Abstract
The effect of RNA silencing in plants can be amplified if the production of secondary small interfering RNAs (siRNAs) is triggered by the interaction of microRNAs (miRNAs) or siRNAs with a long target RNA. miRNA and siRNA interactions are not all equivalent, however; most of them do not trigger secondary siRNA production. Here we use bioinformatics to show that the secondary siRNA triggers are miRNAs and transacting siRNAs of 22 nt, rather than the more typical 21-nt length. Agrobacterium-mediated transient expression in Nicotiana benthamiana confirms that the siRNA-initiating miRNAs, miR173 and miR828, are effective as triggers only if expressed in a 22-nt form and, conversely, that increasing the length of miR319 from 21 to 22 nt converts it to an siRNA trigger. We also predicted and validated that the 22-nt miR771 is a secondary siRNA trigger. Our data demonstrate that the function of small RNAs is influenced by size, and that a length of 22 nt facilitates the triggering of secondary siRNA production.
Keywords: gene silencing, microRNA, transacting siRNA
Small silencing RNAs (sRNAs) in plants and animals, including microRNAs (miRNAs) and small interfering RNAs (siRNAs), play important roles in the development and the response to pathogens and stresses. These RNAs are also valuable tools in functional genomics and biotechnology. The sRNAs associate with ARGONAUTE (AGO) and other proteins in silencing effector complexes, and they bind to a target nucleic acid via Watson–Crick base pairing. In most instances, the silencing is a direct consequence of this interaction, and the AGO effector mediates RNA-mediated DNA or histone methylation, endonucleolytic RNA cleavage, or translational inhibition. In a few instances, an sRNA interaction also triggers the production of secondary siRNAs. The targeted RNA is converted into double-stranded RNA (dsRNA) by RNA-DEPENDENT RNA POLYMERASEs (RDRs), which is then cleaved into the secondary siRNAs by DICER-LIKE (DCL) nucleases (1). Several proteins are known to be required for this process, but until now, the reason why most sRNA interactions do not result in secondary siRNA production was unclear.
The transacting siRNA (tasiRNA) pathway in plants involves secondary siRNA production (2). Noncoding transcripts encoded by TAS1–4 genes serve as the precursors of tasiRNAs (3–5). After miRNA-directed cleavage, part of the remaining transcript is converted into dsRNA by RDR6. DCL4 then cleaves the dsRNA and generates tasiRNAs in a 21-nt phase relative to positions 10 and 11 of the miRNA that defines the site of targeted cleavage. TAS1 and TAS2 are targets of miR173, and their tasiRNAs in turn can target mRNAs for pentatricopeptide repeat (PPR) proteins. In one instance, a small sRNA cascade is initiated by miR173 (6, 7), because a TAS2-derived tasiRNA can itself initiate secondary siRNA production on several PPR mRNAs. The initiator of TAS3 tasiRNA is miR390 (3, 8), and the TAS3 targets are AUXIN RESPONSE FACTOR mRNAs that influence the change from juvenile phase to adult phase, leaf morphology, and lateral root growth (9–12). miR828 initiates TAS4 tasiRNAs that target the mRNAs of MYB transcription factors (5).
A “two-hit” model to explain tasiRNA production is based on the finding that two miRNA target sites are critical for TAS3 tasiRNA production between the two sites (8, 13). One of these sites, where RNA cleavage occurs, could be targeted by any of several different miRNAs; however, the second noncleavable site is functional only if it is the target of miR390 (13). This model is specific for TAS3, and the two-hit requirement is probably related to the specific interaction of miR390 with AGO7 (12). In contrast, TAS1 and TAS2 have only a single miR173 target site, and the AGO protein involved is AGO1 rather than AGO7 (3, 14). RNAs other than TAS precursors also support secondary siRNA production if they have an miR173 target site (14, 15). This finding implies that the determinant of secondary siRNA production is the sRNA initiator rather than the targeted RNA, and it prompted a bioinformatics survey of common features in the miRNAs and tasiRNAs that are triggers of secondary siRNA production. The results reported here reveal that secondary siRNA triggers are represented as 22-nt sRNAs rather than the 21-nt size that is characteristic of most Arabidopsis miRNAs. We also describe experimental results confirming that a size of 22 nt is necessary for an miRNA to trigger secondary siRNA biogenesis on the 3′ cleavage products of their targets.
Results
siRNA Triggers Are Predominantly 22 nt in Size.
Through a combination of literature search and analysis of the Arabidopsis sRNA sequencing data with a phasing algorithm (6), we identified eight miRNAs and one tasiRNA as triggers of secondary siRNA produced from 3′ cleaved transcripts of their targets (Table S1). We also identified miR825* as the potential siRNA trigger at At5g38850, which encodes a TIR-NBS-LRR protein and produces phased siRNA (7).
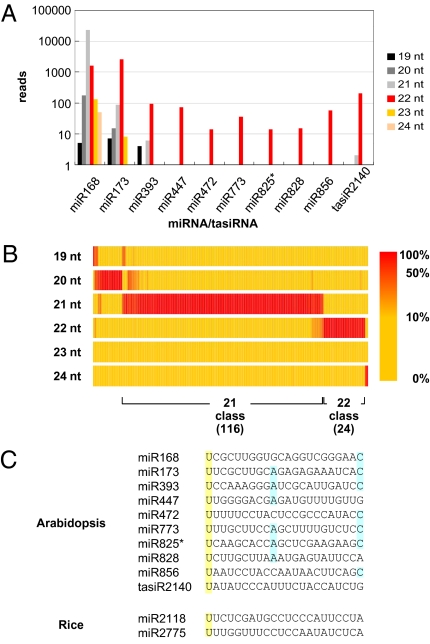
Using Arabidopsis sRNA sequencing datasets derived by 454 or Illumina technology (5, 8, 16–18), we found that the putative miRNA triggers of secondary siRNA production are represented as 22-nt species rather than the normal 21-nt form (Fig. 1A). The MIR168, MIR173, and MIR393 loci also produce other size classes in the 19- to 24-nt range, but for all of these except MIR168, the most abundant form was 22 nt in size (Fig. 1A). Among 163 Arabidopsis MIR loci, only 24 have a dominant 22-nt form (Fig. 1B and Table S2); of these, MIR173 and MIR168 have the highest expression (Fig. 1A and Table S2). Most of the 22-nt dominant miRNAs, including miR780, miR771, and miR848, have been identified recently and lack predicted or validated targets.
Fig. 1.
siRNA triggers are predominantly 22 nt. (A) Size distribution of Arabidopsis siRNA triggers. The abundance of siRNA triggers in size classes from 19 to 24 nt is plotted according to the pooled data of several available sRNA sequencing datasets (5, 8, 16–18). (B) A heat map showing clusters of Arabidopsis miRNAs with sizes enriched mainly as 21 or 22 nt. Each vertical column represents one miRNA species. A gradient of yellow to red represents the frequency (%) of each size class for a given miRNA. The map is organized according to the relative abundance of the sizes (19–24 nt) for each miRNA. (C) Alignment of siRNA triggers. Identical nucleotides and conserved nucleotides among siRNA triggers are highlighted in yellow and blue, respectively.
Among the Arabidopsis tasiRNAs, tasiR2140 [also known as TAS2 3′D6(-)] is the only one known to be associated with secondary siRNA production (6–8). The predicted structure of this TAS2-derived tasiR2140, based on the phase set by miR173, is 21 nt with a 5′ A (3). However, in the AGO1-associated sRNAs (19), tasiR2140 reads were detected mostly in the 22-nt form, with an additional U at the 5′ end relative to the predicted 21-nt tasiR2140. This 22-nt tasiR2140 is likely a functional form based on a 5′ RACE assay of tasiR2140 cleavage products on the target mRNAs (At1g63130 and At1g63080). The sRNA cleavage sites are normally opposite positions 10 and 11 of the sRNA guide molecule, and this position corresponds to the 22-nt form rather than the 21-nt form of tasiR2140 (4, 6). Two rice miRNAs, miR2118 and miR2775, that may direct the 21-nt or 24-nt phased siRNA production in developing inflorescences are also 22 nt in size (Table S1) (20). Thus, a size of 22 nt is a common feature shared by many diverse triggers of secondary siRNA production.
The 22-nt putative triggers of secondary siRNA all have U in the 5′ position (Fig. 1C). However, this is a common feature of miRNAs associated with their loading to AGO1 (19) and is not specific to triggers of secondary siRNAs. Similarly, we found that a preference for A in position 10 of the miRNAs is also a common feature by analyzing 163 Arabidopsis miRNAs, but that this preference is not specific to triggers of secondary siRNAs. Seven of the 12 triggers of secondary siRNAs have C at the 3′ terminal nucleotide, but others have G or A in this position, indicating that this feature is not essential for initiation of secondary siRNAs. Thus, we hypothesized that a size of 22 nt is the primary feature associated with the initiation of secondary siRNA production.
A 22-nt miRNA Triggers Secondary siRNA Production.
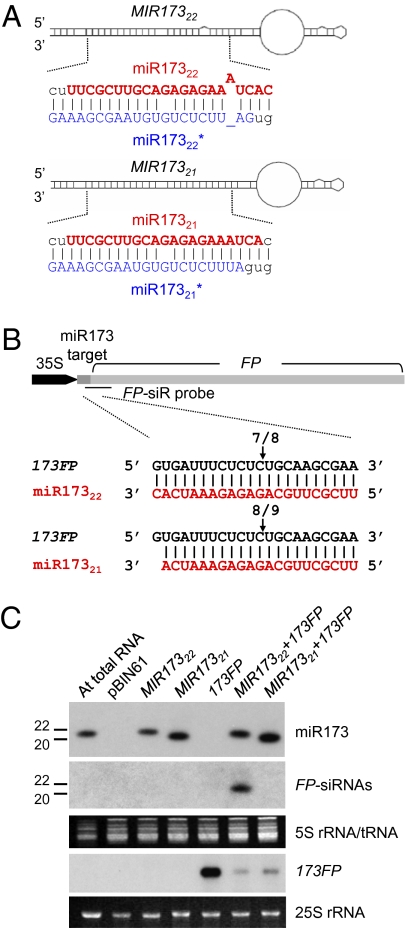
To test this 22-nt hypothesis, we engineered miRNA precursors to change the size of the processed miRNA. With MIR173, this change involved adding an additional nucleotide in miR173* to pair with a bulged base in the WT miR173 duplex. The WT MIR173 precursor is normally cleaved by DCL1, which leaves 21 paired bases between 5′ and 3′ cleavage sites to release the 22-nt miR173 (Fig. 2A). However, with the modified structure, the 21–paired-base rule would cause DCL1 to release a 21-nt miR173; in Fig. 2A, this modified construct is termed MIR17321, and the unmodified construct is termed MIR17322.
Fig. 2.
A size of 22 nt is required for miR173 to trigger secondary siRNA. (A) Predicted foldback structures of MIR17322 and MIR17321 for expressing 22- and 21-nt miR173, respectively. Expected duplexes of miR173 and miR173* are highlighted. (B) Construct of 35S:173FP and cleavage sites identified by 5′ RACE assay. The position of the cleavage site is indicated by an arrow with the proportion of sequenced clones. FP-siR probe (horizontal line) indicates a DNA oligonucleotide probe complementary to the 42-nt region downstream of the predicted cleavage site directed by miR173 for the detection of FP-derived siRNA shown below. (C) Northern blot analysis of miR173, FP-derived siRNA, and 173FP transcripts.
To test the biological activity of the modified MIR173, we used a transient assay system based on infiltration of Agrobacterium cultures transformed with these constructs in Nicotiana benthamiana, in which miR173 is not produced. The MIR constructs were coexpressed with a sensor construct that would be transcribed from the 35S promoter to produce a transcript with a target site of miR173 located at the 5′ end of a truncated GFP coding sequence (FP) (35S:173FP; Fig. 2B).
A 5′ RACE analysis of the transient assay samples confirmed that both miR17322 and miR17321 target the cleavage of 173FP mRNA at the same site (Fig. 2B) and that, correspondingly, accumulation of the full-length 173FP mRNA was lower than when 35S:173FP was expressed alone (Fig. 2C). Northern blot analysis also confirmed that the two MIR173 constructs each produce miRNAs of the expected size; 35S:MIR17322 produces an RNA of the same size as the Arabidopsis endogenous miR173, whereas 35S:MIR17321 produces a smaller species (Fig. 2C). The sequence authenticity of 21-nt miR173 was confirmed by deep-sequencing analysis of an sRNA population in N. benthamiana infiltrated with 35S:MIR17321 and 35S:173FP. Among 19,077 reads of 19- to 24-nt miR173s, 96.9% of the sRNAs are 21 nt with a sequence identical to that shown in the lower panel of Fig. 2A.
Northern blot analysis revealed that FP-derived siRNA could be detected only when 173FP was coinfiltrated with the MIR17322 construct (Fig. 2C). This indicates that the targeted cleavage by both miR17322 and miR17321 is not sufficient for the production of secondary siRNAs. This finding is consistent with our hypothesis that only the 22-nt miR173 can both direct the cleavage of the synthetic sensor RNA and trigger the production of secondary siRNA(s) using the 3′ cleavage fragment of the sensor RNA. These findings also indicate that the miR173 precursor sequence does not give miR17321 the ability to act as a trigger of secondary siRNA.
We also tested the 22-nt hypothesis with miR828 that targets the TAS4 RNA and initiates tasiRNA production (5). However, both miR828 and miR828* are 22 nt, and the modifications of the MIR828 sequence necessary to produce 21-nt miRNA are not obvious. Our alternative approach was to modify the MIR319a foldback (21) so that it would release a 21-nt miR828 without the nucleotide 22. The constructs in this assay, 35S:MIR82822 and 35S:MIR82821 (Fig. S1A), were designed to produce 22-nt and 21-nt miR828 in the transient expression assay, and the sensor construct was 35S:828FP (Fig. S1B). Transient expression of 35S:MIR82822 and 35S:MIR82821 led to production of the predicted 22- and 21-nt miR828s (Fig. S1C). When these constructs were coexpressed with 35S:828FP, the cleavage of the 35S:828FP transcript was at the same site (Fig. S1B), but only miR82822 initiated miRNA-dependent production of FP-siRNA (Fig. S1C). Thus, these results are consistent with the proposed role of 22-nt miRNA in secondary siRNA production.
In a third assay, we aimed to convert a 21-nt miRNA into a 22-nt species with the expectation that this change would be associated with an increased potential to trigger secondary siRNA production. The miR319 is normally expressed as a 21-nt form, and it lacks the ability to trigger secondary siRNA production. By creating a bulge in the miRNA/miRNA* duplex in the foldback sequence of MIR319, this construct should generate a 22-nt miR319 (Fig. 3A). A 5′ RACE analysis confirmed that 319FP transcripts, when coexpressed with 35S:MIR31921 or 35S:MIR31922, are cleaved at the predicted miR319 target site (Fig. 3B). Northern blot analysis demonstrated that these two 35S:MIR319 constructs produce different-sized miRNAs, as predicted (Fig. 3C). However, the electrophoretic migration of these RNAs is anomalous, as has been reported previously (22). We used sequence analysis to confirm the size of the miR319 species; 87.3% of the sequence reads in the 35S:MIR31921 samples were 21 nt, and 61.4% of 35S:MIR31922 were 22 nt (Fig. 3A). Although the 319FP transcript was cleaved in the presence of both 21- and 22-nt miR319, significant FP-siRNA could be detected only when 319FP transcripts were coexpressed with a 22-nt miR319 (Fig. 3C). Deep-sequencing analysis also revealed an obvious increase in FP-siRNAs phased in the register set by miR31922 cleavage site (register 1) or in the 1-nt forward register (register 2) (Fig. 3D). For the first 231 bp downstream of the cleavage site, the normalized abundance [transcripts per 10 million (TP10M)] for phased FP-siRNAs is 9,120 when coexpressed with miR31922 and 1,506 when coexpressed with miR31921. Phase analysis of FP-siRNAs using a previously described algorithm (6) showed a significant P value of 0.00074 when coexpressed with a 22-nt miR319, compared with the P value of 0.00655 when coexpressed with a 21-nt miR319. FP-siRNA production is also triggered by a 22-nt miR319 expressed from the MIR173 and MIR771 foldback constructs 35S:MIR31922(173) and 35S:MIR31922(771) (Figs. S2 and S3). Thus, by adding one extra nucleotide to the 3′ terminus, the 22-nt miR319 acquires the ability to trigger secondary siRNA production. Therefore, the 22-nt signature alone determines whether an miRNA can act as a trigger of secondary siRNA.
Fig. 3.
A change in size from 21 to 22 nt is sufficient to convert miR319 to an siRNA trigger. (A) Predicted foldback structures of MIR31921 and MIR31922 for expressing 21- and 22-nt miR319, respectively. Expected duplexes of miR319 and miR319* are highlighted. Sizes of miR319s produced from MIR31921 or MIR31922 were obtained by small RNA sequencing and shown as percentage (%) of each size class. (B) Construct of 35S:319FP and cleavage sites identified by 5′ RACE assay. The position of the cleavage site is indicated by an arrow with the proportion of sequenced clones. FP-siR probe (horizontal line) indicates a DNA oligonucleotide probe complementary to the 42-nt region downstream of the predicted cleavage site directed by miR319 for the detection of FP-derived siRNA shown below. (C) Northern blot analysis of miR319, FP-derived siRNA, and 319FP transcripts. N. benthamiana miR159 (NbmiR159) has a high sequence similarity with miR319 and likely was cross-hybridized with the miR319 probe. (D) Normalized abundance (TP10M) of FP-derived 21-nt small RNA species was plotted for both sense (S) and antisense (AS) strands of 319FP in N. benthamiana coinfiltrated with 35S:319FP/35S:MIR31921 or 35S:319FP/35S:MIR31922. 21-nt sRNAs in registers (reg.) 1 and 2 corresponding to the miR319-directed cleavage site are highlighted in red.
miR771 Is a New Trigger of Secondary siRNA Production.
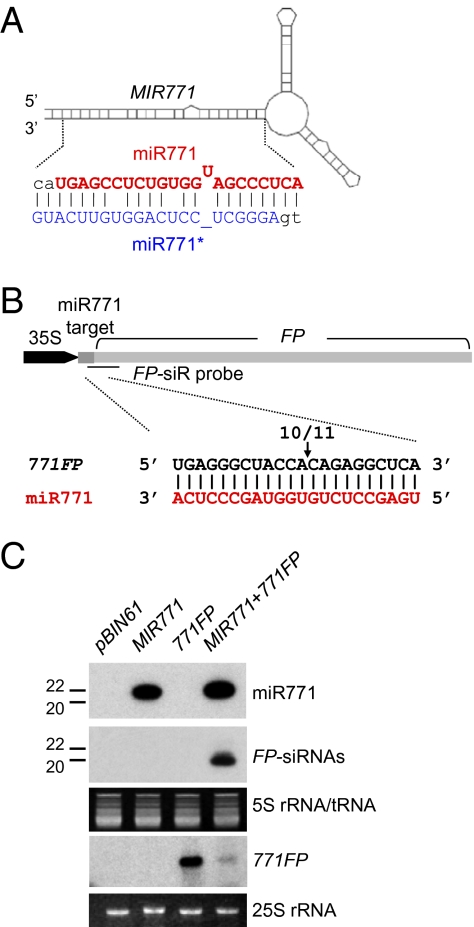
miR771 is one of the 24 Arabidopsis miRNAs that are present predominantly as 22-nt species, and we predicted that it would trigger siRNA production. As shown in Fig. 4, this prediction is confirmed by the finding that the 22-nt miR771 can induce site-specific cleavage of the 771FP transcript and FP-siRNAs are produced. Thus, the 22-nt miR771 is a new trigger of siRNA production.
Fig. 4.
miR771 is a new siRNA trigger. (A) Predicted foldback of MIR771. The expected duplexes of miR771 and miR771* are highlighted. (B) Construct of 35S:771FP and cleavage sites identified by 5′ RACE assay. The position of the cleavage site is indicated by an arrow with the proportion of sequenced clones. FP-siR probe (horizontal line) indicates a DNA oligonucleotide probe complementary to the 42-nt region downstream of the predicted cleavage site directed by miR771 for the detection of FP-derived siRNAs shown below. (C) Northern blot analysis of miR771, FP-derived siRNA, and 771FP transcripts.
Discussion
A Size of 22 nt Is the Key Determinant of siRNA Triggers.
Based on a bioinformatics analysis of sRNA databases, we hypothesized that miRNA or siRNA triggers of secondary siRNA production might have a length of 22 nt rather than the more common 21 nt. The 22-nt miRNAs or siRNAs would direct the cleavage of a target RNA, and the secondary siRNAs would be produced in an RDR-dependent manner from the 3′ cleavage products. We confirmed this hypothesis experimentally by demonstrating that the 22-nt form, but not the 21-nt form, of miR173 and miR828 can initiate siRNA production on the 3′ cleavage product of a sensor RNA, although both can direct cleavage (Fig. 2 and Fig. S1). We further confirmed this finding by demonstrating that an additional nucleotide at the 3′ end converts miR319 into an siRNA trigger (Fig. 3 and Figs. S2 and S3).
We identified eight Arabidopsis miRNA families with 22-nt forms that are known triggers of siRNA production. An additional 13 miRNA families with >50% of the sequence reads as 22-nt species are candidate triggers of secondary siRNA. Our finding that one of these, the 22-nt miR771, is capable of initiating secondary siRNAs confirms that these additional miRNAs are potential siRNA triggers (Fig. 4). Presumably, the secondary siRNAs from these additional 22-nt miRNAs were not detected previously because the target RNA is not expressed in the tissues used to generate the sRNA sequence libraries.
Biogenesis and Action of 22-nt sRNAs.
The biogenesis of 22-nt compared with 21-nt sRNAs might be influenced by the DCL protein involved or by the structure of the sRNA precursor. Examples in which the RNA structure is involved include miR771 and the 22-nt miRNAs listed in Table S1 (except miR828; see below). In all of these examples, the miRNA duplex contains a bulge (miRBase: http://www.mirbase.org/), so that DCL1 generates a 22-nt miRNA and a 21-nt miRNA*.
In other examples, DCL2 generates 22-nt siRNAs from perfectly duplexed precursors. These 22-nt siRNAs might be triggers of secondary siRNAs (23). miR828 is derived from perfectly duplexed 22-nt precursors and might be produced by DCL2, although this possibility has not been tested experimentally. There is good evidence that DCL2 and secondary siRNAs are involved in transgene and viral RNA silencing, however (23–26). We consider a causal connection likely and suggest that the 22-nt small RNAs produced by DCL2 in these systems are the triggers of secondary siRNA production.
The likely involvement of DCL2 in the initiation of secondary siRNA biogenesis could explain why dcl1 dcl4 and dcl1 dcl3 dcl4 mutants grow poorly in greenhouse conditions, whereas dcl1 dcl2 dcl4 and dcl1 dcl2 dcl3 dcl4 mutants are viable (27). In dcl1 dcl4 and dcl1 dcl3 dcl4 genotypes, the DCL2 might generate siRNA triggers using dsRNAs that are normally substrates of DCL1, DCL3, or DCL4. This overproduction of 22-nt sRNAs could then trigger massive overproduction of secondary siRNAs, leading to uncontrolled posttranscriptional silencing in the plant and subsequent impaired plant growth.
The 22-nt forms of miR173, miR828, miR319, miR771, and tasiR2140 all trigger secondary siRNA production after targeted RNA cleavage. These RNAs all have a 5′ U and, like miR173, are likely loaded into a complex with the AGO1 that also is associated with the cleavage-only interactions (14, 19). To explain the different effects of 21- and 22-nt miRNAs bound to AGO1, we propose that the AGO1 protein can adopt different conformations. When bound to the 21-nt miRNA, the protein has a structure that promotes cleavage of the targeted RNA, whereas when the 22-nt miRNA is present, the AGO1 structure promotes target cleavage and recruitment of proteins involved in siRNA biogenesis, including SUPPRESSOR OF GENE SILENCING3 (SGS3) and RDR6. This conformational change is likely to occur because the 5′ and 3′ ends of AGO-bound sRNAs are tethered within defined structural domains of the AGO proteins (28). Thus, an AGO1 bound to 22-nt miRNA is likely to be distorted relative to the form with an associated 21-nt miRNA.
The miR2775 is likely to trigger 24-nt rather than 21-nt phased siRNA production in developing inflorescences of rice (20), but it nevertheless conforms to the 22-nt rule and has a 5′ U (Fig. 1C). An exception to the 22-nt rule is miR390, a 21-nt miRNA, which initiates TAS3 tasiRNA production. Unlike the 22-nt miRNAs that have single site targets, miR390 is effective only when there are two miRNA target sites in TAS3 RNA (8, 13). The TAS3 cleavage product on the 5′ side of the miR390 target site is the secondary siRNA precursor, whereas with TAS1 and TAS2, the siRNAs are produced from the 3′ side of the cleavage site directed by the trigger miRNA. AGO7, rather than AGO1, binds to miR390, and we infer that the variant mechanisms of TAS1 and TAS2 as opposed to TAS3 secondary siRNA biogenesis are somehow related to the different properties of the AGO1 and AGO7 proteins.
Production of 22-nt miRNAs does not always trigger secondary siRNAs; for example, there are both 21-nt and 22-nt species of miR165, miR166, and miR167, but phased secondary siRNAs from their target mRNAs have not been detected. The 22-nt forms of these miRNAs are associated with AGO1 (19) and are more abundant than most known siRNA triggers; however, the proportion of 22-nt species is relatively lower than that of 21-nt species (Table S2). This suggests that the ratio of 22-nt to 21-nt miRNAs, rather than the absolute abundance of 22-nt species of these miRNAs, influences the triggering of secondary siRNAs.
The cell- or tissue-specific triggering of secondary siRNA production also may be regulated by the discrete expression of 21- or 22-nt miRNA species in different cells or tissues. For example, MIR168a generates predominantly a 21-nt mature miRNA and has broad expression in most tissue types (29). In contrast, MIR168b is specifically expressed at the shoot apical meristem and produces similar levels of 21- and 22-nt species (29). The level of 22-nt or the ratio of 22- to 21-nt miR168 may reach the threshold in meristemic cells for miR168 to function as an siRNA trigger.
Future Perspectives.
We previously determined that silencing cascades can be established in plants when miRNAs and siRNAs trigger secondary siRNA production on a long RNA target (6). However, these cascades are of limited extent, and propagation of the cascade is likely constrained. Our present analysis has shown that only a few miRNAs and siRNAs can trigger secondary siRNAs, and that these must be 22 nt long and with a 5′ U so that they associate with AGO1, or otherwise, as in the exceptional example of miR390, they must associate with AGO7.
Further research is needed to establish how a subtle difference between a 21-nt miRNA and a 22-nt miRNA can affect the structural configuration of AGO1 and the recruitment of factors for the production of secondary siRNAs. The biological role of the siRNA cascades is also of interest. In terms of growth and development, some early data suggest that the production of secondary siRNAs might be associated with signaling of positional information (30). These cascades also are likely involved in antiviral defense, given that the 22-nt viral siRNAs are abundant in plants infected with some viruses (31, 32). In these virus-infected plants, the amplification associated with secondary siRNA production is likely important for the effectiveness of RNA silencing for protection against replicating viruses. This amplification contains a potential penalty, however, because misregulation of endogenous gene silencing cascades could have a damaging effect on the gene expression profile in plants. Presumably, plants have mechanisms to ensure that the cascades are effective only at the appropriate time; however, the potential for amplification could aid in the development of efficient gene silencing technologies for virus resistance and manipulation of gene expression in biotechnology and for research applications.
Materials and Methods
Identification of siRNA Triggers.
Loci producing phased siRNAs were identified through a literature search or a phasing analysis of Arabidopsis sRNA sequencing datasets using an algorithm described previously (6). Only 21-nt sRNA sequences were subjected to the phasing analyses, and a cutoff of P ≤ 0.001 was applied. Phased siRNA-producing loci not reported to be targets of known miRNAs or tasiRNAs were searched against known miRNAs and miRNAs*. sRNAs that can direct the cleavage upstream of phased siRNA clusters and set the phase consistent with or 1 nt forward from that of phased siRNAs were collected for further analyses.
Construction of MIRs and Sensors.
For transient expression of miRNAs, sequences containing foldback structures of MIR173, MIR319, MIR828, and MIR771 were amplified from Arabidopsis genomic DNA and inserted into the pBIN61 vector containing the 35S promoter. MIR foldbacks were modified by site-directed mutagenesis to express 21-nt miR173, 21-nt miR828, and 22-nt miR319. For miRNA targets, a 22-nt sequence complementary to the miRNA sequence was fused to a 5′ truncated GFP sequence (FP) and used as the sensor construct. The sequences of MIR foldbacks and sensors are listed in Table S3.
RNA Structure Prediction.
Structures of MIR foldbacks were predicted by mfold v3.2 (33) (http://mfold.bioinfo.rpi.edu/cgi-bin/rna-form1.cgi).
Nicotiana benthamiana Transient Expression.
The procedures for transient expression assay have been described previously (34). Leaves of 3- to 4-wk-old N. benthamiana plants were infiltrated with Agrobacterium strain C58C1 carrying MIR and/or sensor constructs. The input amount of each construct was adjusted according to its expression level. The total input of Agrobacterium was brought to OD600 = 1.0 with use of the empty vector. Infiltrated tissues were collected after 2–3 d for total RNA extraction.
Northern Blot Analysis.
Total RNA of infiltrated tissues was extracted using Trizol reagent (Invitrogen). For the sRNA Northern blot analysis, 3–5 μg of total RNA was separated by 15% denaturing polyacrylamide TBE-urea gels (Invitrogen). DNA oligonucleotides complementary to the miRNAs or the 42-nt region immediately downstream of the predicted cleavage site directed by miRNAs were used as probes to detect the expression of miRNAs or siRNAs from the sensor. For the Northern blot analysis of sensors, 2 μg of total RNA was separated by 1% agarose gel. A 23-nt DNA oligonucleotide complementary to the truncated GFP was used as the probe. The sequences of probes used in this study are listed in Table S3. Probe labeling, hybridization, washing, and signal detection were performed as described previously (6).
sRNA Sequencing.
Total RNA was isolated using the mirVana miRNA Isolation Kit (Ambion) to generate sRNA libraries for sequencing by the Illumina platform.
Validation of Sensor Cleavage.
Modified 5′ RACE with the GeneRacer Kit (Invitrogen) was adopted to validate the cleavage sites on the sensors. The primers used for 5′ RACE are in Table S3.
Supplementary Material
Acknowledgments
We thank the members of the S.-H.W. laboratory and Krys Kelly for helpful discussions. This work was supported by research grants from Academia Sinica (Foresight Project L20-2) and the National Science Council (NSC 97-2311-B-001-003-MY3) (to S.-H.W.). H.-M.C. is the recipient of postdoctoral fellowships from the National Science Council (098-2811-B-001-030) and Academia Sinica. The work at Cambridge was supported by the Gatsby Charitable Foundation and European Research Council Advanced Investigator Grant 233325 REVOLUTION. D.C.B. is a Royal Society Research Professor.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 14945.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1001738107/-/DCSupplemental.
References
- 1.Voinnet O. Use, tolerance and avoidance of amplified RNA silencing by plants. Trends Plant Sci. 2008;13:317–328. doi: 10.1016/j.tplants.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Vaucheret H. MicroRNA-dependent trans-acting siRNA production. Sci STKE. 2005;2005:pe43. doi: 10.1126/stke.3002005pe43. [DOI] [PubMed] [Google Scholar]
- 3.Allen E, Xie Z, Gustafson AM, Carrington JC. MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 4.Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen HM, Li YH, Wu SH. Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc Natl Acad Sci USA. 2007;104:3318–3323. doi: 10.1073/pnas.0611119104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howell MD, et al. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell. 2007;19:926–942. doi: 10.1105/tpc.107.050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 9.Adenot X, et al. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol. 2006;16:927–932. doi: 10.1016/j.cub.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 10.Fahlgren N, et al. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol. 2006;16:939–944. doi: 10.1016/j.cub.2006.03.065. [DOI] [PubMed] [Google Scholar]
- 11.Garcia D, Collier SA, Byrne ME, Martienssen RA. Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr Biol. 2006;16:933–938. doi: 10.1016/j.cub.2006.03.064. [DOI] [PubMed] [Google Scholar]
- 12.Marin E, et al. miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell. 2010;22:1104–1117. doi: 10.1105/tpc.109.072553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montgomery TA, et al. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 14.Montgomery TA, et al. AGO1-miR173 complex initiates phased siRNA formation in plants. Proc Natl Acad Sci USA. 2008;105:20055–20062. doi: 10.1073/pnas.0810241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felippes FF, Weigel D. Triggering the formation of tasiRNAs in Arabidopsis thaliana: The role of microRNA miR173. EMBO Rep. 2009;10:264–270. doi: 10.1038/embor.2008.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gregory BD, et al. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell. 2008;14:854–866. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 18.German MA, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 19.Mi S, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson C, et al. Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res. 2009;19:1429–1440. doi: 10.1101/gr.089854.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18:1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palatnik JF, et al. Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev Cell. 2007;13:115–125. doi: 10.1016/j.devcel.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 23.Mlotshwa S, et al. DICER-LIKE2 plays a primary role in transitive silencing of transgenes in Arabidopsis. PLoS One. 2008;3:e1755. doi: 10.1371/journal.pone.0001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deleris A, et al. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313:68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- 25.Moissiard G, Parizotto EA, Himber C, Voinnet O. Transitivity in Arabidopsis can be primed, requires the redundant action of the antiviral Dicer-like 4 and Dicer-like 2, and is compromised by viral-encoded suppressor proteins. RNA. 2007;13:1268–1278. doi: 10.1261/rna.541307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang XB, et al. RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2010;107:484–489. doi: 10.1073/pnas.0904086107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bouché N, Lauressergues D, Gasciolli V, Vaucheret H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 2006;25:3347–3356. doi: 10.1038/sj.emboj.7601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parker JS. How to slice: Snapshots of Argonaute in action. Silence. 2010;1:3. doi: 10.1186/1758-907X-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaucheret H. AGO1 homeostasis involves differential production of 21-nt and 22-nt miR168 species by MIR168a and MIR168b. PLoS ONE. 2009;4:e6442. doi: 10.1371/journal.pone.0006442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chitwood DH, et al. Pattern formation via small RNA mobility. Genes Dev. 2009;23:549–554. doi: 10.1101/gad.1770009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akbergenov R, et al. Molecular characterization of geminivirus-derived small RNAs in different plant species. Nucleic Acids Res. 2006;34:462–471. doi: 10.1093/nar/gkj447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie Z, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:e104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voinnet O, Rivas S, Mestre P, Baulcombe D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 2003;33:949–956. doi: 10.1046/j.1365-313x.2003.01676.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.