Abstract
Canonical animal microRNAs (miRNAs) are generated by sequential cleavage of precursor substrates by the Drosha and Dicer RNase III enzymes. Several variant pathways exploit other RNA metabolic activities to generate functional miRNAs. However, all of these pathways culminate in Dicer cleavage, suggesting that this is a unifying feature of miRNA biogenesis. Here, we show that maturation of miR-451, a functional miRNA that is perfectly conserved among vertebrates, is independent of Dicer. Instead, structure-function and knockdown studies indicate that Drosha generates a short pre-mir-451 hairpin that is directly cleaved by Ago2 and followed by resection of its 3′ terminus. We provide stringent evidence for this model by showing that Dicer knockout cells can generate mature miR-451 but not other miRNAs, whereas Ago2 knockout cells reconstituted with wild-type Ago2, but not Slicer-deficient Ago2, can process miR-451. Finally, we show that the mir-451 backbone is amenable to reprogramming, permitting vector-driven expression of diverse functional miRNAs in the absence of Dicer. Beyond the demonstration of an alternative strategy to direct gene silencing, these observations open the way for transgenic rescue of Dicer conditional knockouts.
Keywords: Slicer, gene suppression, miRNA reprogramming
MicroRNAs (miRNAs) are ∼22-nt regulatory RNAs that collectively serve essential functions in higher eukaryotes (1). Conserved biogenesis machinery governs the production of canonical miRNAs in invertebrate and vertebrate cells (2). Primary miRNA (pri-miRNA) transcripts containing one or more hairpin structures are first cleaved in the nucleus by the Drosha RNase III enzyme and its dsRNA binding partner DiGeorge syndrome critical region gene 8 (DGCR8)/Pasha to yield ∼55–70 nt precursor miRNA (pre-miRNA) hairpins. These are cleaved again in the cytoplasm by the Dicer RNase III enzyme to yield a miRNA/miRNA* duplex, from which one strand matures in a complex with an Argonaute (Ago) protein. The miRNA guides the Ago complex to target transcripts, often bearing 7-nt complements to the 5′ end of the miRNA, for destabilization and/or translational inhibition (3). Mammalian genomes encode four Ago-class proteins, of which Ago2 is uniquely capable of directly cleaving highly complementary targets (4, 5), a process popularly termed slicing.
Since the elucidation of this framework, several alternative pathways for miRNA biogenesis have emerged. For example, mirtrons are short hairpin introns that exploit the splicing machinery to generate pre-mirRNA hairpins, thereby bypassing Drosha cleavage (6–8). Functional miRNAs can also be generated from certain small nucleolar RNAs (9, 10) and tRNAs (11, 12), presumably through Drosha-independent mechanisms. The viral miRNA precursor miR-M1-7 from murine γ-herpesvirus 68 fuses a tRNA to an miRNA hairpin and is processed by tRNase-Z (13). Endogenous shRNAs (endo-shRNAs) are also independent of the canonical nuclear processing machinery, and their hairpin termini might be defined by RNA polymerase III (11).
Common to these sundry biogenesis pathways is cleavage of an intermediate precursor by Dicer and routing of the mature small RNA into an Argonaute, suggesting that these are defining features of miRNA-class regulatory RNAs. In this study, we show that the maturation of the highly conserved vertebrate miR-451 bypasses Dicer, and instead requires direct cleavage of its precursor hairpin through Ago2 Slicer activity. We exploit these properties of mir-451 as a flexible platform for Dicer-independent, vector-mediated expression of miRNAs.
Results
Atypical Conservation and Pattern of Small RNA Reads Derived from mir-451.
Many animal miRNA loci are clustered, and this property identified a conserved hairpin christened mir-451, located 100 bp downstream of mir-144 (14). As is typical for conserved miRNA genes, mir-144 exhibits a saddle-shaped pattern of divergence in which its terminal loop exhibits many more nucleotide substitutions than its stems and for which the miRNA* is slightly less constrained than the miRNA (15, 16). In sharp contrast, the terminal loop of mir-451 is invariant from human to fish, whereas specific nucleotides in the stem are variable (Fig. 1 A and B).
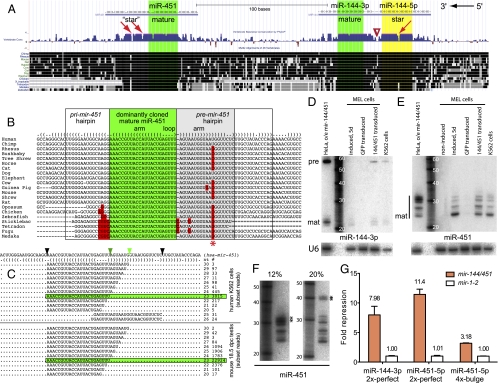
Fig. 1.
Atypical conservation and pattern of small RNAs from mir-451. (A) mir-144/mir-451 region from the University of California Santa Cruz Genome Browser; mature miRNAs (green) are encoded by the bottom strand. mir-144 exhibits a typical evolutionary pattern with far greater divergence in the terminal loop (triangle) than in mature miRNA or star arm (yellow). mir-451 is more conserved in its terminal loop than its 3′ hairpin arm. (B) Vertebrate mir-451 alignments illustrate the constrained terminal loop; asterisk marks a variable position in the 3′ hairpin. (C) Dominant miR-451 reads are 23 nt (green highlighted box), but larger species up to 30 nt (light green triangle) are observed in multiple species; rare reads are derived from the 3p arm. Data were analyzed from GSM494811 (K562) and GSM433295 (mouse testis); see Dataset S1 for full analysis of these and other miR-451–containing libraries. (D) mir-144 generates typical pre-miRNA hairpin and mature miRNA. (E) mir-451 generates small RNAs ranging from 22 to 24 nt to >30 nt. Endogenous mir-144 and mir-451 were detected in K562 cells or MEL cells induced with HMBA; these miRNAs are not expressed by uninduced MEL or GFP-transduced MEL cells. Ectopic miRNAs were generated by transfection of HeLa cells with mir-144/451 plasmid or transduction of MEL cells with mir-144/451 lentivirus. (F) The two upper bands (**) in miR-451 blots exhibit differential mobility in different acrylamide gels from ∼32 to >40 nt. (G) Sensor assays in HeLa cells transfected with mir-144/451 show repression of miR-144-3p and miR-451 perfect targets and of miR-451 seed target. Sensor values were normalized against mir-1-2 construct; SDs from quadruplicate assays are shown.
The unusual conservation features of mir-451 are linked to an atypical pattern of small RNA reads. Its dominantly cloned species extend across the terminal loop into the complementary side of the hairpin (17), and even longer reads were observed. We found these to be reproducible and conserved characteristics of mir-451. In addition to dominant 23-nt reads typical of miRNAs, a population of 24- to 30-nt reads extending well into the complementary hairpin arm is produced by mir-451 in human (18), mouse (19–21), dog (22), and chicken (23) (Fig. 1C and Dataset S1). In contrast to its 3′ heterogeneity, the 5′ terminus of miR-451 was precisely defined in all species examined (Dataset S1).
We confirmed the atypical sizes of mir-451–derived species with Northern analysis of human K562 cells and murine erythroleukemia (MEL) cells induced with hexamethylene bisacetamide (HMBA). By comparison, its partnered gene mir-144 generated an ∼58-nt pre-miRNA and an ∼22-nt mature miRNA, as expected for a canonical miRNA gene (Fig. 1D). In contrast, a probe antisense to miR-451 detected a series of bands extending past 30 nt (Fig. 1E). This was recapitulated by lentiviral transduction of murine mir-144/451 into uninduced MEL cells or by transfection of a human mir-144/mir-451 plasmid into HeLa cells. Notably, the >30-nt bands detected by miR-451 probe exhibited variable mobility from ∼32 to 42 nt according to gel percentage and temperature (Figs. 1F and Fig. S1). Previous studies noted that short hairpins can exhibit atypical mobility even under denaturing conditions (24), suggesting that these bands correspond to pre-mir-451 hairpins (Fig. 1B).
Despite its atypical properties, miR-451 regulates erythroid development and represses endogenous transcripts (25–27). We verified the regulatory activity of miR-451 using model luciferase constructs containing perfectly complementary sites or bulged miR-451 target sites. Both targets were repressed by a mir-451/mir-144 construct relative to a noncognate miRNA expression construct (mir-1-2) in HeLa cells, with severalfold greater repression of the perfect targets than the bulged targets (Fig. 1G). Thus, miR-451 has the regulatory capacity of a typical miRNA.
Structural Requirements for Biogenesis and Function of miR-451.
To gain insight into the atypical biogenesis of miR-451, we investigated structurally variant precursors (Fig. 2A). We first tested the role of the stem basal to the duplex involving mature miR-451, which is broadly conserved and resembles the lower stem of canonical pri-miRNAs. Structurally nonconservative mutations abrogated its ability to repress miR-451 sensor (Fig. 2B), implicating processing by Drosha/DGCR8. In addition, libraries from pro- and pre-B cells (21) contained reads with heterogeneous 5′ ends and precise 3′ ends that directly abutted mature miR-451 (Dataset S1); such “moR” reads are diagnostic of Drosha processing (28, 29). As well, human blood cells (18), mouse testis (20), and dog lymphocytes (22) express rare RNAs mapping downstream of mature miR-451 (Fig. 1C and Dataset S1). Many of these extend to a position constituting a 2-nt 3′ overhang to the strict 5′ end of miR-451 (Fig. 2A, black arrowheads), consistent with RNase III cleavage. These are probably not related to miR-451 biogenesis, because they overlap within the terminal loop of pre-mir-451. They might result from breakage within the unstructured loop, perhaps reminiscent of tRNA cleavage within anticodon loops (30). In any case, these data support the existence of an ∼42-nt pre-mir-451 hairpin produced by Drosha/DGCR8 cleavage (Figs. 1C and 2A), as indicated by Northern analysis (Fig. 1F).
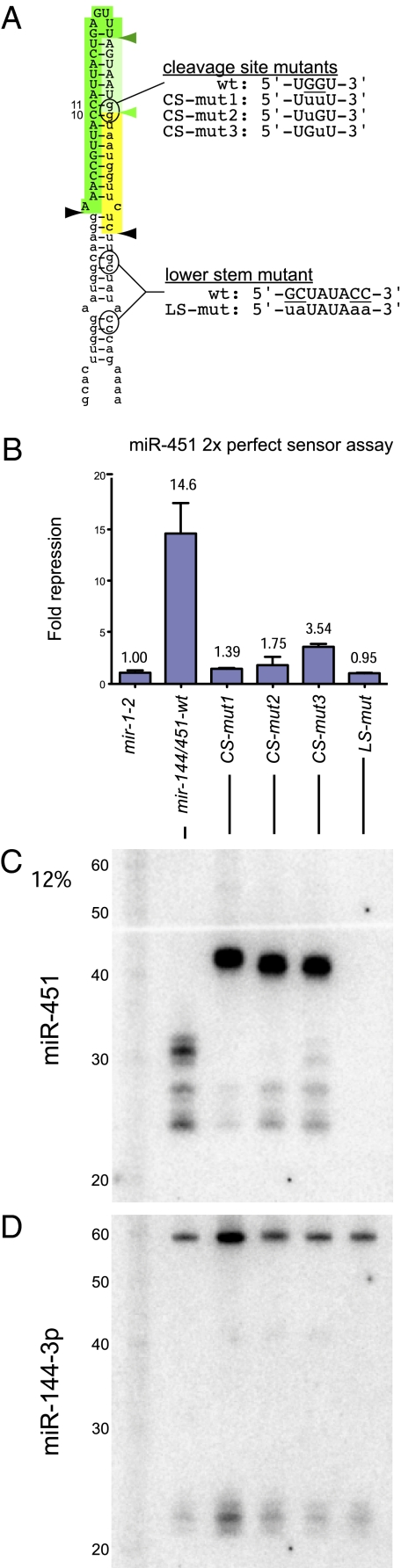
Fig. 2.
Structural requirements for miR-451 biogenesis. (A) Schematic of human mir-451 hairpin. Site mutants were generated in the context of a functional mir-144/451 plasmid. (B) Regulatory activity of mir-451 variants was assayed using a 2× perfect sensor normalized to its activity in the presence of mir-1-2 (Fig. 1G). Pairing at the putative cleavage site across from positions 10 and 11 from the 5′ end of mature miR-451 is essential for miR-451 activity as is the integrity of the lower stem. (C) Northern analysis reveals that the three cleavage site mutants accumulate >40-nt species; the lower stem mutant did not produce any short RNA species. (D) Stripping and reprobing reveal the accumulation of pre-mir-144 and mature miR-144 from the different structural variants, providing a loading control.
The highly paired pre-mir-451 stem called to mind the capacity of mammalian Ago2 to cleave highly paired canonical pre-miRNAs (31). In principle, slicing programmed by the 5′ end of pre-mir-451 would generate 30-nt miR-451 species (Figs. 1C and 2A, light green arrowheads). We tested the consequence of disrupting pairing at the 10th and 11th nucleotides across from the 5′ end of miR-451. The single mutants were strongly compromised for activity, whereas the double mutant was almost completely functionally inactive (Fig. 2B). This supported the notion that Ago2 cleavage mediates its biogenesis.
Northern analysis of these variants was informative. Although the wild-type construct produced the characteristic series of bands, the three constructs mutated at the putative Ago2 cleavage site strongly accumulated >40-nt bands (Fig. 2C) corresponding in size to Drosha-cleaved pre-mir-451 hairpins (Fig. 2A, highlighted region). Point mutations in atypically migrating hairpins restore linear mobility (24), suggesting a rationale for their more expected mobility compared with wild type. The levels of mature miR-451 paralleled the sensor assays: the double-cleavage site mutant produced almost none, whereas the single mutants produced slightly more miR-451; the lower-stem mutant yielded neither pre-mir-451 nor shorter matured RNAs. The various mir-451 mutations did not substantially affect the biogenesis of mir-144 present on these constructs, which served as an internal control to these experiments (Fig. 2D). Taken together, these tests implied Drosha cleavage and Ago2 slicing in the biogenesis of miR-451.
Drosha-Dependent, Dicer-Independent Biogenesis of miR-451.
We followed these structural studies with genetic manipulation of miRNA processing factors by introducing siRNAs to HeLa cells. The knockdown efficiencies were confirmed by qPCR analysis (Fig. S2). Consistent with the inability of the pri-mir-451 lower-stem mutant to be matured (Fig. 2B), HeLa cells depleted for Drosha or its partner DGCR8 were compromised in their ability to generate mir-451 intermediates or mature miR-451 (Fig. 3A). We also probed against its “star” arm (that is, the 3p arm complementary to mature miR-451). We expected this to label the pre-mir-451 hairpin but not the Ago2-cleaved hairpin, because the latter contains <9-nt complementarity to the probe. We observed hybridization to bands of apparent ∼35-nt length in 16% gels (labeled pre), which comigrated with the larger bands detected by miR-451 probe (Fig. 3A). This implied that these species contain the full 42-nt mir-451 hairpin sequence.
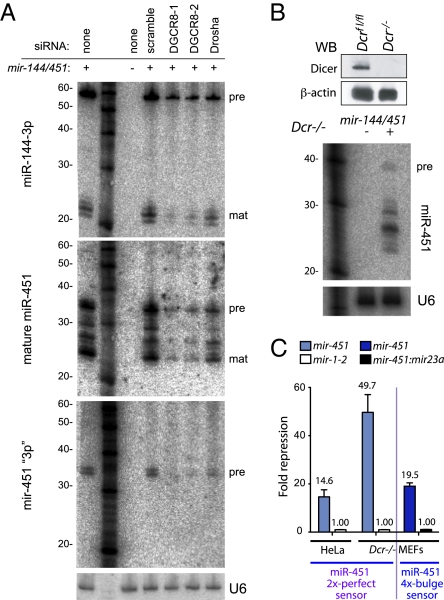
Fig. 3.
Maturation of miR-451 requires Drosha complex but not Dicer. (A) siRNA-mediated knockdown of DGCR8 and Drosha showed their requirement for biogenesis of miR-144 and miR-451 produced by mir-144/mir-451 construct. A probe against the 3p arm of mir-451 hairpin defines pre-mir-451 hairpin bands (pre), which are substantially decreased on knockdown of DGCR8 and Drosha. The miR-451 blot was stripped and reprobed for U6 as a loading control. (B) Western blot verifies the absence of Dicer in a viable Dicer−/− MEF cell line. Transfection of mir-144/mir-451 into these cells yielded mature miR-451 small RNAs. We used 16% gels, but blots in B were run at 500 V, whereas blots in A were run at 250 V, causing differential migration of the pre-miRNA hairpin (Fig. S1). (C) The mir-144/mir-451 construct was highly active in Dicer−/− MEFs, both on perfect and bulged sensors. Quadruplicate assays were performed, and SDs were plotted; tests in HeLa were normalized to mir-1-2, whereas tests in Dicer−/− were normalized to mir-144/451 construct in which miR-451 was reprogrammed with miR-23a-3p.
The 42-nt Drosha-cleaved mir-451 hairpin does not possess sufficient duplex to be a Dicer substrate, suggesting that its maturation might bypass Dicer. We performed stringent tests using a viable line of mouse embryonic fibroblasts (MEFs) bearing the floxed allele of Dicer lacking exon 22 (32). These cells do not express Dicer protein (Fig. 3B) and are arrested for endogenous miRNA biogenesis at the pre-miRNA stage. Nevertheless, transfection of mir-144/451 construct into Dicer−/− MEFs yielded processed small RNAs (Fig. 3B) with strong repressive capacity, both on perfect and bulged miR-451 sensors (Fig. 3C). These data show a Dicer-independent, miRNA-class regulatory RNA in animal cells.
Ago2 Slicer Activity Is Essential for Maturation of miR-451.
Conditional knockout of mAgo2 in the hematopoietic system reduces the level of many miRNAs (33). However, among miRNAs expressed in bone marrow, miR-451 was uniquely susceptible to loss of Ago2. We isolated RNA from wild-type and Ago2−/− bone marrow (reconstituted in lethally irradiated recipients from Ago2[fl/fl]; MxCre donors) and analyzed their small RNAs by microarray. Not only was miR-451 the most abundant miRNA in wild-type bone marrow, its nearly complete absence (>400-fold lower) in the Ago2 mutant made it by far the most highly depleted miRNA (Fig. 4A; see raw and processed data in Dataset 3). We used Northern analysis to confirm the complete loss of intermediate and mature forms of miR-451 in Ago2−/− bone marrow, which instead accumulated pre-mir-451 (Fig. 4B).
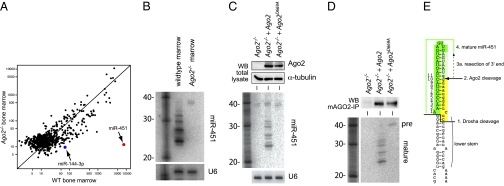
Fig. 4.
Ago2 Slicer activity mediates miR-451 maturation. (A) Microarray profiling of wild-type and Ago2−/− bone marrow revealed that miR-451 is the highest-expressed miRNA in bone marrow and uniquely deficient in the absence of Ago2. (B) Northern blot verifies loss of processed miR-451 intermediates in Ago2−/− marrow. (C Top) Western blot verification of Ago2−/− MEFs reconstituted with control virus, Ago2-expressing virus, or Ago2[D669A] virus. (Bottom) Transfection of mir-144/mir-451 construct into this panel of cells shows that Ago2 Slicer function is strictly required for production of mature miR-451. (D Upper) Ago2 proteins were immunoprecipitated from the panel of reconstituted Ago2−/− MEFs. (Lower) Associated small RNAs were analyzed by Northern blot and probed for miR-451; Ago2[D669A] cannot mature miR-451. (E) Model for mir-451 processing.
The strict Ago2 dependency of miR-451 was consistent with the inefficient maturation of pri-mir-451 cleavage-site mutants (Fig. 2C). However, these data do not exclude that changes in the erythroid compartment of Ago2 mutants might contribute to loss of miR-451 (33) nor do they directly address the requirement of Ago2 Slicer activity. We tested this using a panel of Ago2−/− MEFs reconstituted with control MigR retrovirus or retroviruses carrying wild-type mAgo2 or catalytically inactive (D669A) mAgo2 (33). Western blots showed equivalent Ago2 expression levels in the reconstituted cells (Fig. 4C Top), and the mir-451/mir-144 construct was efficiently expressed as assessed by quantitative reverse transcription-polymerase chain reaction across the primary transcript (Fig. S3). However, only Ago2−/− cells reconstituted with wild-type Ago2 could mature miR-451 from its hairpin precursor (Fig. 4C Middle).
We examined the state of mir-451 intermediates in wild-type and catalytic-dead Ago2 proteins by immunoprecipitating Ago2 followed by Northern analysis. As seen in Fig. 4D, wild-type Ago2 contained matured miR-451 species, whereas Ago2[D669A] associated exclusively with the hairpin precursor. Finally, to account for the presence of pre-mir-451 in the absence of Ago2, we checked whether the hairpin was incorporated into other Agos. We observed that Ago1 immunoprecipitated with pre-mir-451 but not matured miR-451 species (Fig. S4). This confirmed that Ago2–mediated slicing is essential for the biogenesis of miR-451 and cannot be substituted by other Agos. We hypothesize that hairpin cleavage renders its 3′ end amenable to resection by ribonuclease(s) to generate 23- to 24-nt mature miR-451 (Fig. 4E).
mir-451 Backbone Confers Dicer-Independent Expression of Other miRNAs.
Many studies have used Dicer knockouts to assess biological processes that are dependent on miRNAs. For example, maternal and zygotic (MZ) loss of zebrafish Dicer impairs clearance of maternal transcripts during the embryonic maternal–zygotic transition (34). This was demonstrably due to miRNAs in the miR-430 family, because injection of synthetic miR-430 into MZ-Dicer embryos rescued early development. However, transgenic rescue of miRNAs in Dicer mutant tissue has not been possible, because miRNA biogenesis generally requires Dicer.
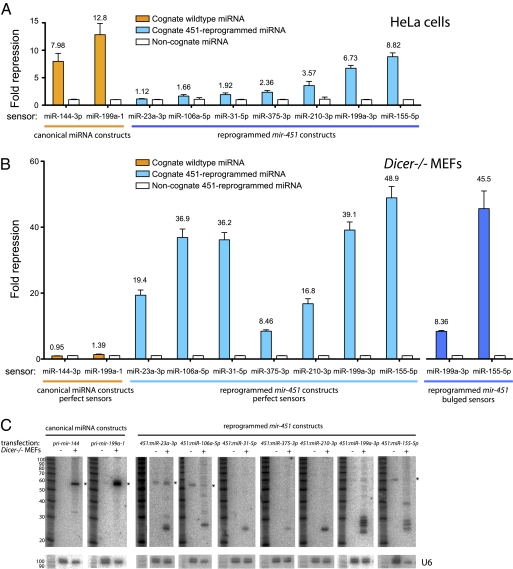
We therefore investigated whether pri-mir-451 was amenable to reprogramming. This was not necessarily possible, given the strict conservation of miR-451 primary sequence (Fig. 1B). Still, we replaced the human mir-451 hairpin with a variety of miRNA sequences, while maintaining its secondary structure, and performed initial tests of perfect sensors in HeLa cells. Encouragingly, we observed significant repression by several miRNAs reprogrammed into the mir-451 backbone (Fig. 5A). With miR-199-3p, the activity of the reprogrammed construct was comparable to its endogenous pri-miRNA context, yielding ∼7-fold repression compared with a noncognate miRNA (mir-1-2). In total, six of seven reprogrammed constructs were active, although they varied in their magnitude of target repression.
Fig. 5.
The mir-451 backbone confers Dicer-independent expression of other miRNAs. (A) mir-451 hairpins were reprogrammed with other miRNAs and tested against perfect sensors in HeLa (blue bars). Activities of canonical miRNA constructs are shown for comparison (orange bars); all tests were normalized to mir-1-2 as a noncognate miRNA (white bars). (B) Canonical miRNA constructs are essentially inactive in Dicer−/− MEFs (orange bars), whereas reprogrammed mir-451 hairpins are highly active (blue bars). We also observed strong repression of bulged sensors (dark blue bars), indicating that the 5′ ends of miRNAs from reprogrammed constructs were defined accurately. Data were normalized to sensor activity in the presence of 451:miR-23a-3p as a noncognate functional control miRNA; 451:miR-199a-3p was used as a control for the reprogrammed miR-23a-3p sensor test. Quadruplicate assays were performed, and SDs were plotted. (C) Northern analysis confirms the failure of Dicer−/− MEFs to process ectopic canonical miRNAs. In contrast, short RNAs were produced from reprogrammed mir-451 backbones in Dicer−/− MEFs; analysis of untransfected cells reveals the accumulation of some endogenous pre-miRNA species (asterisks).
We challenged these reprogrammed constructs to function in Dicer−/− MEFs, in which canonical miRNA constructs are nonfunctional (Fig. 5B). We observed robust activity (20- to 50-fold target repression) of all constructs in the absence of Dicer, including ones whose activity was marginal in HeLa cells. Moreover, reprogrammed mir-451 constructs could strongly repress bulged targets (Fig. 5B), providing evidence for the accurate definition of 5′ ends of reprogrammed miRNAs in Dicer knockout cells as with miR-451 (Fig. 3C). Although canonical miRNAs are inactive in these mutant cells, we could perform meaningful normalizations by comparing reprogrammed constructs with a functional reprogrammed 451:miR-23a-3p construct (whose activity was, in turn, normalized to functional 451:miR-199a-3p).
Ago2 can directly use some pre-miRNAs and other long RNAs as guide molecules, and their loading is potentiated in the absence of Dicer (35). Therefore, sensor tests do not necessarily report on Dicer-independent miRNA production. To address this, we analyzed Dicer−/− MEFs transfected with reprogrammed mir-451 constructs and observed accumulation of small RNAs in all cases (Fig. 5C). Some constructs generated a ladder of bands, as with miR-451, whereas others predominantly accumulated a single species. Further analysis of these constructs may provide insight into the nature and efficiency of the resection activity on different Ago2-cleaved hairpin substrates.
Finally, comparing these constructs in Dicer−/− MEFs and HeLa cells, we observed several instances where small RNA maturation was apparently increased in the absence of Dicer (Fig. S5). It is possible that lack of Ago2 loading of endogenous Dicer-dependent substrates (i.e., miRNAs and/or siRNAs) might enhance biogenesis and activity of miR-451–based constructs. However, other differences in small RNA pathway status or gene expression could well contribute to the distinct activity of these constructs in different cells; thus, additional tests of a competition model for Ago2 loading are necessary. In any case, these data provide strict evidence that the mir-451 backbone permits diverse, functional, processed miRNAs to be generated in Dicer knockout cells.
Discussion
Our studies show that the highly conserved vertebrate pre-mir-451 hairpin is independent of Dicer and instead, is reliant on direct cleavage by Ago2. This differs from its role in generating Ago2-cleaved precursor miRNAs, in which Ago2 was proposed to cleave certain well-paired pre-miRNAs before Dicer cleavage, perhaps facilitating removal of passenger-strand products (31). In the case of mir-451, slicing of its short pre-miRNA hairpin is absolutely prerequisite for further 3′ trimming to generate the mature miRNA and cannot be supported by nonslicing Ago proteins. This adds to other endogenous processes in mammalian cells, including regulation of highly or perfectly complementary targets by certain miRNAs (36) and endogenous siRNAs (11, 19, 37) that may collectively underlie the retention of cleavage activity by vertebrate Ago2.
While our work was under review, the Hannon and Giraldez groups reported similar evidence regarding the Drosha-dependent, Dicer-independent, Ago2-mediated biogenesis of miR-451 in mammalian (38) and zebrafish (39) systems. Our biogenesis data are consistent with their data and extend the capacity of this alternative biogenesis pathway for small RNA expression. Beyond the analysis of synthetic mir-451–based RNA hairpins (38, 39), we provide broad evidence for flexible reprogramming of mir-451 using DNA vectors. In particular, we showed efficient processing of an artificial mir-451–based construct into miRNA-sized species that could regulate both perfect and bulged targets, even in Dicer knockout cells. These data provide proof of principle that the mir-451 backbone may permit transgenic expression of individual miRNAs in systemic or conditional murine Dicer knockouts (32, 40–42), opening the potential for genetic rescues in the animal.
We have not yet identified a nuclease responsible for resection of the 3′ end of Ago2-cleaved mir-451. A nuclease is inferred to down-regulate pre-miRNAs after their 3′ uridylation by terminal uridylyltransferase 4 (43), but its identity has similarly been elusive. Still, the collected studies suggest that nucleases targeting the 3′ ends of small RNA precursors have both negative and positive roles for miRNA biogenesis. We also note that the efficient function of miR-451 and many reprogrammed miRNAs in Dicer−/− MEFs indicates that Dicer is not essential for substrate loading into Ago2. In light of the discovery that functional Dicer-independent “primal RNAs” associate with Schizosaccharomyces pombe Ago1 (44), direct loading of RNAs into Argonaute proteins may be an ancient strategy.
Materials and Methods
Primers used for cloning, quantitative polymerase chain reaction, and blotting are listed in Dataset S2. Methods for Northern analysis, cell culture and luciferase sensor assays (6), Dicer mutant (32), and Ago2 mutant (33) were previously described. Microarray experiments was performed by LC Sciences. Detailed methods are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Jidong Liu (Sloan-Kettering Institute, New York) for Ago plasmids, Gunter Meister (Universität Regensburg, Regensburg, Germany) for Ago antibodies, Michael Chen for mapping some of the small RNA data, Katsutomo Okamura for critical comments, and Alexander Tarakhovsky (The Rockefeller University, New York) for support and discussion. We are grateful to the many laboratories that enabled our studies by depositing their published small RNA datasets into public databases. Work in E.C.L.'s group was supported by the Burroughs Wellcome Fund (1004721), the Alfred Bressler Scholars Fund, the Starr Cancer Consortium (I3-A139), and the National Institutes of Health (R01-GM083300).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006432107/-/DCSupplemental.
References
- 1.Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: Shared themes amid diversity. Nat Rev Genet. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 3.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 4.Meister G, et al. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 5.Liu J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 6.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ender C, et al. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 10.Taft RJ, et al. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole C, et al. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bogerd HP, et al. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral MicroRNAs. Mol Cell. 2010;37:135–142. doi: 10.1016/j.molcel.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altuvia Y, et al. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33:2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai EC, Tomancak P, Williams RW, Rubin GM. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42.1–20. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berezikov E, et al. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 17.Nelson PT, et al. A novel monoclonal antibody against human Argonaute proteins reveals unexpected characteristics of miRNAs in human blood cells. RNA. 2007;13:1787–1792. doi: 10.1261/rna.646007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaz C, et al. Analysis of microRNA transcriptome by deep sequencing of small RNA libraries of peripheral blood. BMC Genomics. 2010;11:288. doi: 10.1186/1471-2164-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tam OH, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vagin VV, et al. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 2009;23:1749–1762. doi: 10.1101/gad.1814809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuchen S, et al. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity. 2010;32:828–839. doi: 10.1016/j.immuni.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedländer MR, et al. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 23.Glazov EA, et al. A microRNA catalog of the developing chicken embryo identified by a deep sequencing approach. Genome Res. 2008;18:957–964. doi: 10.1101/gr.074740.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirao I, Nishimura Y, Tagawa Y, Watanabe K, Miura K. Extraordinarily stable mini-hairpins: electrophoretical and thermal properties of the various sequence variants of d(GCGAAAGC) and their effect on DNA sequencing. Nucleic Acids Res. 1992;20:3891–3896. doi: 10.1093/nar/20.15.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papapetrou EP, Korkola JE, Sadelain M. A genetic strategy for single and combinatorial analysis of miRNA function in mammalian hematopoietic stem cells. Stem Cells. 2010;28:287–296. doi: 10.1002/stem.257. [DOI] [PubMed] [Google Scholar]
- 26.Dore LC, et al. A GATA-1-regulated microRNA locus essential for erythropoiesis. Proc Natl Acad Sci USA. 2008;105:3333–3338. doi: 10.1073/pnas.0712312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rasmussen KD, et al. The miR-144/451 locus is required for erythroid homeostasis. J Exp Med. 2010;207:1351–1358. doi: 10.1084/jem.20100458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruby JG, et al. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi W, Hendrix D, Levine M, Haley B. A distinct class of small RNAs arises from pre-miRNA-proximal regions in a simple chordate. Nat Struct Mol Biol. 2009;16:183–189. doi: 10.1038/nsmb.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell. 2009;138:215–219. doi: 10.1016/j.cell.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 31.Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 32.Yi R, et al. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet. 2006;38:356–362. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
- 33.O'Carroll D, et al. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev. 2007;21:1999–2004. doi: 10.1101/gad.1565607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giraldez AJ, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 35.Tan GS, et al. Expanded RNA-binding activities of mammalian Argonaute 2. Nucleic Acids Res. 2009;37:7533–7545. doi: 10.1093/nar/gkp812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 38.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010; 465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cifuentes D, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010; 328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci USA. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muljo SA, et al. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bernstein E, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 43.Heo I, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 44.Halic M, Moazed D. Dicer-independent primal RNAs trigger RNAi and heterochromatin formation. Cell. 2010;140:504–516. doi: 10.1016/j.cell.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.