Abstract
The origin recognition complex (ORC) is a DNA replication initiator protein also known to be involved in diverse cellular functions including gene silencing, sister chromatid cohesion, telomere biology, heterochromatin localization, centromere and centrosome activity, and cytokinesis. We show that, in human cells, multiple ORC subunits associate with hetereochromatin protein 1 (HP1) α- and HP1β-containing heterochromatic foci. Fluorescent bleaching studies indicate that multiple subcomplexes of ORC exist at heterochromatin, with Orc1 stably associating with heterochromatin in G1 phase, whereas other ORC subunits have transient interactions throughout the cell-division cycle. Both Orc1 and Orc3 directly bind to HP1α, and two domains of Orc3, a coiled-coil domain and a mod-interacting region domain, can independently bind to HP1α; however, both are essential for in vivo localization of Orc3 to heterochromatic foci. Direct binding of both Orc1 and Orc3 to HP1 suggests that, after the degradation of Orc1 at the G1/S boundary, Orc3 facilitates assembly of ORC/HP1 proteins to chromatin. Although depletion of Orc2 and Orc3 subunits by siRNA caused loss of HP1α association to heterochromatin, loss of Orc1 and Orc5 caused aberrant HP1α distribution only to pericentric heterochromatin-surrounding nucleoli. Depletion of HP1α from human cells also shows loss of Orc2 binding to heterochromatin, suggesting that ORC and HP1 proteins are mutually required for each other to bind to heterochromatin. Similar to HP1α-depleted cells, Orc2 and Orc3 siRNA-treated cells also show loss of compaction at satellite repeats, suggesting that ORC together with HP1 proteins may be involved in organizing higher-order chromatin structure and centromere function.
Keywords: centromere, origin recognition complex, pericentric heterochromatin, chromatin structure
The initiation of DNA replication in eukaryotic cells is a highly coordinated process that is intimately linked to chromatin organization. The accurate duplication of DNA is governed by many proteins including the origin recognition complex (ORC) (1, 2). Binding of ORC to replication origins is a key step in initiation of DNA replication, leading to the recruitment of other prereplication complex (pre-RC) proteins including Cdc6, Cdt1, and the MCM2-7. Unlike yeast ORC, human ORC binds to chromatin in a nonsequence-specific manner (3). In human cells, Orc2–5 forms the core ORC, and Orc1 is transiently associated with this complex (4–7). During the G1 to S phase transition, human Orc1 dissociates from chromatin, is ubiquitylated and degraded by ubiquitin-mediated proteolysis, and is reloaded to the chromatin at the M-G1 transition when new pre-RCs are formed (5, 7–9). The remaining ORC subunits are dynamically associated with chromatin throughout the remainder of the cell cycle (5, 6).
In addition to their role in DNA replication, ORC subunits have also been implicated in transcriptional gene silencing in Saccharomyces cerevisiae (10), heterochromatin formation in Drosophila, mouse, and humans (11–15), sister chromatid cohesion in S. cerevisiae, Schizosaccharomyces pombe, and Xenopus (16–18), ribosome biogenesis (19), centrosome biology in human and mouse (15, 20, 21), mitotic chromosome function in human and Xenopus (15, 22, 23), cytokinesis in human cells and Drosophila (24, 25), regulation of dendrite development in postmitotic neurons (26), and neural transmission and synaptic function in Drosophila (27) (see reviews in refs. 28, 29).
The human Orc2 subunit is associated with heterochromatin in a cell cycle-dependent manner, being present only at the centromeres during late G2 and mitosis (15, 30). Furthermore, Orc2 biochemically interacts indirectly with the heterochromatin protein 1 (HP1) in mammalian, Drosophila, and Xenopus cells (14, 15, 31, 32). Depletion of Orc2 or Orc3 in HeLa cells results in a mitotic arrest with abnormally condensed chromosomes, lack of chromosome alignment at the metaphase plate, and spindle defects (15).
The interaction between ORC and heterochromatic proteins like HP1α and β and ORC binding to centromeres suggests a direct role for ORC subunits in chromosome organization and for chromosome condensation and segregation during mitosis (33–35). HP1 is a heterochromatin-associated protein that localizes to pericentromeres and telomeres, has a dose-dependent effect on gene silencing, and has a critical role in heterochromatin formation and maintenance (36). To address the role of ORC/HP1 in heterochromatin organization and mitotic progression, we systematically analyzed the role of individual ORC subunits in HP1 binding and recruitment to heterochromatic foci in human cells. We find that ORC and HP1 are required for maintenance of each other's association with heterochromatin and identify a role for ORC in maintenance of higher-order chromatin structure at centromeres. Interestingly, different ORC subunits play differential roles in heterochromatin structure and function.
Results
Multiple ORC Subunits Localize to Heterochromatin.
Immunofluorescence studies showed that human and mouse Orc2 localizes in a cell cycle-regulated manner to many diffuse foci in the nucleus, HP1-containing heterochromatic loci, and centrosomes and centromeres (Fig. 1 B’–D’) (11, 15, 20, 21, 30). In addition to Orc2, other ORC subunits including Orc1, Orc3, and Orc5 localize at the Orc2- and HP1-associated heterochromatin in MCF7 cells (Fig. 1). In human cells, Orc2 coimmunoprecipitates with HP1α and β (15), and in Drosophila embryonic nuclei, HP1 colocalizes with Orc2 at heterochromatin (14). Because other ORC subunits also colocalize with HP1α in human cells, it suggests an evolutionary conserved function for ORC at heterochromatin.
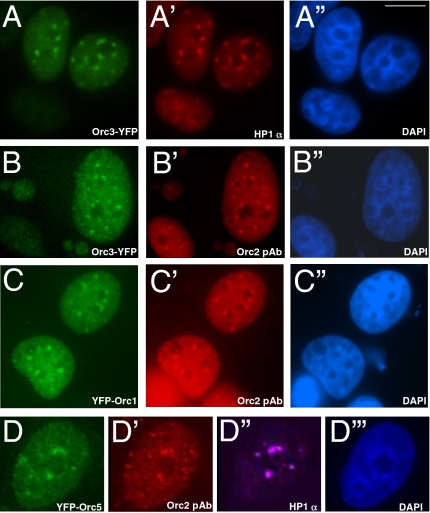
Fig. 1.
Multiple ORC subunits localize with HP1 to heterochromatic foci. YFP fusion constructs were transfected into MCF7 cells, and endogenous proteins were detected by indirect immunofluorescence. Orc3-YFP colocalizes with endogenous HP1α at heterochromatic foci (A, A’, and A”). Endogenous Orc2 and Orc3-YFP colocalize at the heterochromatic foci (B, B’, and B”). YFP-Orc1 colocalizes with endogenous Orc2 at the heterochromatic foci (C, C’, and C”). YFP-Orc5 showed colocalization with both endogenous Orc2 and HP1α at the heterochromatic foci in MCF7 cells (D, D’, D”, and D”’). Chromatin was stained with DAPI in blue. (Scale bar, 5 μm.)
Differential Binding of ORC Subunits to Heterochromatin.
A large proportion of the HP1 protein pool displays a remarkably dynamic exchange at heterochromatin (37–39). Because the human Orc1, Orc2, Orc3, and Orc5 subunits localized to heterochromatin, we compared the mobility of YFP-tagged ORC subunits with YFP-HP1α at heterochromatin by fluorescence recovery after photo-bleaching (FRAP) analysis (Fig. 2A). A defined heterochromatic region in the nucleus was irreversibly bleached of the YFP signal, and the recovery kinetics of fluorescence intensity into the bleached region were assessed. Orc2 and Orc3 had very similar and complete kinetics of fluorescence recovery, as observed for HP1α. However, Orc1 showed very different and incomplete recovery kinetics, suggesting that Orc1 chromatin association is unique among ORC subunits tested, perhaps reflecting different chromatin interaction (Fig. 2A). Thus, by FRAP analysis, it is clear that ORC proteins, especially Orc2 and Orc3, assemble and disassemble from the heterochromatic foci in a dynamic fashion like the bulk of HP1α protein, but Orc1 seems to exchange less frequently, if at all.
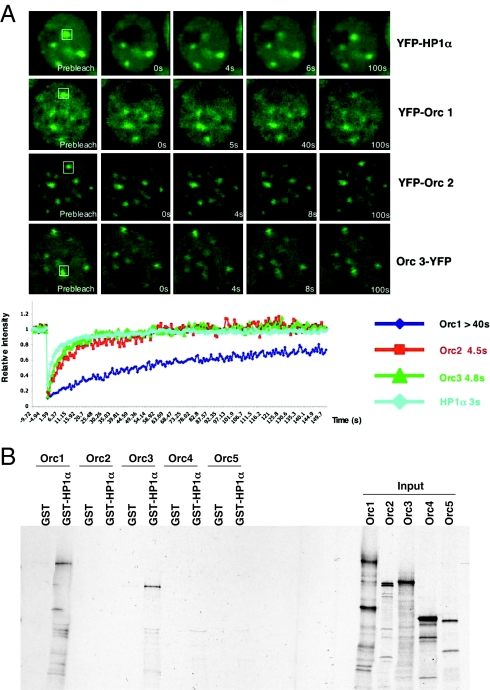
Fig. 2.
ORC dynamics at the heterochromatin. (A) FRAP kinetics of various ORC subunits at the heterochromatic foci in MCF7 cells. Similar FRAP kinetics of ∼3–4 s t1/2 for YFP-HP1α, YFP-Orc2, and Orc3-YFP were observed at heterochromatic foci, whereas YFP-Orc1 showed >40 s t1/2 at heterochromatin. (B) GST pull-down assays using GST or GST-HP1α beads and incubating with in vitro transcribed and translated mRNA from individual ORC subunits shows direct binding of both human Orc1 and Orc3 to HP1α. The input is shown on Right.
Orc1 and Orc3 Directly Bind to HP1α.
In Drosophila and Xenopus, HP1 was found to interact with the N-terminal domain of the Orc1 subunit (14). In human cells, Orc1 interacts with HP1; however, Orc1 is degraded after entry into S phase (7–9, 40) at the time when Orc2, 3, and 5 continue to show restricted association to centric heterochromatin. The localization of some ORC subunits at heterochromatin in the latter one-half of the cell-division cycle, when Orc1 is greatly reduced or not present, suggests that another ORC subunit must associate with heterochromatin through an interaction that is independent of Orc1. This interaction could be either direct or through an ORC/HP1-associated protein (41). To resolve this paradox, we carried out GST pull-down assays to determine if other ORC subunits bind to HP1. The results show that both human Orc1 and Orc3 independently bind to HP1α (Fig. 2B and Fig. S1).
Orc3 Contains Two Domains That Interact with HP1α.
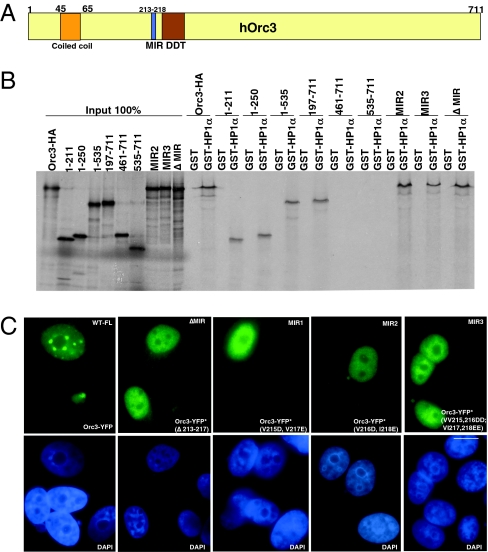
The region of the Orc1 protein that binds to HP1α has been mapped as partially overlapping with the bromo-associated homology domain of Orc1 (aa151-269) (32). To map the region of Orc3 that interacts with HP1α, various mutants of Orc3 were generated in a coupled in vitro transcription-translation vector and tested for binding to GST or GST-HP1α (Fig. 3 A and B and Fig. S2). Two independent domains in Orc3, a coiled-coil domain at the N terminus and a second region containing a MOD1-interacting region (MIR; 213-218aa) (42), were identified, both of which directly bound to HP1α. The GST pull down was conducted at 100 mM NaCl conditions (Fig. 3B). The interaction of HP1 with the MIR domain was reduced at higher salt conditions (150 mM NaCl condition); however, the interaction with the coiled-coil domain remained unaffected. The Orc3 MIR domain was similar to a HP1-binding domain found in chromatin assembly factor 1 (CAF1) (42). The consensus sequence PXVHH was found in human Orc3 and was found to be highly conserved in all animal and plant species analyzed (Fig. 3A and Fig. S3 A and B). Fluorescent-tagged constructs of Orc3 harboring MIR mutants were generated, and their expression and localization were analyzed in human MCF7 cells after transient transfections (Fig. 3C and Fig. S3C). All MIR mutants failed to localize to the heterochromatin, showing that, although Orc3 can interact with HP1 in the absence of the MIR domain, the MIR domain is critical in vivo. Other mutations in the coiled-coil region, converting lysine residues spaced four amino acids apart to acidic residues alone, did not affect Orc3-HP1 interaction, but when combined with a MIR domain deletion, they abolished the interaction between Orc3 and HP1 (Fig. S2B). Interestingly, these coiled-coil mutations within the context of the short amino-terminal fragment of Orc3 (amino acids 1–133) did not abolish the interaction between HP1 in a GST-HP1α pull-down assay, but within the context of full-length Orc3, they were required along with the MIR domain. Thus, the interaction between Orc3 and HP1α is complex but consistent with FRAP experiments showing that HP1α (37–39), Orc2, and Orc3 (Fig. 2A) all have a t1/2 in the range of 3–4 s, suggesting that they may be recruited to and maintained on heterochromatin as a single complex.
Fig. 3.
Mapping the Orc3- and HP1α-interaction domain. (A) Schematic representation of predicted domains or motifs in Orc3 protein. (B) Various deletion and mutant Orc3 constructs were in vitro transcribed/translated and used in GST pull-down assays with GST alone or GST-HP1α in 100 mM NaCl. The MIR mutants are Mir2, V216D, I218E; Mir3, VV215,216DD, VI217,218EE, or a Mir-domain deletion. All Orc3 constructs were tagged with the HA epitope. Orc3 has two HP1α-interacting regions, one is the coiled-coil region at the N terminus and the second is the MIR-containing region. (C) Immunolocalization of Orc3-YFP and Orc3-YFP* (MIR mutants) in human cells after transfection into MCF7 cells. (Scale bar, 5 μm.)
Depletion of Individual ORC Subunits Disrupts HP1α Localization.
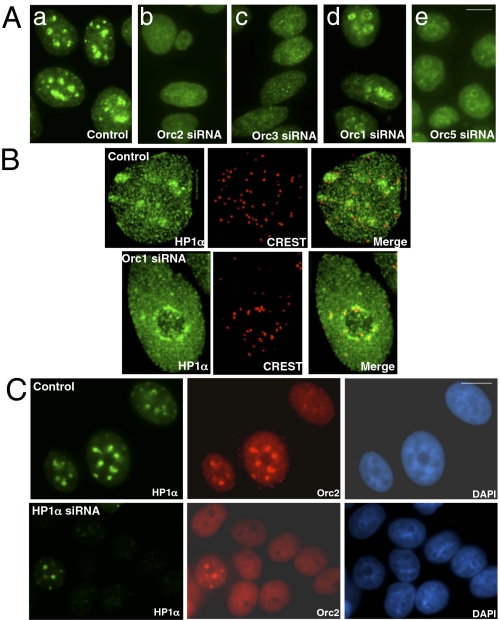
To address the role of individual ORC subunits in HP1 localization to heterochromatin, a RNAi approach (43) was used to determine the consequences of loss of specific ORC subunits in human cells (Fig. S4). Whereas HP1α labeling in control HeLa cells localized to pronounced heterochromatic foci (Fig. 4Aa), in Orc1- (Fig. 4Ad) and Orc5-depleted (Fig. 4Ae) cells, HP1 localized to a ring-like distribution around the periphery of the nucleoli (Fig. S5). Orc1- and Orc5-depleted cells also showed clustering of centromeres (CREST localization) that colocalized with HP1α around the nucleolar periphery, suggesting a change in chromatin organization in the absence of these two ORC subunits (Fig. 4B). In contrast, cells treated with siRNA that depleted Orc2 and Orc3 showed a predominantly homogenous labeling of HP1α, although some residual small foci remained (>60% of the cells) (Fig. 4 Ab and Ac and Fig. S5). Mutations in Drosophila Orc2 also disrupt HP1 association with heterochromatin (31). Our observations and results from Drosophila on the interaction between ORC and HP1 (14, 31, 41) suggest that ORC facilitates recruitment of HP1 to heterochromatin or ORC stabilizes the association of HP1 at heterochromatin. Interestingly, displacement of HP1 from heterochromatin using deacetylase inhibitors has shown that Orc1 continues to associate with heterochromatin under these conditions (32). In contrast, treatment of human cells with siRNA targeted to HP1α resulted in the loss of Orc2 association with heterochromatin (Fig. 4C), suggesting that Orc2 (with Orc3) and HP1 may be recruited to the chromatin as a single complex and require each other for their stable association to heterochromatin.
Fig. 4.
Depletion of ORC results in abnormal distribution of HP1α protein. (A) Depletion of individual ORC subunits results in aberrant organization of HP1α proteins. Depletion of Orc2 (Ab) and Orc3 (Ac) from human HeLa cells results in loss of HP1 from heterochromatic foci and redistribution as homogenous pool. Depletion of Orc1 (Ad) and Orc5 (Ae) results in the redistribution of HP1 to the pericentric heterochromatin, mostly around the nucleolar periphery. (B) Luciferase siRNA-treated cells showing HP1α-positive foci (green) and CREST-labeling centromere (red). In contrast, the Orc1 siRNA-treated cells showed redistribution of HP1α foci and clustering of the centromeres around the nucleolar periphery. (C) Recruitment of Orc2 to heterochromatin is also HP1α-dependent. HP1α siRNA-treated cells show loss of Orc2 binding to prominent heterochromatic foci. Note that one HP1α-positive cell continues to show binding of Orc2 to the heterochromatic foci. Chromatin was stained with DAPI (blue). (Scale bar, 5 μm.)
Loss of ORC Proteins Results in Abnormal Compaction of Satellite Repeats.
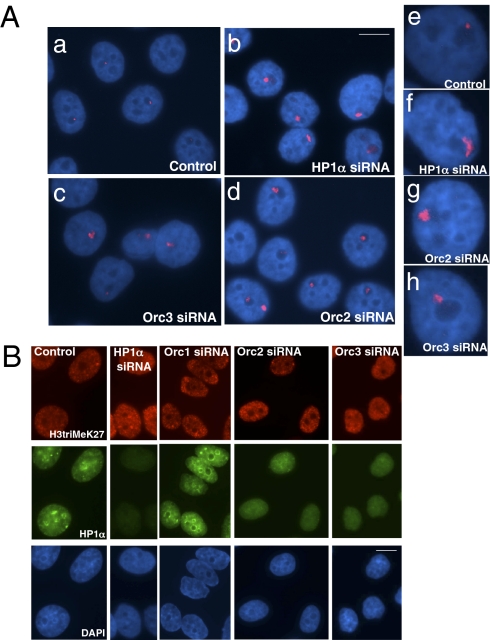
Centromeric chromatin is highly compacted and consists of transcriptionally repressed satellite repeats (44, 45). HP1 has been shown to concentrate mainly at pericentromeric heterochromatin (30). Depletion of the Orc2 subunit of ORC resulted in defects in HP1 localization to heterochromatin and also, abnormally condensed chromosomes during mitosis (Fig. 4) (15). HP1/Swi6 has been shown to be required for the recruitment of cohesin to centromeric regions, promoting proper chromosome segregation (46). We, therefore, examined the effect of ORC subunit depletion on chromatin compaction at specific satellite repeats at a centromeric region in human cells. Specific chromosome 9 satellite probes (Qbiogene) were used for DNA FISH in HeLa cells depleted of Orc2 or Orc3 with siRNAs. In cells targeted with a control luciferase siRNA, chromosome 9 satellite appeared as discrete, single compact foci in the majority of cells (Fig. 5 Aa and Ae) (85 ± 4%), whereas in cells treated with HP1α siRNA, the repeated DNA decompacted in about one-half of the cells (Fig. 5 Ab and Af) (45 ± 5%). Similar to HP1α-depleted cells, Orc2 siRNA-treated (Fig. 5 Ad and Ag) (37 ± 6% cells) and Orc3 siRNA-treated (Fig. 5 Ac and Ah) (35 ± 4%) cells showed loss of compaction of the chromosome 9 satellite repeat region.
Fig. 5.
ORC proteins are required for HP1-associated heterochromatin assembly. (A) Loss of ORC proteins results in abnormal compaction of satellite repeats. DNA FISH using a chromosome 9 satellite repeat probe in HeLa cells depleted of individual ORC subunits Orc3 (Ac) and Orc2 (Ad) results in loss of chromatin compaction at chromosome 9 satellite repeats. Similar observation was made for HP1α siRNA-treated cells (Ab). (Scale bar, 5 μm.) Higher-resolution images showing decondensed chromatin associated with chromosome 9 satellite repeat in cells depleted of HP1α (Af), Orc2 (Ag), and Orc3 (Ah) siRNA-treated cells. Note the single compacted DNA FISH signal in luciferase siRNA-treated cells (Aa and Ae). (B) ORC depletion does not affect Polycomb-associated heterochromatin. Localization of Polycomb-associated repressive mark; trimethylation of K27 on H3 was analyzed in cells depleted of HP1α, Orc1, Orc2, or Orc3. No change in the K27 trimethylation on H3 was observed, whereas the HP1α proteins continue to show abnormal distribution on ORC-depleted cells. (Scale bar, 10 μm.)
ORC Depletion Does Not Affect Polycomb Association with Heterochromatin.
ORC proteins associate with heterochromatic proteins like HP1 in a cell cycle-dependent manner. Whether ORC subunits play a more global role in establishment of heterochromatic structures in the mammalian cell nucleus was addressed by studying the effect of ORC depletion on other repressed chromatin states such as in Polycomb-associated heterochromatic regions (47). Polycomb-related proteins associate with pericentromeric heterochromatin and play important roles in facultative heterochromatin regulation and transcriptional repression, including association with ORC in plants (48, 49). To determine whether ORC subunits are involved in the recruitment or maintenance of various Polycomb factors to heterochromatic regions, we used a RNAi approach to deplete individual ORC subunits from human cells and study the localization of Polycomb-associated repressive marks, including trimethylation of K27 on histone H3 (Fig. 5B), components of the PRC1;Bmi1 (Fig. S6), and PRC2; EZH2 (Fig. S7). No significant changes were observed in the localization of Polycomb-associated proteins and chromatin modification after ORC depletion.
Discussion
In human cells, ORC subunits display a dynamic association with chromatin during the cell-division cycle (5, 7–9, 40). Orc1 associates with chromatin before other ORC subunits during telophase of rapidly proliferating cells and remains bound during G1 phase (8, 32, 50). Available evidence suggests that the entire ORC is assembled during G1 phase, although subcomplexes exist (5, 6); however, Orc1 is degraded after the G1 to S phase transition, and Orc2 and Orc3 gradually are displaced from chromatin during S phase, ending up exclusively at centric (and in some cells, also telomeric) heterochromatin in mitosis (15). A principal role for ORC is in establishing pre-RCs; however, 30% of human cells that had Orc2 or Orc3 depleted were arrested in mitosis with abnormally condensed chromosomes and defects in chromosome segregation (15), similar to the observation that Orc2 mutation in Drosophila cause defects in mitosis (14, 34, 51). Orc2 and Orc3 are associated with constitutive heterochromatin and HP1 in Drosophila cells (14, 41). We have shown that multiple ORC subunits are associated with heterochromatin, but Orc1 is only associated in G1 phase before it is degraded. Thus, it was of interest to find that ORC has multiple ways to associate with HP1 and heterochromatin, one involving Orc1 (14, 32) and the other involving a multidomain interaction with Orc3 that is independent of Orc1. The interactions between Orc3 and HP1 involve a conserved MIR domain (PXVHH) that is also found in other HP1-interacting proteins such as CAF-1, where the MIR motif interacts with the HP1 shadow chromodomain (42, 52), thereby allowing the chromodomain of HP1 to bind to histone H3 trimethyl-K9 residues found at heterochromatin (53). Depletion of ORC subunits did not qualitatively effect the histone H3 trimethyl-K9 mark at heterochromatin, suggesting that this histone mark is not sufficient to recruit HP1 to heterochromatin, a conclusion previously reported (54). Recent ChIP experiments in HCT116 cells have shown partial loss of H3K9me3 at telomeres after Orc2 depletion (55).
Depletion of ORC or HP1α by RNAi revealed mutual dependence for recruitment or maintenance of both ORC and HP1 on heterochromatin, but in the case of ORC, it depended on which ORC subunit was depleted. Recently, the hMis14 protein that binds to HP1 through a MIR domain (PXVHH motif) was shown to be required for maintenance or recruitment of HP1 to the heterochromatic inner centromere, but in this case, the two proteins interacted in interphase and not during mitosis (56). We suggest that there may be a complex involving a number of proteins and/or RNA (55), including ORC, that has dynamic association with heterochromatin and either recruits or maintains HP1 at heterochromatin. ORC and HP1 are also known to interact in a complex with the HP1/ORC associated protein (HOAP) protein in Drosophila (41), and thus, HOAP, ORC, and possibly other proteins may form a larger complex of proteins that are required for heterochromatin function.
The observation that ORC has multiple HP1-interacting domains also explains why, when different ORC subunits were depleted, there were differential effects on HP1 localization to different constitutive heterochromatic loci. Depletion of Orc2 or Orc3 caused disruption of HP1, possibly leading to compromised gene silencing, sister-chromatid cohesion, and centromere function in mitosis (15, 41, 51, 53, 57). In contrast, depletion of Orc1 and Orc5 resulted in loss of HP1 from large heterochromatin foci but not from centric heterochromatin that surrounds the nucleoli in human cells, the loci where Orc2 and Orc3 are located during mitosis (15, 30). Moreover, we also found differential stability of ORC subunits in FRAP experiments, with Orc1 stably bound to heterochromatin, but Orc2, Orc3, and HP1 having a short 3- to 4-s half-life. Because Orc1 is most likely the DNA-recognition protein in ORC, as it is in S. cerevisiae, it is possible that Orc1 first localizes to the DNA in telophase and recruits Orc2, Orc3, and other ORC subunits along with HP1; then, heterochromatin function is dependent on the dynamic association of HP1, Orc2, Orc3, and perhaps other proteins or RNA. We suggest that Orc2 and Orc3 have activities in addition to DNA replication.
We demonstrated that Orc2 and HP1 are required for condensation of centric heterochromatic satellite repeats. Decondensation of heterochromatin at centromeres may cause genome instability, as shown in Drosophila and mammalian cells that have lost HP1 (51, 53, 57, 58). For example, the Drosophila Orc2 k43 mutant allele has been shown to have mitotic defects (34), but this was attributed to defects in DNA replication. Alternatively, we suggest that the instability of satellite repeat heterochromatin at centromeres and subsequent problems with chromosome segregation are major causes of genome instability in Orc2 mutant cells. In addition, the Drosophila Orc2 mutants show suppression of position-effect variegation (PEV), consistent with the suggestion that ORC plays a role in formation or maintenance of heterochromatin (14). Recent work has also pointed to the role of ORC through an interaction with the Shelterin complex, including telomeric repeat-containing RNAs, in telomere structure and maintenance and in heterochromatin organization (55). Studies on S. pombe have shown that Swi6, the functional homolog of HP1 involved in centromere activity and mating-type gene silencing (59), is required for the recruitment of cohesin to centromeric regions and for proper chromosome segregation (46). Yeast two-hybrid analysis has suggested the interaction of Swi6 and Orc5 in S. pombe (59). Although trimethylation of histone H3 on lysine 9 and chromo domain-containing HP1-like proteins is absent in S. cerevisiae, the involvement of ORC in concert with the silent information regulator proteins for transcriptional gene silencing may be similar to its role in PEV in Drosophila. In S. cerevisiae, Orc2 depletion has been shown to delay progression through mitosis because of a defect in sister chromatid cohesion, and mutants in the Orc5 subunit arrest with completely replicated DNA in mitosis (12, 18, 60). The pre-RC has also been shown to be critical for recruitment of Xenopus Scc2 and cohesin to chromatin (61), suggesting that pre-RC components play important roles in higher-order chromatin organization. All this evidence suggests the involvement of ORC in the maintenance of constitutive heterochromatin (review in ref. 29). How these activities of ORC relate to its role at origins of DNA replication and which may themselves function as chromosome-organization elements require continued investigation.
Methods
Cell Culture and RNA Interference.
HeLa cells were grown in DMEM containing low glucose (Gibco/Life Technologies) supplemented with penicillin-streptomycin and 10% FBS (HyClone). MCF7 and U2OS were grown under standard conditions (ATCC). RNAi was carried out as described previously (43). The small interfering RNAs from Orc1 (62), Orc2 and Orc3 (15), HP1α (CCUGAGAAAAACUUGGAUU), and control luciferase (43) were synthesized by Dharmacon. siRNA was delivered into the cells at a final concentration of 100 nM using oligofectamine (Invitrogen). Cells were transfected three times at a gap of 24–30 h and analyzed for immunoblotting and immunofluorescence.
Immunofluorescence and Antibodies.
Cells were fixed for 15 min in 2.0% formaldehyde in PBS (pH 7.4) followed by 0.5% triton-X treatment for 7 min on ice. Immunofluorescence was carried out using standard procedures. Cells were examined using a Zeiss Axioplan 2i fluorescence microscope or Axioimager (Carl Zeiss Inc.) equipped with Chroma filters (Chroma Technology). OpenLab software (Improvision) or Axiovision software was used to collect digital images from a Hamamatsu ORCA cooled CCD camera. Antibodies used were anti-Orc2 pAb205, anti-HP1α (Chemicon), anticentromere (Sigma), and anti-K27 H3 pAb (Upstate).
DNA FISH.
HeLa cells were preextracted in cytoskeletal buffer (CSK: 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 10 mM Pipes at pH 6.8) containing 0.5% Triton-X 100 for 5 min on ice and fixed with 3.7% freshly prepared formaldehyde for 15 min at room temperature. The cells were washed in 1× PBS (pH 7.2) and heat denatured in 70% formamide and 2× SSC at 72 °C for 5 min followed by hybridization with labeled chromosome 9-specific satellite probe (Q-biogene) in 2× SSC, 50% formamide, 10% dextran sulfate, yeast tRNA, and Cot-1 DNA overnight at 37 °C as previously described (63).
FRAP Analysis.
MCF7 cells were grown on 40-mm diameter glass coverslips and transfected with YFP-Orc1, YFP-Orc2, Orc3-YFP, or YFP-HP1α. The coverslip was transferred into a FCS2 live-cell chamber (Bioptechs) at 37 °C. FRAP experiments were performed on a LSM510 using a 63× NA 1.4 Planapochromat oil-immersion objective (Zeiss) with a 488-nm laser line. Laser power was kept at 1.2% to prevent oversaturation. For FRAP analysis, a heterochromatic foci was photobleached for 250–300 ms with argon laser set to maximum power at 100% transmission; 10 prebleach and 400 postbleach frames were recorded for each series. Zeiss LSM510 software was used for quantification of fluorescence intensities (mean fluorescence intensity of an area). Using the Excel program, the mean fluorescence intensities of the bleached and unbleached region for each time point were background subtracted and normalized to the mean of the 10 prebleach values. These values were divided by the respective total nuclear fluorescence to correct for change of nuclear fluorescence. For each construct, 6–10 nuclei were analyzed. One-half times of recovery were calculated from the curves.
GST Pull-Down Assay.
To generate [35S]methionine-labeled proteins, 1.5 μg plasmid DNA was used as a template in the TNT-coupled reticulocyte lysate system as per supplier's instructions (Promega). [35S]methionine-Redivue was purchased from Amersham Biosciences; 4 μg purified protein (GST or GST-HP1α) was diluted in pull-down assay buffer (25 mM Tris-Cl, pH 7.5, 50 mM KCl, 10% glycerol, 0.02% Nonidet P-40, 0.1 mM EDTA, 5 mM magnesium acetate, 5 mM β-mercaptoethanol). Radioactively labeled ORC subunits or Orc3 mutants were generated using the TNT-coupled reticulocyte lysate system (Promega) and added to the reaction. The pull down was done at 4 °C for 3 h, and resin with bound proteins was washed three times in buffer containing 100–150 mM KCl, boiled, and analyzed by SDS/PAGE followed by phosphorimaging analysis. The screen was then processed in a FLA-5100 imaging system (Fuji) to visualize the proteins.
Supplementary Material
Acknowledgments
We thank the late Dr. Régine Losson (University of Strasbourg, Strasbourg, France) for providing HP1 reagents. We also thank Patty Wendel, Paula Bubulya, and Pam McCloskey for expert technical help. This work was supported by National Science Foundation Award 0843604 (to S.G.P.), National Cancer Institute Grant CA13106 (to B.S.) and a special fellowship from the Leukemia and Lymphoma Society and University of Illinois start-up funds (to S.G.P.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009945107/-/DCSupplemental.
References
- 1.Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 2.Dutta A, Bell SP. Initiation of DNA replication in eukaryotic cells. Annu Rev Cell Dev Biol. 1997;13:293–332. doi: 10.1146/annurev.cellbio.13.1.293. [DOI] [PubMed] [Google Scholar]
- 3.Vashee S, et al. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 2003;17:1894–1908. doi: 10.1101/gad.1084203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dhar SK, Delmolino L, Dutta A. Architecture of the human origin recognition complex. J Biol Chem. 2001;276:29067–29071. doi: 10.1074/jbc.M103078200. [DOI] [PubMed] [Google Scholar]
- 5.Siddiqui K, Stillman B. ATP-dependent assembly of the human origin recognition complex. J Biol Chem. 2007;282:32370–32383. doi: 10.1074/jbc.M705905200. [DOI] [PubMed] [Google Scholar]
- 6.Vashee S, Simancek P, Challberg MD, Kelly TJ. Assembly of the human origin recognition complex. J Biol Chem. 2001;276:26666–26673. doi: 10.1074/jbc.M102493200. [DOI] [PubMed] [Google Scholar]
- 7.Kreitz S, Ritzi M, Baack M, Knippers R. The human origin recognition complex protein 1 dissociates from chromatin during S phase in HeLa cells. J Biol Chem. 2001;276:6337–6342. doi: 10.1074/jbc.M009473200. [DOI] [PubMed] [Google Scholar]
- 8.Méndez J, et al. Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol Cell. 2002;9:481–491. doi: 10.1016/s1097-2765(02)00467-7. [DOI] [PubMed] [Google Scholar]
- 9.Tatsumi Y, Ohta S, Kimura H, Tsurimoto T, Obuse C. The ORC1 cycle in human cells: I. Cell cycle-regulated oscillation of human ORC1. J Biol Chem. 2003;278:41528–41534. doi: 10.1074/jbc.M307534200. [DOI] [PubMed] [Google Scholar]
- 10.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 11.Auth T, Kunkel E, Grummt F. Interaction between HP1alpha and replication proteins in mammalian cells. Exp Cell Res. 2006;312:3349–3359. doi: 10.1016/j.yexcr.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 12.Ehrenhofer-Murray AE, Gossen M, Pak DT, Botchan MR, Rine J. Separation of origin recognition complex functions by cross-species complementation. Science. 1995;270:1671–1674. doi: 10.1126/science.270.5242.1671. [DOI] [PubMed] [Google Scholar]
- 13.Gerbi SA, Strezoska Z, Waggener JM. Initiation of DNA replication in multicellular eukaryotes. J Struct Biol. 2002;140:17–30. doi: 10.1016/s1047-8477(02)00538-5. [DOI] [PubMed] [Google Scholar]
- 14.Pak DT, et al. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell. 1997;91:311–323. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- 15.Prasanth SG, Prasanth KV, Siddiqui K, Spector DL, Stillman B. Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J. 2004;23:2651–2663. doi: 10.1038/sj.emboj.7600255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gillespie PJ, Hirano T. Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr Biol. 2004;14:1598–1603. doi: 10.1016/j.cub.2004.07.053. [DOI] [PubMed] [Google Scholar]
- 17.Kato H, et al. Schizosaccharomyces pombe Orc5 plays multiple roles in the maintenance of genome stability throughout the cell cycle. Cell Cycle. 2008;7:1085–1096. [PubMed] [Google Scholar]
- 18.Shimada K, Gasser SM. The origin recognition complex functions in sister-chromatid cohesion in Saccharomyces cerevisiae. Cell. 2007;128:85–99. doi: 10.1016/j.cell.2006.11.045. [DOI] [PubMed] [Google Scholar]
- 19.Du YC, Stillman B. Yph1p, an ORC-interacting protein: Potential links between cell proliferation control, DNA replication, and ribosome biogenesis. Cell. 2002;109:835–848. doi: 10.1016/s0092-8674(02)00773-0. [DOI] [PubMed] [Google Scholar]
- 20.Hemerly AS, Prasanth SG, Siddiqui K, Stillman B. Orc1 controls centriole and centrosome copy number in human cells. Science. 2009;323:789–793. doi: 10.1126/science.1166745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stuermer A, et al. Mouse pre-replicative complex proteins colocalise and interact with the centrosome. Eur J Cell Biol. 2007;86:37–50. doi: 10.1016/j.ejcb.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Cuvier O, Lutzmann M, Méchali M. ORC is necessary at the interphase-to-mitosis transition to recruit cdc2 kinase and disassemble RPA foci. Curr Biol. 2006;16:516–523. doi: 10.1016/j.cub.2006.01.059. [DOI] [PubMed] [Google Scholar]
- 23.Cuvier O, Stanojcic S, Lemaitre JM, Mechali M. A topoisomerase II-dependent mechanism for resetting replicons at the S-M-phase transition. Genes Dev. 2008;22:860–865. doi: 10.1101/gad.445108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chesnokov IN, Chesnokova ON, Botchan M. A cytokinetic function of Drosophila ORC6 protein resides in a domain distinct from its replication activity. Proc Natl Acad Sci USA. 2003;100:9150–9155. doi: 10.1073/pnas.1633580100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prasanth SG, Prasanth KV, Stillman B. Orc6 involved in DNA replication, chromosome segregation, and cytokinesis. Science. 2002;297:1026–1031. doi: 10.1126/science.1072802. [DOI] [PubMed] [Google Scholar]
- 26.Huang Z, Zang K, Reichardt LF. The origin recognition core complex regulates dendrite and spine development in postmitotic neurons. J Cell Biol. 2005;170:527–535. doi: 10.1083/jcb.200505075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinto S, et al. latheo encodes a subunit of the origin recognition complex and disrupts neuronal proliferation and adult olfactory memory when mutant. Neuron. 1999;23:45–54. doi: 10.1016/s0896-6273(00)80752-7. [DOI] [PubMed] [Google Scholar]
- 28.Chesnokov IN. Multiple functions of the origin recognition complex. Int Rev Cytol. 2007;256:69–109. doi: 10.1016/S0074-7696(07)56003-1. [DOI] [PubMed] [Google Scholar]
- 29.Sasaki T, Gilbert DM. The many faces of the origin recognition complex. Curr Opin Cell Biol. 2007;19:337–343. doi: 10.1016/j.ceb.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 30.Craig JM, et al. Analysis of mammalian proteins involved in chromatin modification reveals new metaphase centromeric proteins and distinct chromosomal distribution patterns. Hum Mol Genet. 2003;12:3109–3121. doi: 10.1093/hmg/ddg330. [DOI] [PubMed] [Google Scholar]
- 31.Huang DW, et al. Distinct cytoplasmic and nuclear fractions of Drosophila heterochromatin protein 1: Their phosphorylation levels and associations with origin recognition complex proteins. J Cell Biol. 1998;142:307–318. doi: 10.1083/jcb.142.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lidonnici MR, et al. Subnuclear distribution of the largest subunit of the human origin recognition complex during the cell cycle. J Cell Sci. 2004;117:5221–5231. doi: 10.1242/jcs.01405. [DOI] [PubMed] [Google Scholar]
- 33.Cimini D, Mattiuzzo M, Torosantucci L, Degrassi F. Histone hyperacetylation in mitosis prevents sister chromatid separation and produces chromosome segregation defects. Mol Biol Cell. 2003;14:3821–3833. doi: 10.1091/mbc.E03-01-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loupart ML, Krause SA, Heck MS. Aberrant replication timing induces defective chromosome condensation in Drosophila ORC2 mutants. Curr Biol. 2000;10:1547–1556. doi: 10.1016/s0960-9822(00)00844-7. [DOI] [PubMed] [Google Scholar]
- 35.Melcher M, et al. Structure-function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation, and mitotic progression. Mol Cell Biol. 2000;20:3728–3741. doi: 10.1128/mcb.20.10.3728-3741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwon SH, Workman JL. The heterochromatin protein 1 (HP1) family: Put away a bias toward HP1. Mol Cells. 2008;26:217–227. [PubMed] [Google Scholar]
- 37.Cheutin T, et al. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 2003;299:721–725. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- 38.Festenstein R, et al. Modulation of heterochromatin protein 1 dynamics in primary Mammalian cells. Science. 2003;299:719–721. doi: 10.1126/science.1078694. [DOI] [PubMed] [Google Scholar]
- 39.Schmiedeberg L, Weisshart K, Diekmann S, Meyer Zu Hoerste G, Hemmerich P. High- and low-mobility populations of HP1 in heterochromatin of mammalian cells. Mol Biol Cell. 2004;15:2819–2833. doi: 10.1091/mbc.E03-11-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DePamphilis ML. The ‘ORC cycle’: A novel pathway for regulating eukaryotic DNA replication. Gene. 2003;310:1–15. doi: 10.1016/s0378-1119(03)00546-8. [DOI] [PubMed] [Google Scholar]
- 41.Shareef MM, Badugu R, Kellum R. HP1/ORC complex and heterochromatin assembly. Genetica. 2003;117:127–134. doi: 10.1023/a:1022963223220. [DOI] [PubMed] [Google Scholar]
- 42.Murzina N, Verreault A, Laue E, Stillman B. Heterochromatin dynamics in mouse cells: Interaction between chromatin assembly factor 1 and HP1 proteins. Mol Cell. 1999;4:529–540. doi: 10.1016/s1097-2765(00)80204-x. [DOI] [PubMed] [Google Scholar]
- 43.Elbashir SM, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 44.Bouzinba-Segard H, Guais A, Francastel C. Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc Natl Acad Sci USA. 2006;103:8709–8714. doi: 10.1073/pnas.0508006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- 47.Simon JA, Tamkun JW. Programming off and on states in chromatin: Mechanisms of Polycomb and trithorax group complexes. Curr Opin Genet Dev. 2002;12:210–218. doi: 10.1016/s0959-437x(02)00288-5. [DOI] [PubMed] [Google Scholar]
- 48.Saurin AJ, et al. The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J Cell Biol. 1998;142:887–898. doi: 10.1083/jcb.142.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collinge MA, Spillane C, Köhler C, Gheyselinck J, Grossniklaus U. Genetic interaction of an origin recognition complex subunit and the Polycomb group gene MEDEA during seed development. Plant Cell. 2004;16:1035–1046. doi: 10.1105/tpc.019059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prasanth SG, Méndez J, Prasanth KV, Stillman B. Dynamics of pre-replication complex proteins during the cell division cycle. Philos Trans R Soc Lond B Biol Sci. 2004;359:7–16. doi: 10.1098/rstb.2003.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pflumm MF, Botchan MR. Orc mutants arrest in metaphase with abnormally condensed chromosomes. Development. 2001;128:1697–1707. doi: 10.1242/dev.128.9.1697. [DOI] [PubMed] [Google Scholar]
- 52.Thiru A, et al. Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. EMBO J. 2004;23:489–499. doi: 10.1038/sj.emboj.7600088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vermaak D, Malik HS. Multiple roles for heterochromatin protein 1 genes in Drosophila. Annu Rev Genet. 2009;43:467–492. doi: 10.1146/annurev-genet-102108-134802. [DOI] [PubMed] [Google Scholar]
- 54.Stewart MD, Li J, Wong J. Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Mol Cell Biol. 2005;25:2525–2538. doi: 10.1128/MCB.25.7.2525-2538.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell. 2009;35:403–413. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kiyomitsu T, Iwasaki O, Obuse C, Yanagida M. Inner centromere formation requires hMis14, a trident kinetochore protein that specifically recruits HP1 to human chromosomes. J Cell Biol. 2010;188:791–807. doi: 10.1083/jcb.200908096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kellum R, Alberts BM. Heterochromatin protein 1 is required for correct chromosome segregation in Drosophila embryos. J Cell Sci. 1995;108:1419–1431. doi: 10.1242/jcs.108.4.1419. [DOI] [PubMed] [Google Scholar]
- 58.Inoue A, Hyle J, Lechner MS, Lahti JM. Perturbation of HP1 localization and chromatin binding ability causes defects in sister-chromatid cohesion. Mutat Res. 2008;657:48–55. doi: 10.1016/j.mrgentox.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 59.Matsuda K, et al. Yeast two-hybrid analysis of the origin recognition complex of Saccharomyces cerevisiae: Interaction between subunits and identification of binding proteins. FEM Yeast Res. 2007;7:1263–1269. doi: 10.1111/j.1567-1364.2007.00298.x. [DOI] [PubMed] [Google Scholar]
- 60.Dillin A, Rine J. Roles for ORC in M phase and S phase. Science. 1998;279:1733–1737. doi: 10.1126/science.279.5357.1733. [DOI] [PubMed] [Google Scholar]
- 61.Takahashi TS, Yiu P, Chou MF, Gygi S, Walter JC. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat Cell Biol. 2004;6:991–996. doi: 10.1038/ncb1177. [DOI] [PubMed] [Google Scholar]
- 62.Ohta S, Tatsumi Y, Fujita M, Tsurimoto T, Obuse C. The ORC1 cycle in human cells: II. Dynamic changes in the human ORC complex during the cell cycle. J Biol Chem. 2003;278:41535–41540. doi: 10.1074/jbc.M307535200. [DOI] [PubMed] [Google Scholar]
- 63.Xing Y, Johnson CV, Moen PT, Jr., McNeil JA, Lawrence J. Nonrandom gene organization: Structural arrangements of specific pre-mRNA transcription and splicing with SC-35 domains. J Cell Biol. 1995;131:1635–1647. doi: 10.1083/jcb.131.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.