Abstract
In the absence of treatment, most HIV-1-infected humans develop AIDS. However, a minority are long-term nonprogressors, and resistance is associated with the presence of particular HLA-B*27/B*57 molecules. In contrast, most HIV-1-infected chimpanzees do not contract AIDS. In comparison with humans, chimpanzees experienced an ancient selective sweep affecting the MHC class I repertoire. We have determined the peptide-binding properties of frequent chimpanzee MHC class I molecules, and show that, like HLA-B*27/B*57, they target similar conserved areas of HIV-1/SIVcpz. In addition, many animals appear to possess multiple molecules targeting various conserved areas of the HIV-1/SIVcpz Gag protein, a quantitative aspect of the immune response that may further minimize the chance of viral escape. The functional characteristics of the contemporary chimpanzee MHC repertoire suggest that the selective sweep was caused by a lentiviral pandemic.
Keywords: HIV/SIV, peptide-binding, selection, selective sweep
In evolutionary terms, chimpanzees and humans are each other's closest living relatives (1), and both species are susceptible to infection with HIV type 1 (HIV-1), which in particular individuals can lead to acquired immunodeficiency syndrome (AIDS). The HIV-1 pandemic in humans arose from multiple zoonotic events caused by the transmission of the chimpanzee-derived simian immunodeficiency virus (SIVcpz) (2). Natural infections with various SIVcpz strains have been documented in Central and East African chimpanzee populations (3). Recently, AIDS-like immunopathology was reported for some animals of the East African subspecies (4). However, naturally infected chimpanzees of the same subspecies that have been monitored carefully during captivity over long periods of time—at our center—do not develop any signs of AIDS, despite relatively high viral plasma loads (5). Moreover, AIDS-like disease did not emerge in West African chimpanzees after the experimental introduction of a natural SIVcpz strain (5). Hence, chimpanzees are susceptible to HIV-1/SIVcpz infection but are relatively resistant to progress toward AIDS (6, 7). On the contrary, most human subjects develop AIDS upon HIV-1 infection. However, cohort studies have established the existence of individuals who have been infected for more than 17 years but have not progressed to AIDS; these are the long-term nonprogressors/elite controllers. Resistance has been attributed to infections with defective viruses or to mutations in the chemokine (C-C motif) receptor 5 of the host, the primary coreceptor for viral entry (8, 9). Furthermore, cytotoxic T-cell (CTL) responses play a prominent role in controlling intracellular viral infections. The T-cell receptor on such cells may recognize major histocompatibility complex (MHC) class I molecules complexed with a viral peptide, and subsequently the CTL will lyse the infected cell. In humans and chimpanzees, these molecules are designated HLA- and Patr-A, -B, and -C, respectively, and they display abundant levels of polymorphism (10). Resistance to progression to AIDS in human subjects is strongly associated with the presence of particular HLA-B*27/B*57 molecules, in conjunction with specific killer-cell inhibitory receptors (11, 12). Genome-wide association studies have further substantiated the prominent role of the MHC class I region in controlling HIV-1 replication (13, 14).
We have reported several lines of genetic evidence that chimpanzees experienced a selective sweep that targeted the Patr class I repertoire, and hypothesized that this was caused by HIV-1/SIVcpz or a related ancestral retrovirus (15–17). The sweep, dated to ≈2–3 million years ago, also matches the time frame with respect to when the chimpanzee genome might have been exposed to retroviral integrations (18). We wished to investigate whether the selective sweep was caused by HIV-1/SIVcpz or a closely related ancestor, and whether this repertoire skewing resulted in the preferential selection of Patr molecules that are similar to AIDS-resistant molecules in human individuals, such as HLA-B*27/B*57. For this purpose, we have determined the peptide-binding motifs of Patr molecules that occur at a high frequency in a wild-caught chimpanzee population. Based on this information, the HIV-1/SIVcpz proteomes were scanned for potential CTL epitopes, which were subsequently tested in peptide-binding studies. Attention was focused on the Gag (p24) protein as a model system, as adaptive immune responses to this viral protein are thought to play a prominent role in the control of viral replication (19–21). In addition, cellular immune-response data were reevaluated in the context of the information gained from the peptide-binding studies. The results demonstrate that most chimpanzees from the population investigated possess at least one Patr molecule that can bind analogous conserved areas of the HIV-1/SIVcpz Gag protein, as does HLA-B*27/B*57.
Results
Definition of Peptide-Binding Motifs.
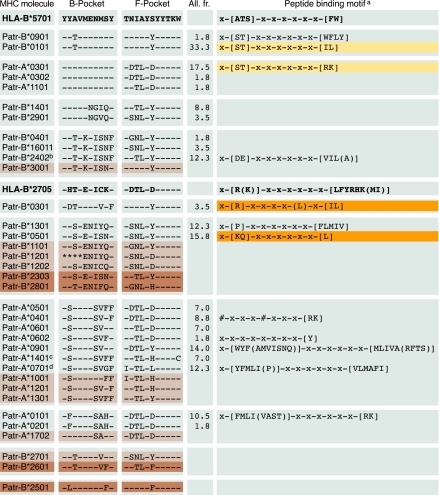
The peptide-binding site of MHC class I molecules generally accommodates nonamer peptides. The second residue of the peptide (numbered from the N terminus) is often fixed as an anchor in the B pocket, whereas the C terminus is bound to the F pocket (22). HLA and Patr class I gene products display polymorphism and, as a consequence, different molecules possess distinct peptide-binding motifs (10). The Patr-A and -B molecules present in the population studied were clustered based on the sharing of similar B and F pockets (Fig. 1). The molecules with the highest frequency within a given cluster (Patr-A*0301, -B*0101, -B*0301, and -B*0501) were selected for the determination of their peptide-binding motifs. For this purpose, single-molecule-expressing cell lines were constructed. Subsequently, the Patr molecules were isolated by immunopurification, followed by extraction of the pool of natural bound peptides. We then identified the amino acid sequence of the isolated peptides by tandem mass spectrometry. Binding motifs were assembled from the isolated 8-, 9-, and 10-mer peptides (Tables S1 and S2 and Fig. S1).
Fig. 1.
Chimpanzee class I molecules clustered based on B- and F-pocket similarities. The B and F pocket of HLA-B*5701 are taken as consensus. A dash indicates identity to the consensus, an amino acid replacement is represented by the conventional one-letter code, and an asterisk indicates the position where the amino acid is unknown. The allele frequency (All. fr.) figures of the wild-caught founder population have been provided. Molecules depicted in light and dark brown are from chimpanzees of the Central and East African subspecies, respectively. Dark orange represents unreported peptide-binding motifs, whereas light orange highlights the motifs that are in agreement with data reported by another research team (23, 24). The binding motifs for HLA-B*2705 and -B*5701 as well as previously identified Patr-binding motifs are provided (23–25, 43). aThe conventional one-letter code identifies the preferred, or in brackets the tolerated, residues at the corresponding positions. bPatr-B*2402 has an identical B and a similar F pocket (–TL-F—–) as Patr-B*2401. Binding motif shown is from Patr-B*2401. cMolecule present in West and East African subspecies. dMolecule present in West, Central, and East African subspecies. The # marks additional primary anchors.
The motifs elucidated for Patr-A*0301 and -B*0101 share their B-pocket anchor with HLA-B*5701 (Fig. 1), in accordance with data generated by others (23, 24). For Patr-A*0301, the F-pocket anchor is similar to HLA-A*03/11, whereas Patr-B*0101 prefers hydrophobic amino acids at this position, a phenomenon shared with different HLA molecules (25). The as yet unreported motifs for Patr-B*0301 and -B*0501 were either similar to those described for particular HLA-B*27 molecules or were identical to those of HLA-B*3902/B*4801, respectively (25). With the current data, the total number of peptide-binding motifs defined for Patr class I molecules is 12. This further broadens our ability to understand and study the MHC–peptide interactions in chimpanzees.
Prediction and Selection of Potential HIV-1/SIVcpz Gag Epitopes.
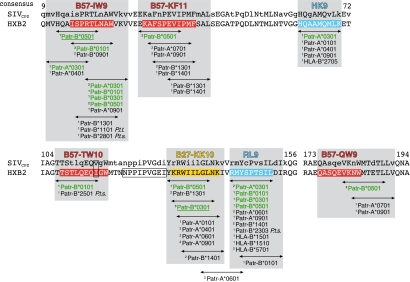
The peptide-binding motifs allowed us to scan the entire Gag protein for potential epitopes using a database (26). We paid particular attention to those areas of Gag that are also recognized by HLA-B*27/B*57, as these epitopes may be crucial for the control of viral replication in humans (19, 27). These CTL epitopes have been designated IW9, KF11, TW10, KK10, and QW9 (Fig. 2). Based on the binding motifs, Patr-A*0301 and -B*0101 are predicted to recognize two peptides that overlap with the IW9 epitope (Fig. 2). In addition, Patr-B*0101 is predicted to bind a TW10-related epitope as well. The scan for Patr-B*0301 revealed that it can target overlapping areas of IW9 and KK10, whereas Patr-B*0501 can recognize different areas such as IW9, KF11, KK10, and QW9 (Fig. 2). Taken together, 10 different Gag peptides were selected for further analysis of their capacity to act as ligands for the four Patr molecules (Fig. 2 and Fig. S2).
Fig. 2.
Map of potential HIV-1 Gag CTL epitopes recognized by Patr class I molecules. The Gag sequence of HXB2 is taken as consensus, with the consensus of SIVcpz above (http://www.hiv.lanl.gov). A lowercase letter in the SIVcpz consensus indicates a variable position. HLA-B*27 and -B*57 CTL epitopes are marked in yellow and red, respectively. Potential 9-mer epitopes are indicated by an arrow, with the relevant Patr molecule(s) mentioned. Molecules marked in green indicate the predicted MHC/peptide combinations that are tested in peptide-binding studies. Molecules specific for Central (P.t. troglodytes, P.t.t.) or East African chimpanzees (P.t. schweinfurthii, P.t.s.) are indicated. The predicted binding affinities using the NetMHCpan 2.0 algorithm are indicated by 1high, 2intermediate, or 3low. The arrows above the underscored Patr molecules indicate previously reported CTL epitopes (30). The two potential epitopes, HK9 and RL9, identified with the NetMHCpan 2.0 algorithm are marked in blue. The control peptide, NPPIPVGEI, is boxed. The box in the B57-TW10 epitope indicates covered variation (Fig. S2).
Subsequently, an additional algorithm was applied as a tool to verify the binding affinity of the selected peptides (28, 29). Moreover, this algorithm was used to search for other potential CTL epitopes. Two peptides, HK9 and RL9, were chosen, as they showed high binding level scores for various Patr molecules (Fig. 2). In humans, CTL responses directed against HK9 and RL9 have been observed in conjunction with the restriction elements HLA-A*11 and -B*52 or -A*02, respectively (http://www.hiv.lanl.gov). The Gag-derived peptide NPPIPVGEI and a Gag-unrelated peptide EEALQAFTY, both predicted to have no or a very low binding affinity for each of the four molecules, were selected as controls.
Peptide-Binding Studies.
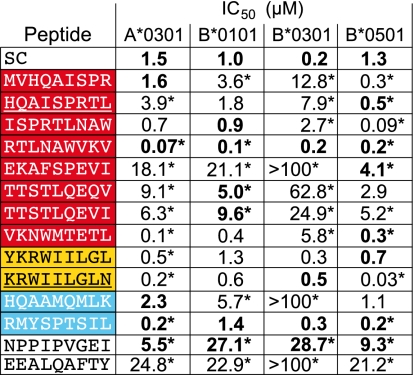
To determine the binding affinity of the selected Gag peptides, a cell-based peptide-binding competition assay was developed. Each Patr molecule has its specific biotin-labeled indicator peptide against which the binding affinity of the different selected peptides was determined. The binding affinity of the indicator peptide itself was verified using its unlabeled peptide as a competitor, henceforth referred to as the standardized competitor (SC). Dose-inhibition curves for the peptides predicted to bind to Patr-A*0301, -B*0101, -B*0301, and -B*0501 were derived from pooled data from five individual experiments (Fig. S3), and all showed a high or intermediate affinity to the relevant Patr molecules (Fig. 3, bold). The intrinsic binding affinity of the SC for Patr-B*0301 was high in comparison with the binding affinity of the SCs for the other three molecules. Because the peptides RTLNAWVKV and KRWIILGLN bound with a similar high affinity to Patr-B*0301 as its SC, this implicates a high binding affinity for these peptides as well. Moreover, for HK9, tested for Patr-A*0301, we observed a binding affinity similar to that of the SC, whereas RL9 has disparate binding affinities for the different molecules.
Fig. 3.
IC50 (μM) values of the different competitor peptides tested for Patr-A*0301, -B*0101, -B*0301, and -B*0501. The IC50 values in bold were determined from regression curves (Fig. S3) derived from pooled data from five individual experiments. For all other conceivable combinations, the IC50 values were derived from pooled data from three individual experiments. Previously described CTL epitopes are underscored (30). The color codes of the peptides correspond to the colors in Fig. 2. SC, standardized competitor (a peptide similar to the biotin-labeled indicator peptide). An asterisk indicates a significantly higher or lower binding affinity than the SC.
In addition, the selected peptides were tested in all other conceivable combinations with the four Patr molecules. The results, derived from pooled data from three individual experiments, are summarized (Fig. 3 and Fig. S2B). Based on a peptide-binding motif, one would expect that each MHC class I molecule can bind a restricted spectrum of peptides. Indeed, we found that for most additional tested Patr/peptide combinations, the peptides bound with a lower binding affinity than that of the corresponding SC. However, for a few Patr/peptide combinations, a higher binding affinity than that of the corresponding SC was observed: for example, Patr-A*0301 in combination with KRWIILGLN (Fig. 3). Such additional specific binding was not predicted based on the peptide-binding motifs. Apparently, however, some of the Patr class I molecules can bind a broader range of peptides that map to conserved areas of Gag. The molecular basis of this phenomenon is not yet understood. Finally, the two control peptides were found to bind with a low binding affinity to all four Patr molecules.
In conclusion, the data illustrate that Patr-B*0301 and -B*0501, each previously described to act as restriction elements for peptides in the KK10 and IW9 areas (30), respectively, also have the capacity to bind other peptides mapping to areas targeted by HLA-B*27/B*57. Overall, the four Patr molecules investigated seem to be able to bind peptides with a high binding affinity, mapping to areas that overlap the IW9, KK10, and QW9 CTL epitopes. Hence, the majority of the peptides that were selected as potential epitopes indeed represent good binders for the respective molecules.
Evaluation of CTL Data.
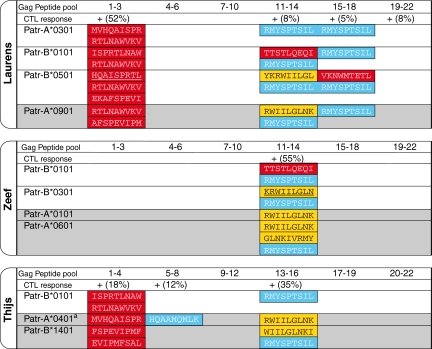
The actual binding of a peptide to an MHC molecule does not warrant that immune recognition by effector cells will take place. On the basis of the present findings, we reevaluated the cellular immune responses for three chimpanzees (Laurens, Zeef, and Thijs) belonging to an HIV-1-positive cohort (31). For Laurens, polyclonal CTL responses to four different Gag pools were detected (Fig. 4). The highest percentage of specific lysis was detected for Gag pool 1–3, and was at that time ascribed to epitope HQAISPRTL (part of the IW9 area) presented in the context of Patr-B*0501 (30). However, the present data illustrate that other molecules expressed by Laurens (Patr-A*0301 and -B*0101) can bind peptides overlapping with HQAISPRTL or the KF11 epitope, and could therefore contribute to the polyclonal CTL response to Gag pool 1–3 (Fig. 4). Moreover, the molecules expressed by Laurens were found to bind the RL9 peptide and peptides overlapping the TW10, KK10, and QW9 epitopes, which may contribute to the polyclonal CTL responses observed for Gag pools 11–14 and 15–18 (Fig. 4). For Zeef, only a response to Gag pool 11–14 was observed (Fig. 4). Initially, this response was ascribed to KRWIILGLN (part of the KK10 area) presented in the context of Patr-B*0301 (30). The results suggest that the combination of Patr-B*0101 and peptide TTSTLQEQI (part of the TW10 area) could contribute to the high specific lysis observed for peptide pool 11–14 as well. Finally, for Thijs, responses to three different Gag pools were detected (Fig. 4), although at that time the restriction elements were not understood. Thijs does not possess Patr-B*0301 and -B*0501, responsible for the CTL responses in Zeef and Laurens, respectively. This implies that other molecules (for instance, Patr-B*0101) are also able to recognize conserved areas (Fig. 4).
Fig. 4.
Analysis of cellular immune responses in three chimpanzees belonging to an experimentally infected HIV-1 cohort. The peptides present in the Gag peptide pools are summarized in Fig. S4. A plus sign indicates that a CTL response to the Gag pool was observed, and the percentage-specific lysis is given in brackets (31). The Patr-A and -B molecules present on the haplotypes of chimpanzees Laurens, Zeef, and Thijs are given. The relevant MHC/peptide combinations that probably contribute to the previously observed Gag-pool-specific CTL responses are given, and the color codes of the peptides correspond to the colors in Fig. 2. Previously described CTL epitopes are underscored (30). A gray background indicates data obtained using only the NetMHCpan 2.0 algorithm. aAnimal is homozygous for this molecule.
The absence of CTL responses to certain Gag pools in Laurens, Zeef, and Thijs is most likely explained by the fact that these areas do not contain peptides that can bind to the molecules present in these particular animals. Alternatively, CTL-precursor frequencies may be too low to detect in the polyclonal assays used. In contrast, these areas can be recognized by various HLA molecules (http://www.hiv.lanl.gov). Thus, these results suggest that many of the CTL responses observed in chimpanzees may be directed to conserved areas of the Gag protein, which are also targeted by the HLA-B*27/B*57 molecules in humans.
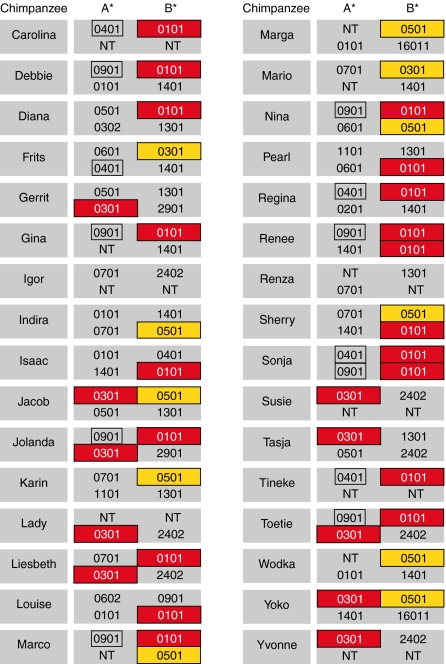
Peptide-Binding Results in the Context of Patr Class I Haplotypes.
A haplotype is defined as the unique combination of MHC alleles segregating together on a chromosome. MHC region genes are codominantly expressed and, like humans, chimpanzees possess one A and one B gene per haplotype. The peptide-binding study results have been superimposed on the chimpanzee haplotypes. First, it was noted that most animals possess at least one molecule with the ability to bind a conserved area of Gag that is targeted by HLA-B*27 and/or -B*57 in humans (Fig. 5). Second, some animals encode multiple class I molecules, in cis (e.g., Jacob and Yoko) or trans configuration (e.g., Jolanda, Liesbeth, and Marco), which can bind similar and/or disparate conserved HIV-1 Gag peptides as HLA-B*27/B*57. These observations imply that the quality of the Patr molecule to bind conserved areas of Gag may play an important role in the protection against the development of AIDS. However, the fact that particular animals encode multiple class I molecules able to bind conserved Gag peptides suggests that, in chimpanzees, perhaps also a quantitative feature is operative. In conclusion, the majority of the founder animals possess at least one Patr molecule that, like HLA-B*27/B*57, is able to bind peptides from similar conserved areas of Gag, and as such could respond to an HIV-1/SIVcpz infection in a similar way as described for HLA-B*27/B*57-positive human long-term nonprogressors.
Fig. 5.
MHC class I haplotypes of the founder chimpanzees. Molecules that bind conserved Gag peptides overlapping the areas targeted only by HLA-B*57 are presented in red, whereas orange indicates molecules binding peptides overlapping the areas targeted by HLA-B*27 and -B*57. Boxed molecules are predicted to bind HLA-B*27/B*57-like overlapping conserved Gag peptides based on the NetMHCpan 2.0 algorithm. NT indicates not typed, because data are no longer accessible.
Discussion
We have investigated whether the selective sweep targeting the Patr class I repertoire was possibly caused by HIV-1/SIVcpz (or a closely related retrovirus) and has resulted in the preferential selection of molecules that confer resistance to AIDS. The peptide-binding motifs of four frequent Patr class I molecules were defined, and we found that these molecules, like the AIDS-protective HLA-B*27 and -B*57 molecules, bind peptides derived from conserved areas of the HIV-1/SIVcpz Gag protein. Ninety-four percent of the wild-caught West African chimpanzees studied possess at least one of these four Patr class I molecules. Moreover, many chimpanzees express several molecules that can bind multiple peptides originating from various conserved Gag regions, which suggests that chimpanzees may have developed a “double-lock” strategy. In humans, the importance of heterozygous advantage with regard to AIDS resistance is also applicable (32), and one would expect that heterozygous individuals that contain both HLA-B*27 and -B*57 progress more slowly to AIDS or behave as elite controllers (33). Accordingly, three chimpanzees of an experimentally infected HIV-1 cohort were shown to display broadly reactive CTL responses to conserved areas of HIV-1. Taken together, the results suggest that the chimpanzee MHC class I repertoire was skewed to favor the survival of animals whose Patr molecules recognized conserved areas of Gag, a property shared with HLA-B*27/B*57. This may be due to the sharing of similar peptide-binding motifs between humans and chimpanzees, but also to the ability of some Patr molecules to target overlapping areas of the Gag protein, which are conserved across different strains. As such, the present report significantly extends the information that was gathered in an earlier CTL study (30). Apart from Patr-B, a Patr-A molecule was also shown to bind peptides from conserved Gag areas similar to those targeted by HLA-B*27/B*57, and as such may contribute to controlling HIV-1/SIVcpz viral load. In comparison with HLA-A, Patr-A molecules have more promiscuous peptide-binding motifs, allowing the presentation of a greater spectrum of peptides (Fig. 1). Nonetheless, one should bear in mind that we only determined the peptide-binding motifs for four Patr molecules. Whether the picture that emerges may become more prominent could be addressed when peptide-binding and cellular data become available for other molecules. To verify this assumption, the Gag protein was scanned with the NetMHCpan 2.0 algorithm (28) for two additional molecules: Patr-A*0401 and -A*0901. High- or intermediate-affinity peptides overlapping the IW9 and KK10 epitopes were identified (Fig. 2), and indeed CTL response to Gag pools covering these peptides was observed (Fig. 4). The incorporation of the data for Patr-A*0401 and -A*0901 into the haplotype list further strengthens both the qualitative and quantitative features of the immune response to an HIV-1/SIVcpz infection (Fig. 5). However, it has to be taken into consideration that it is likely that other Patr molecules may not be able to target conserved areas of Gag similar to those targeted by HLA-B*27/B*57.
Immunodominant CTL responses to Gag found in HLA-B*27/B*57-positive human long-term nonprogressors and HIV-1-infected chimpanzees may play a crucial role in the control of viral replication (19, 27, 30). The Gag protein may represent one of the Achilles’ heels of HIV-1/SIVcpz. However, from a mechanistic point of view, it is not clearly understood how this works (27), because not all HLA-B*27- or -B*57-positive individuals can control an HIV-1 infection. Although heterozygous advantage may have an additional impact, other host and viral factors may play a role as well. In this respect, it is worth mentioning that we do not want to claim that the MHC is the only genetic region that explains the relative resistance to AIDS in chimpanzees, because other genes, such as the chemokine (C-C motif) receptor 5 and CD4, show signs of selection and may have been affected by a sweep as well (34, 35). Furthermore, it was shown recently that rhesus macaques that are elite controllers in AIDS vaccine studies possess T-cell responses to different viral proteins, primarily Vif and Nef, restricted by the Mamu-B*08 or -B*17 molecule (36, 37). These observations illustrate that, at least in rhesus macaques, T-cell responses to different viral proteins could play a prominent role in elite control. Therefore, the present study, in which we focused on different conserved Gag epitopes, may only represent a part of our understanding how chimpanzees may control viral replication. Follow-up studies are needed to sort out the role of other viral proteins in controlling the development of AIDS in chimpanzees.
A recent report indicated that naturally infected East African animals contract AIDS (4). The fact that chimpanzees can develop AIDS was shown earlier for one animal that was found to contain a recombinant virus that emerged after a coinfection with three different HIV-1 isolates (7). After transfer of this virus isolate to other chimpanzees, it caused CD4+ T-cell depletion and increased plasma viral loads as well, indicating that specific HIV-1 isolates are able to provoke AIDS or AIDS-like symptoms. SIVcpz strains isolated from naturally infected chimpanzees appear to represent a mosaic of Old World monkey virus segments (38). Chimpanzees and their ancestors were most likely infected by preying upon different monkey species known to have been infected with several types of SIV strains. It is plausible that new recombinant SIVcpz strains are generated occasionally (39), and that some of these strains have pathogenic characteristics. Nonhuman primate species have been challenged by SIV infections over long periods of time, and must have developed ways to control the development of AIDS (15, 40), like the AIDS-resistant West African chimpanzees studied here. There is evidence that the selective sweep or subsequent selection processes must have been more prominent in West African chimpanzees than in other chimpanzee populations (34, 35, 41). This may be due to population separation, and may be dependent on the monkey species being preyed upon and on their respective SIV infections (42). As a consequence, repertoires could have been edited in slightly different manners. Nevertheless, the broad picture that is emerging suggests that most chimpanzees possess MHC class I molecules that bind conserved epitopes in Gag, similar to those targeted by AIDS-resistant HLA-B*27/B*57-positive humans. The present functional characteristics of the skewed MHC class I repertoire in chimpanzees suggest that the ancient selective sweep was caused by a lentiviral pandemic. As a consequence, most chimpanzees seem to be able to cope with retroviral infections such as HIV-1/SIVcpz, like AIDS-resistant HLA-B*27/B*57-positive humans.
Materials and Methods
Patr-A- and -B-Transfected Cell Lines.
Constructs containing the relevant Patr-A or -B sequences were individually transfected into K562 cells (which lack HLA expression) by electroporation. Details are described in SI Materials and Methods.
Determination of Peptide-Binding Motifs.
K562 Patr-A- or -B-positive transfectants were cultured in IMDM supplemented with G418 and 5% FCS up to 1 × 1010 cells. Subsequently, the cells were lysed. Sepharose beads covalently linked to monoclonal antibody W6/32 were used to preclear the lysate. The beads were then washed, and in a final step the MHC-peptide complexes were eluted with 10% acetic acid in water. High molecular mass material (MHC molecules) was removed through Centriprep filtration. The peptide pool was prefractionated on a C18 RP-HPLC system (Dr. Maisch GmbH), and was subsequently analyzed by nano-HPLC-tandem mass spectrometry with an LTQ-FT (Thermo Electron). Details are described in SI Materials and Methods.
Selection and Synthesis of Indicator and Competitor Peptides.
Details are described in SI Materials and Methods.
Cell-Based Peptide-Binding Competition Assays and CTL Assays.
A plate-based cell-based peptide-binding competition assay was developed to measure the binding affinity of the different peptides using time-resolved fluorescence as read-out (see also Fig. S5). The full protocol is described in SI Materials and Methods. Previously described CTL assays were reevaluated (30, 31).
Calculation of Percentage Inhibition, IC50 Values, and Statistical Analysis.
Details are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank D. Devine for editing the manuscript, H. van Westbroek for preparing the figures, Drs. J. Bajramovic, D. Davis, and G. Koopman for their suggestions and critical reading of the manuscript, Prof. Dr. P. van den Elsen (Leiden University Medical Center, Leiden, The Netherlands) for kindly providing the pcDNA/neo vector and MC1061/P3 Escherichia coli, Dr. B. Faber for purification and concentration of monoclonal antibody W6/32, and Dr. D. Stepniak for assistance with peptide isolation. This study was supported by the Biomedical Primate Research Centre.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009136107/-/DCSupplemental.
References
- 1.Fujiyama A, et al. Construction and analysis of a human-chimpanzee comparative clone map. Science. 2002;295:131–134. doi: 10.1126/science.1065199. [DOI] [PubMed] [Google Scholar]
- 2.Hahn BH, Shaw GM, De Cock KM, Sharp PM. AIDS as a zoonosis: Scientific and public health implications. Science. 2000;287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- 3.Santiago ML, et al. SIVcpz in wild chimpanzees. Science. 2002;295:465. doi: 10.1126/science.295.5554.465. [DOI] [PubMed] [Google Scholar]
- 4.Keele BF, et al. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature. 2009;460:515–519. doi: 10.1038/nature08200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heeney JL, et al. Transmission of simian immunodeficiency virus SIVcpz and the evolution of infection in the presence and absence of concurrent human immunodeficiency virus type 1 infection in chimpanzees. J Virol. 2006;80:7208–7218. doi: 10.1128/JVI.00382-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heeney JL, Dalgleish AG, Weiss RA. Origins of HIV and the evolution of resistance to AIDS. Science. 2006;313:462–466. doi: 10.1126/science.1123016. [DOI] [PubMed] [Google Scholar]
- 7.Novembre FJ, et al. Development of AIDS in a chimpanzee infected with human immunodeficiency virus type 1. J Virol. 1997;71:4086–4091. doi: 10.1128/jvi.71.5.4086-4091.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirchhoff F, Greenough TC, Brettler DB, Sullivan JL, Desrosiers RC. Absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N Engl J Med. 1995;332:228–232. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- 9.Samson M, et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 10.Parham P, Ohta T. Population biology of antigen presentation by MHC class I molecules. Science. 1996;272:67–74. doi: 10.1126/science.272.5258.67. [DOI] [PubMed] [Google Scholar]
- 11.Kiepiela P, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 12.Martin MP, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalmasso C, et al. ANRS Genome Wide Association 01 Distinct genetic loci control plasma HIV-RNA and cellular HIV-DNA levels in HIV-1 infection: The ANRS Genome Wide Association 01 study. PLoS One. 2008;3:e3907. doi: 10.1371/journal.pone.0003907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fellay J, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Groot NG, et al. Reduced MIC gene repertoire variation in West African chimpanzees as compared to humans. Mol Biol Evol. 2005;22:1375–1385. doi: 10.1093/molbev/msi127. [DOI] [PubMed] [Google Scholar]
- 16.de Groot NG, et al. Pinpointing a selective sweep to the chimpanzee MHC class I region by comparative genomics. Mol Ecol. 2008;17:2074–2088. doi: 10.1111/j.1365-294X.2008.03716.x. [DOI] [PubMed] [Google Scholar]
- 17.de Groot NG, et al. Evidence for an ancient selective sweep in the MHC class I gene repertoire of chimpanzees. Proc Natl Acad Sci USA. 2002;99:11748–11753. doi: 10.1073/pnas.182420799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yohn CT, et al. Lineage-specific expansions of retroviral insertions within the genomes of African great apes but not humans and orangutans. PLoS Biol. 2005;3:e110. doi: 10.1371/journal.pbio.0030110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borghans JA, Mølgaard A, de Boer RJ, Keşmir C. HLA alleles associated with slow progression to AIDS truly prefer to present HIV-1 p24. PLoS One. 2007;2:e920. doi: 10.1371/journal.pone.0000920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez-Picado J, et al. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J Virol. 2006;80:3617–3623. doi: 10.1128/JVI.80.7.3617-3623.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rolland M, et al. Broad and Gag-biased HIV-1 epitope repertoires are associated with lower viral loads. PLoS One. 2008;3:e1424. doi: 10.1371/journal.pone.0001424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falk K, Rötzschke O, Stevanović S, Jung G, Rammensee HG. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351:290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 23.Sidney J, et al. Detailed characterization of the peptide binding specificity of five common Patr class I MHC molecules. Immunogenetics. 2006;58:559–570. doi: 10.1007/s00251-006-0131-4. [DOI] [PubMed] [Google Scholar]
- 24.Sidney J, et al. Characterization of the peptide-binding specificity of the chimpanzee class I alleles A 0301 and A 0401 using a combinatorial peptide library. Immunogenetics. 2007;59:745–751. doi: 10.1007/s00251-007-0243-5. [DOI] [PubMed] [Google Scholar]
- 25.Marsh SGE, Parham P, Barber LD. The HLA FactsBook. San Diego: Academic; 2000. [Google Scholar]
- 26.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanović S. SYFPEITHI: Database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 27.Goulder PJ, Watkins DI. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat Rev Immunol. 2008;8:619–630. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoof I, et al. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009;61:1–13. doi: 10.1007/s00251-008-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pérez CL, et al. Broadly immunogenic HLA class I supertype-restricted elite CTL epitopes recognized in a diverse population infected with different HIV-1 subtypes. J Immunol. 2008;180:5092–5100. doi: 10.4049/jimmunol.180.7.5092. [DOI] [PubMed] [Google Scholar]
- 30.Balla-Jhagjhoorsingh SS, et al. Conserved CTL epitopes shared between HIV-infected human long-term survivors and chimpanzees. J Immunol. 1999;162:2308–2314. [PubMed] [Google Scholar]
- 31.Balla-Jhagjhoorsingh SS, et al. Protection from secondary human immunodeficiency virus type 1 infection in chimpanzees suggests the importance of antigenic boosting and a possible role for cytotoxic T cells. J Infect Dis. 2001;184:136–143. doi: 10.1086/322019. [DOI] [PubMed] [Google Scholar]
- 32.Carrington M, et al. HLA and HIV-1: Heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 33.Bailey JR, et al. Transmission of human immunodeficiency virus type 1 from a patient who developed AIDS to an elite suppressor. J Virol. 2008;82:7395–7410. doi: 10.1128/JVI.00800-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hvilsom C, et al. Genetic subspecies diversity of the chimpanzee CD4 virus-receptor gene. Genomics. 2008;92:322–328. doi: 10.1016/j.ygeno.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 35.MacFie TS, Nerrienet E, de Groot NG, Bontrop RE, Mundy NI. Patterns of diversity in HIV-related loci among subspecies of chimpanzee: Concordance at CCR5 and differences at CXCR4 and CX3CR1. Mol Biol Evol. 2009;26:719–727. doi: 10.1093/molbev/msp016. [DOI] [PubMed] [Google Scholar]
- 36.Mothé BR, et al. Characterization of the peptide-binding specificity of Mamu-B*17 and identification of Mamu-B*17-restricted epitopes derived from simian immunodeficiency virus proteins. J Immunol. 2002;169:210–219. doi: 10.4049/jimmunol.169.1.210. [DOI] [PubMed] [Google Scholar]
- 37.Valentine LE, et al. Infection with “escaped” virus variants impairs control of simian immunodeficiency virus SIVmac239 replication in Mamu-B*08-positive macaques. J Virol. 2009;83:11514–11527. doi: 10.1128/JVI.01298-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bailes E, et al. Hybrid origin of SIV in chimpanzees. Science. 2003;300:1713. doi: 10.1126/science.1080657. [DOI] [PubMed] [Google Scholar]
- 39.Leitner T, Dazza MC, Ekwalanga M, Apetrei C, Saragosti S. Sequence diversity among chimpanzee simian immunodeficiency viruses (SIVcpz) suggests that SIVcpzPts was derived from SIVcpzPtt through additional recombination events. AIDS Res Hum Retroviruses. 2007;23:1114–1118. doi: 10.1089/aid.2007.0071. [DOI] [PubMed] [Google Scholar]
- 40.Sodora DL, et al. Toward an AIDS vaccine: Lessons from natural simian immunodeficiency virus infections of African nonhuman primate hosts. Nat Med. 2009;15:861–865. doi: 10.1038/nm.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wooding S, et al. Contrasting effects of natural selection on human and chimpanzee CC chemokine receptor 5. Am J Hum Genet. 2005;76:291–301. doi: 10.1086/427927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liégeois F, et al. Full-length genome characterization of a novel simian immunodeficiency virus lineage (SIVolc) from olive Colobus (Procolobus verus) and new SIVwrcPbb strains from western red Colobus (Piliocolobus badius badius) from the Tai Forest in Ivory Coast. J Virol. 2009;83:428–439. doi: 10.1128/JVI.01725-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKinney DM, et al. Identification of five different Patr class I molecules that bind HLA supertype peptides and definition of their peptide binding motifs. J Immunol. 2000;165:4414–4422. doi: 10.4049/jimmunol.165.8.4414. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.