was eluted from a narrow-bore C18 column with a methanol gradient as described in SI Materials and Methods and assayed for alkalinizing activity. The active fraction (50–51) was pooled for mass spectral analysis. (C) MALDI mass spectral analysis of the pooled fractions from Fig.1B contained a major mass peak at 1,416.8 Da. (D) The 1,416.83 mass peak was subjected to MS/MS (Fig. S1), and the fragmentation spectra revealed a 12-aa peptide.
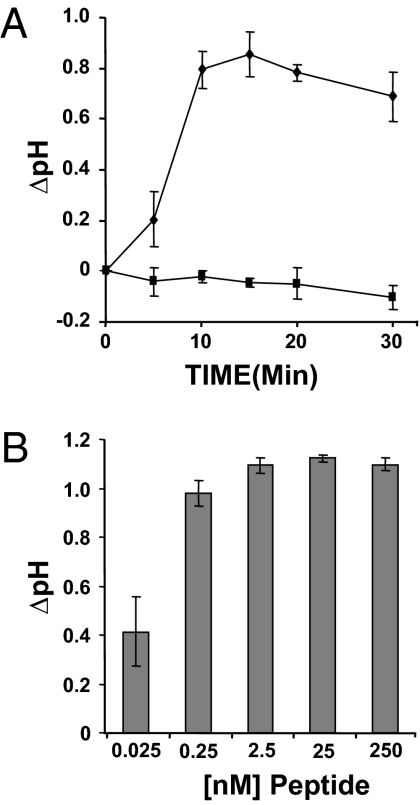
The synthetic soybean peptide was capable of producing a pH change within 10 min and a maximal alkalinizing response in 15 min (Fig. 2A). The peptide was active at extremely low concentrations with a half-maximal response of <0.25 nM (Fig. 2B). The response time and concentration were comparable to previously isolated defense peptides (16, 21, 22).
Fig. 2.
Alkalinization assay kinetics of the synthetic peptide. (A) Cultured soybean cells [strain: PI 553039 (Davis)] were used 4 d after subculture. Aliquots of peptide or H2O (10 μL) were added to 1 mL of suspension cells on an orbital shaker at 160 rpm. The pH of the suspension cell media was recorded at various time points. (B) Aliquots of peptide (10 μL) were added to 1 mL of cells at various concentrations, and the pH of the suspension cell media was recorded after 15 min. Experiments were done in triplicate from three separate flasks of soybean suspension cells. Error bars indicate SD.
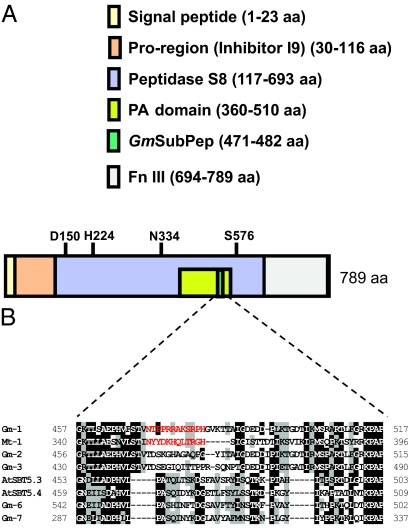
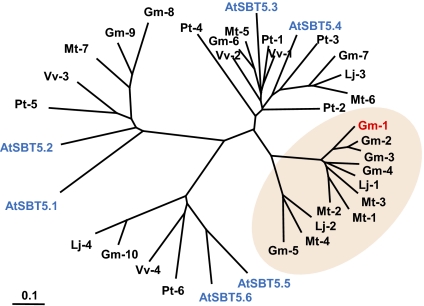
The peptide sequence identified in the soybean genome database was derived from a gene designated Glyma18g48580, which is predicted to code for a subtilisin-like protease (subtilase). Therefore, we have named the peptide Glycine max Subtilase Peptide (GmSubPep). Because the predicted coding sequence of Glyma18g48580 in the database was incomplete, a start codon was predicted from the genomic DNA sequence by comparisons with other subtilases, and the full-length coding region of Glyma18g48480 was amplified by RT-PCR and sequenced (Fig. S2). Glyma18g48580 contains all of the characteristic domains found in plant subtilases, including a signal sequence for secretion to the outside of the cell, a proprotein region, a peptidase domain, a protease-associated (PA) domain located within the peptidase S8 region, and a C-terminal fibronectin III (Fn III) domain (Fig. 3A) (23, 24). The amino acids required for formation of the catalytic triad, Asp-150, His-224, and Asn-334, along with the essential serine at the catalytic site (Ser-576), are conserved (Fig. 3A). The sequence of the biologically active peptide was found within the PA domain of Glyma18g48580, in a region that is unique to a small group of putative soybean [Glyma18g48530 (Gm-1), Glyma18g48490 (Gm-2), and Glyma09g37910 (Gm-3)] subtilases (Fig. 3B). Although the BLAST search indicated that the most similar proteins to Glyma18g48580 in Arabidopsis are AtSBT5.3 (AIR3/At2g04160) and AtSBT5.4 (At5g59810), more similar proteins to AtSBT5.3 and AtSBT5.4 were predicted in the soybean genome database. Phylogenetic analysis revealed that Glyma18g48580 (Gm-1) and its homologs in legume plants form a distinct group with no apparent Arabidopsis, poplar (Populus trichocarpa), or grape (Vitis vinifera) ortholog in subtilase subfamily 5 (Fig. 4). Only one EST sequence (Mt-1) contained a sequence similar to GmSubPep, and this was aligned with GmSubPep with a sequence of “NYYDKHQLTRGH,” containing three matches and three similar amino acids. The peptide was synthesized, but no alkalinizing activity was found.
Fig. 3.
Bioactive peptide is located within Glyma18g48580, which encodes a putative subtilase. (A) The predicted subtilase protein is 789 aa in length, and the peptide sequence is located within the PA domain within the peptidase S8 region. The catalytic triad consists of D150, H224, and N334, with the active site serine at position 576. (B) The peptide-containing region of Glyma18g48580.1 (Gm-1) is compared with the closest subtilase homologs from soybean [Gm-2 (Glyma18g48530.1), Gm-3 (Glyma18g48490.1), and with Gm-6 (Glyma05g28500.1) and Gm-7 (Glyma07g39990.1)], Medicago trancatula [Mt-1 (ABN05911)], and Arabidopsis [AtSBT5.3 (At2g04160) and AtSBT5.4 (At5g59810)]. The isolated peptide and a peptide sequence with similarity to GmSubPep from Mt-1 are indicated in red.
Fig. 4.
Phylogenetic tree of subtilases closely related to the Arabidopsis SBT5 subfamily. The soybean subtilase (Glyma18g48580) containing the bioactive peptide is indicated in red (Gm-1), and the members of the Arabidopsis subtilase 5 family are indicated in blue. The group of Gm-1-related subtilases that seems to be uniquely present in legumes is indicated by pink shading. The protein sequences were obtained from Plant GDB (http://www.plantgdb.org/), Phytozome (http://www.phytozome.net/), and the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/). Abbreviations for genes: Arabidopsis thaliana (AtSBT5.1, At1g20150.1; AtSBT5.2, At1g20160.1; AtSBT5.3, At2g04160.1; AtSBT5.4, At5g59810.1; AtSBT5.5, At5g45640.1; AtSBT5.6, At5g45650.1), Glycine max (Gm-1, Glyma18g48580.1; Gm-2, Glyma18g48530.1; Gm-3, Glyma18g48490.1; Gm-4, Glyma09g37910.1; Gm-5, Glyma14g05230.1; Gm-6, Glyma05g28500.1; Gm-7, Glyma07g39990.1; Gm-8, Glyma06g02500.1; Gm-9, Glyma04g02440.1; Gm-10, Glyma13g29470.1), Medicago truncatula (Mt-1; ABN05911, Mt-2, AC133779_25.4; Mt-3, AC151423_9.5; Mt-4, CR936328_23.4; Mt-5, CT573401_13.4; Mt-6, AC175049_27.4; Mt-7, CT967316_16.4), Lotus japonicus (Lj-1, LjSGA_007130.1; Lj-2, chr2.CM0826.300.nd; Lj-3, chr4.LjT24N17.30.nc; Lj-4, chr6.CM0118.310.nc), Vitis vinifera (Vv-1, GSVIVT00029050001, Vv-2, GSVIVT00033477001; Vv-3, GSVIVT00014361001; Vv-4, GSVIVT00007221001), and Populus trichocarpa (Pt-1, XP_002320540.1; Pt-2, XP_002299062.1; Pt-3, XP_002308740.1; Pt-4, XP_002299063.1; Pt-5, XP_002300693.1; Pt-6, XP_002317314.1).
Among the suspension-cultured cells tested from a wide array of species, only the suspension cells produced from Glycine max were capable of producing an alkalinizing response to GmSubPep (Fig. S3), further suggesting the uniqueness of the peptide sequence in Glyma18g48580. Additionally, when GmSubPep was synthesized with an additional amino acid on either the N-terminal or C-terminal end corresponding to the amino acid coded for by the gene sequence, a significant decrease in alkalinizing activity was observed, indicating that the isolated 12-aa peptide is correctly processed (Fig. S4).
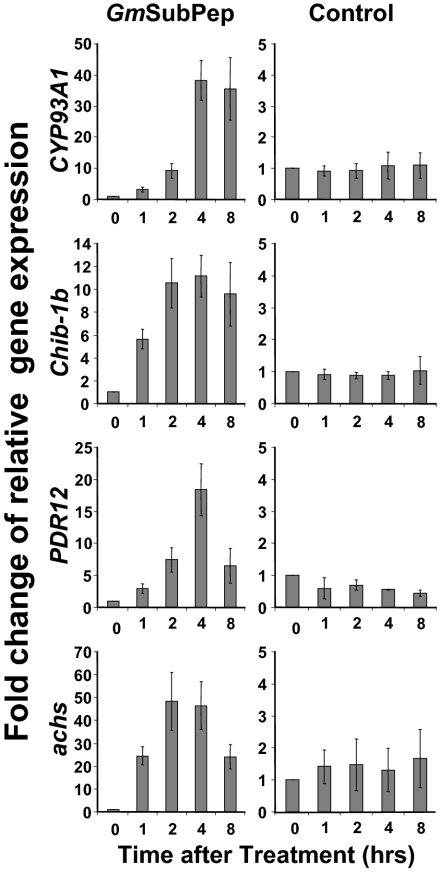
To elucidate the possible involvement of GmSubPep in soybean defense responses, expression analysis of soybean defense-related genes were conducted using quantitative RT-PCR (qRT-PCR) with GmSubPep or the control peptide, systemin, which only induces defense gene expression in the Solanaceae family (Fig. 5). In cells treated with GmSubPep, a cytochrome P450 gene, Cyp93A1, involved in phytoalexin synthesis (25), was induced to 40 times the expression level of the control peptide, and a pathogenesis-related gene, chitinase 1b-1 (Chib-1b) (26), was induced to 12 times control levels. A salicylic acid–inducible ATP-binding cassette transporter, PDR12 (27), was induced to 18 times the expression level of the control peptide, and chalcone synthase (achs) (28), involved in phytoalexin production, was induced to 50 times the control level. The induction was rapid, with a significant induction of Chib-1b and achs in 1 h and induction of Cyp93A1 and PDR12 in 2 h. All four of the genes had maximal expression between 2 and 4 h and began to decline at 8 h.
Fig. 5.
Time course of relative expression levels of defense-related genes in soybean suspension cells in response to GmSubPep as analyzed by real-time RT-PCR. RNA was isolated from suspension cells at various times after addition of either GmSubPep or a control peptide at a final concentration of 25 nM as described in Materials and Methods. The gene expression levels are indicated relative to expression at 0 h. Cyp93A1, a cytochrome P450 gene involved in phytoalexin synthesis; Chib-1b, chitinase gene, a pathogenesis-related gene; PDR12, a salicylic acid–inducible ATP-binding cassette transporter; achs, chalcone synthase, involved in phytoalexin production. ELF1B was amplified as an internal control to normalize the RNA level in each sample. Error bars indicate SD from three biological replicates.
To characterize the precursor gene of GmSubPep, Glyma18g48580 gene expression was analyzed using qRT-PCR. The expression levels of Glyma18g48580 were similar in all actively growing plant tissues, except in mature, lower leaves, where Glyma18g48580 expression was barely detectable (Fig. S5). Hormones that are known regulators of plant defense gene expression, methyl jasmonate, methyl salicylate, and ethephon, did not induce the expression of Glyma18g48580 when compared with two genes, Cyp93A1 and Chib-1b, known to be induced by either wounding or elicitors of defense responses (Fig. S6). In contrast to Glyma18g48580 expression, the genes for prosystemin, proHypSys, and PROPEP have all been shown to be induced by methyl jasmonate and wounding. It may be possible that the subtilase coded for by Glyma18g48580 is activated upon pathogen attack or wounding, thus regulation of any protease activity, as well as the peptide signaling activity, would occur through posttranscriptional mechanisms. Thus, neither would be dependent upon up-regulated transcription of Glyma18g48580.
Recently, the first crystal structure of a plant subtilase revealed that the PA domain protrudes from the subtilase and is involved in dimerization of the subtilase (24). Because GmSubPep seems to be an evolutionary insertion in the PA domain for a small family of proteases, GmSubPep may be positioned in an easily accessible region for cleavage from the subtilase for defense signaling.
The subtilase is predicted to be secreted to the apoplast, placing it in contact with components of fungal or bacterial invasions. The high constitutive expression in developing tissues and lack of induction by defense-related phytohormones suggests involvement in apoplastic processes unrelated to defense, in contrast to some extracellular proteases (29–31). It is conceivable that the subtilase is activated upon attack and that in the ensuing battle with foreign proteins (virulence factors of the invading pathogen) GmSubPep is released to signal the first line of defense, somewhat like vertebrate defense-related proteins that can also stimulate host immune responses (32). Whether Glyma18g48580 interacts specifically with an invading species or has a function in other plant processes remains to be determined.
Herein we report the discovery of GmSubPep, a 12-aa peptide that induces defense gene expression. The activity of GmSubPep resembles that of other peptidic defense signaling peptides in plants, but unlike these known peptides, GmSubPep is derived from an extracellular protease. Two different approaches to elucidating plant peptide ligands have been used, the first being a genetic approach, as exemplified by both the S-locus cysteine-rich (SCR) incompatibility ligands (33) and the CLAVATA3/CLAVATA1 receptor–ligand relationship for differentiation at the shoot apical meristem (34). The second elucidation method is biochemical, whereby peptides are purified from plant extracts and bioassayed, as was the case for the systemins (15), AtPeps (16), and the phytosulfokines (35). Although a genetic dissection of function may have revealed a defense phenotype attributable to the subtilase, the discovery of the unknown peptide signal within the subtilase with a completely different function would have been difficult. Biochemical methodology, established to screen for ligands of membrane-bound receptors, was necessary for the discovery of this unique mechanism of plant defense.
GmSubPep has similarities to the unique inceptin peptide that is localized to the chloroplast and is only released from the chloroplastic ATP synthase protein upon ingestion by an herbivore (18, 19). GmSubPep also has similarities to the hydroxyproline-rich systemins by its presumed localization in the apoplast where, when proteolyzed from the subtilase, it interacts with a yet-to-be-identified membrane-bound receptor to initiate defense signaling. The specific role of GmSubPep will be revealed when the conditions/factors necessary for release of the peptide from its precursor are obtained. Isolation of the receptors and production of mutant/transgenic plants lacking signaling capabilities will be of foremost importance for demonstrating a role for GmSubPep.
Materials and Methods
Alkalinization Assay.
Soybean suspension cells (varieties A3525 and Davis) were maintained in Murashige and Skoog medium as previously described with tobacco cells (21). Cultures were maintained by transferring 2.5–5 mL of cells to 40 mL of media every 7 d and shaking at 160 rpm. Soybean suspension cells were used 4–6 d after transfer. Before assaying for alkalinizing activity, a flask of cells was aliquoted into 24-well cell culture cluster plates (1 mL per well) and allowed to equilibrate at 160 rpm until the pH of the cells ceased to decline (≈2 h). Aliquots of HPLC fractions or purified peptide (1–10 μL) were added, and the pH was recorded after 15 min.
Plant Material and Growth Conditions.
For peptide isolation, soybean plants, Glycine max, variety A3525, were grown in growth chambers (18 h light at 28 °C and 6 h dark at 18 °C, 300 μmol photons m−2 s−1) for ≈4 wk. The plants were sprayed with methyl jasmonate as previously described (21, 35). After 15 h, the leaves were collected, ground in liquid nitrogen, and stored at −20 °C until use. The same conditions were used for collection of RNA samples after treatments, except the plants were used at 3 wk.
Peptide Isolation, Analysis, and Synthesis.
The peptide isolation was done as previously described with modifications (21, 36). The complete method is described in SI Materials and Methods. MALDI spectra were obtained on an Applied Biosystems 4800 TOF/TOF mass spectrometer with 200 Hz Nd-YAG laser. Concentrated samples in aqueous solution were mixed 1:1 with matrix solution (α-cyano-4-hydroxycinnamic acid, 6 mg/mL in 50:50 acetonitrile: 0.25% trifluoroacetic acid in water) and air dried. Calibrated MS spectra (±0.02 Da) were obtained as the summations of 4,000 laser shots, whereas MS/MS spectra (±0.1 Da) were summations of 10,000 laser shots. Peptides were synthesized by N-(9-fluorenyl) methoxycarbonyl chemistry by solid-phase techniques using an Applied Biosystems model 431 synthesizer and purified by reversed-phase HPLC. Peptide stocks (2.5 mM in distilled water) were checked for purity and for correctness with the predicted mass on a Finnigan LC/Q mass spectrometer using direct injection.
Wounding, Hormone, and Peptide Treatments.
Three-week-old Glycine max (variety A3525) plants having six to eight expanded leaves were used in wounding experiments, performed under growth chamber conditions consisting of 18 h light at 28 °C and 6 h dark at 18 °C (300 μmol photons m−2 s−1). For each plant, the top four leaves were wounded across the midvein using a hemostat. Time-course experiments were performed in which the wounded leaves were collected at 0, 0.5, 1, 2, 4, and 8 h after the mechanical injury. The corresponding leaves from unwounded plants served as controls for each time point. The leaf samples were immediately frozen in liquid nitrogen and kept at −80 °C until use. For treatment with defense gene inducers, plants were sprayed with solutions of either 625 μM MeJA, 2 mM methyl salicylate, or 7 mM ethephon: all in double-distilled H2O containing 0.1% Triton X-100. Control plants were sprayed with 0.1% Triton X-100. The leaf samples were collected in triplicate for time-course experiments after spraying as above and immediately frozen in liquid nitrogen and kept at −80 °C until use. The leaf material was ground to a fine powder in a mortar and pestle with liquid N2, and total RNA was isolated with TRIZOL reagent (Invitrogen) according to the manufacturer's protocol.
Suspension cells (variety: Davis) were used for determining the induction of genes by GmSubPep. Cells were grown as described above. At 4 d, either GmSubPep or a control peptide, systemin, were added to a final concentration of 25 nM. Three-milliliter aliquots were removed at 0, 1, 2, 4, and 8 h and were filtered through a #4 Whatman filter paper. The cells were immediately frozen in liquid nitrogen and stored at −80 °C until use. RNA samples were prepared as described above.
Computer Analyses.
Domain prediction was performed by the Pfam program (37) and by comparison with tomato (Solanum lycopersicum) subtilase 3 (SlSBT3), whose crystal structure has recently been determined (24). Subcellular localization was predicted by the WoLF PSORT (38) and the TargetP 1.1 programs (39). Phylogenetic analysis was conducted by the Clustal W program (40). The phylogenetic tree was drawn by the TreeView program (41).
PCR Analysis.
Five micrograms of total RNA was reverse transcribed using SuperScript II (Invitrogen), and the resulting cDNA was used for amplifying the full-length coding sequence of Glyma18g48580 by PCR with SubPep-F (5′-TATGCCTGACAAGCAATTCG-3′) and SubPep-R (5′-CTCGACCAATTTGGGAATTT-3′) primers. For the gene expression analysis, 1 μg of total RNA was subjected to the RT reaction using the DyNAmo cDNA Synthesis Kit (Finnzymes) according to the manufacturer's protocol. The cDNA was diluted five times with H2O and then subjected to real-time qPCR analysis using the DyNAmo SYBR Green qPCR kit (Finnzymes) and the Mx3000P (Stratagene). Primers used in qRT-PCR are listed in Table S1. Primers for elongation factor 1-β (ELF1B) were included as an internal control for normalization in all experimental runs. DNA sequences [ELF1B (Glyma02g44460), CYP93A1 (D83968), Chib-1b (AB007127), Gmachs1 (X54644), and PDR12 (AM261476)] were obtained from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/), Phytozome (http://www.phytozome.net/), and PlantGDB (http://www.plantgdb.org/).
Supplementary Material
Acknowledgments
We thank Sue Vogtman for growing plants; Dr. William Siems for assistance in obtaining MS analyses; Dr. Gerhard Munske for peptide synthesis; Dr. John Browse for advice and support; and Drs. Andreas Schaller, Gregg Howe, and Alisa Huffaker for critical reading of the manuscript. All experiments were performed in the laboratory of Dr. Clarence A. Ryan and supported by National Science Foundation Grant IBN 0090766 (to C.A.R.), the Charlotte Y. Martin Foundation (C.A.R.), Monsanto Company, and the Washington State University College of Agriculture, Human, and Natural Resources Sciences.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
4Deceased October 7, 2007.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007568107/-/DCSupplemental.
References
- 1.Heil M. Damaged-self recognition in plant herbivore defence. Trends Plant Sci. 2009;14:356–363. doi: 10.1016/j.tplants.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Schweizer P, Felix G, Buchala A, Muller C, Metraux JP. Perception of free cutin monomers by plant cells. Plant J. 1996;10:331–341. [Google Scholar]
- 3.Darvill AG, Albersheim P. Phytoalexins and their elicitors—a defense against microbial infection in plants. Annu Rev Plant Physiol Plant Mol Biol. 1984;35:243–275. [Google Scholar]
- 4.Ryan CA. Oligosaccharide signalling in plants. Annu Rev Cell Biol. 1987;3:295–317. doi: 10.1146/annurev.cb.03.110187.001455. [DOI] [PubMed] [Google Scholar]
- 5.Howe GA, Jander G. Plant immunity to insect herbivores. Annu Rev Plant Biol. 2008;59:41–66. doi: 10.1146/annurev.arplant.59.032607.092825. [DOI] [PubMed] [Google Scholar]
- 6.Wu J, Baldwin IT. Herbivory-induced signalling in plants: Perception and action. Plant Cell Environ. 2009;32:1161–1174. doi: 10.1111/j.1365-3040.2009.01943.x. [DOI] [PubMed] [Google Scholar]
- 7.Boller T, Felix G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 8.Felix G, Duran JD, Volko S, Boller T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999;18:265–276. doi: 10.1046/j.1365-313x.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- 9.Kunze G, et al. The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell. 2004;16:3496–3507. doi: 10.1105/tpc.104.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hahlbrock K, et al. Oligopeptide elicitor-mediated defense gene activation in cultured parsley cells. Proc Natl Acad Sci USA. 1995;92:4150–4157. doi: 10.1073/pnas.92.10.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van den Ackerveken GF, Vossen P, De Wit PJGM. The AVR9 race-specific elicitor of Cladosporium fulvum is processed by endogenous and plant proteases. Plant Physiol. 1993;103:91–96. doi: 10.1104/pp.103.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamoun S. Nonhost resistance to Phytophthora: novel prospects for a classical problem. Curr Opin Plant Biol. 2001;4:295–300. doi: 10.1016/s1369-5266(00)00176-x. [DOI] [PubMed] [Google Scholar]
- 13.Kendra DF, Hadwiger LA. Characterization of the smallest chitosan oligomer that is maximally antifungal to Fusarium solani and elicits pisatin formation in Pisum sativum. Exp Mycol. 1984;8:276–281. [Google Scholar]
- 14.Ryan CA, Pearce G, Scheer J, Moura DS. Polypeptide hormones. Plant Cell. 2002;14(Suppl):S251–S264. doi: 10.1105/tpc.010484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryan CA, Pearce G. Systemins: A functionally defined family of peptide signals that regulate defensive genes in Solanaceae species. Proc Natl Acad Sci USA. 2003;100(Suppl 2):14577–14580. doi: 10.1073/pnas.1934788100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huffaker A, Pearce G, Ryan CA. An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc Natl Acad Sci USA. 2006;103:10098–10103. doi: 10.1073/pnas.0603727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huffaker A, Ryan CA. Endogenous peptide defense signals in Arabidopsis differentially amplify signaling for the innate immune response. Proc Natl Acad Sci USA. 2007;104:10732–10736. doi: 10.1073/pnas.0703343104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmelz EA, et al. Fragments of ATP synthase mediate plant perception of insect attack. Proc Natl Acad Sci USA. 2006;103:8894–8899. doi: 10.1073/pnas.0602328103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmelz EA, Engelberth J, Alborn HT, Tumlinson JH, 3rd, Teal PEA. Phytohormone-based activity mapping of insect herbivore-produced elicitors. Proc Natl Acad Sci USA. 2009;106:653–657. doi: 10.1073/pnas.0811861106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Felix G, Boller T. Systemin induces rapid ion fluxes and ethylene biosynthesis in Lycopersicon peruvianum cells. Plant J. 1995;7:381–389. [Google Scholar]
- 21.Pearce G, Moura DS, Stratmann J, Ryan CA. Production of multiple plant hormones from a single polyprotein precursor. Nature. 2001;411:817–820. doi: 10.1038/35081107. [DOI] [PubMed] [Google Scholar]
- 22.Pearce G, Moura DS, Stratmann J, Ryan CA. RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc Natl Acad Sci USA. 2001;98:12843–12847. doi: 10.1073/pnas.201416998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cedzich A, et al. The protease-associated domain and C-terminal extension are required for zymogen processing, sorting within the secretory pathway, and activity of tomato subtilase 3 (SlSBT3) J Biol Chem. 2009;284:14068–14078. doi: 10.1074/jbc.M900370200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ottmann C, et al. Structural basis for Ca2+independence and activation by homodimerization of tomato subtilase 3. Proc Natl Acad Sci USA. 2009;106:17223–17228. doi: 10.1073/pnas.0907587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki G, et al. Induction of a novel cytochrome P450 (CYP93 family) by methyl jasmonate in soybean suspension-cultured cells. FEBS Lett. 1996;383:83–86. doi: 10.1016/0014-5793(96)00229-3. [DOI] [PubMed] [Google Scholar]
- 26.Watanabe A, et al. Molecular cloning and ethylene-inducible expression of Chib1 chitinase from soybean (Glycine max (L.) Merr.) Biosci Biotechnol Biochem. 1999;63:251–256. doi: 10.1271/bbb.63.251. [DOI] [PubMed] [Google Scholar]
- 27.Eichhorn H, Klinghammer M, Becht P, Tenhaken R. Isolation of a novel ABC-transporter gene from soybean induced by salicylic acid. J Exp Bot. 2006;57:2193–2201. doi: 10.1093/jxb/erj179. [DOI] [PubMed] [Google Scholar]
- 28.Akada S, Kung SD, Dube SK. The nucleotide sequence of gene 1 of the soybean chalcone synthase multigene family. Plant Mol Biol. 1991;16:751–752. doi: 10.1007/BF00023443. [DOI] [PubMed] [Google Scholar]
- 29.Krüger J, et al. A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science. 2002;296:744–747. doi: 10.1126/science.1069288. [DOI] [PubMed] [Google Scholar]
- 30.Xia Y, et al. An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO J. 2004;23:980–988. doi: 10.1038/sj.emboj.7600086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tornero P, Conejero V, Vera P. Primary structure and expression of a pathogen-induced protease (PR-P69) in tomato plants: Similarity of functional domains to subtilisin-like endoproteases. Proc Natl Acad Sci USA. 1996;93:6332–6337. doi: 10.1073/pnas.93.13.6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tani K, et al. Defensins act as potent adjuvants that promote cellular and humoral immune responses in mice to a lymphoma idiotype and carrier antigens. Int Immunol. 2000;12:691–700. doi: 10.1093/intimm/12.5.691. [DOI] [PubMed] [Google Scholar]
- 33.Schopfer CR, Nasrallah ME, Nasrallah JB. The male determinant of self-incompatibility in Brassica. Science. 1999;286:1697–1700. doi: 10.1126/science.286.5445.1697. [DOI] [PubMed] [Google Scholar]
- 34.Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- 35.Matsubayashi Y, Sakagami Y. Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proc Natl Acad Sci USA. 1996;93:7623–7627. doi: 10.1073/pnas.93.15.7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearce G, et al. Isolation and characterization of hydroxyproline-rich glycopeptide signals in black nightshade leaves. Plant Physiol. 2009;150:1422–1433. doi: 10.1104/pp.109.138669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36(Database issue):D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horton P, et al. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007;35(Web Server issue):W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 40.Larkin MA, et al. ClustalW and ClustalX version 2. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 41.Page RDM. TreeView: An application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.