Abstract
Gene targeting by homologous recombination in embryonic stem cells is extensively used to generate specific mouse mutants. However, most mammalian species lack tools for targeted gene manipulation. Since double-strand breaks strongly increase the rate of homologous recombination at genomic loci, we explored whether gene targeting can be directly performed in zygotes by the use of zinc-finger nucleases. Here we report that gene targeting is achieved in 1.7–4.5% of murine one-cell embryos upon the coinjection of targeting vectors with zinc-finger nucleases, without preselection. These findings enable the manipulation of the mammalian germ line in a single step in zygotes, independent of ES cells.
Gene targeting by homologous recombination in ES cells is widely used to mutate and modify the mouse genome. Thereby the mouse is established as the most commonly used animal model system (1). With the exception of mice, most other mammalian species lack tools for targeted gene manipulation because functional ES cell lines are not established. Gene targeting in somatic cells combined with nuclear transfer has been used as an alternative for the engineering of livestock species, but represents an elaborate task (2, 3).
Zinc-finger nucleases (ZFN) link a DNA binding domain of the zinc-finger type to the nuclease domain of Fok I and enable the induction of double-strand breaks (DSBs) at preselected genomic sites (4). DSBs closed by the error-prone, nonhomologous end-joining (NHEJ) DNA repair pathway frequently exhibit nucleotide deletions and insertions at the cleavage site. This technology has been applied to introduce knockout mutations into the germ line of rats and zebrafish by the expression of ZFNs in early embryos that target coding sequences (5–8).
It has been initially shown by using the yeast homing endonuclease I-SceI that the induction of DSBs at genomic insertions of I-SceI recognition sites increases the rate of homologous recombination (HR) in mammalian cells by several orders of magnitude (9). Artificially designed ZFNs further increased the ability to generate site-specific double-strand breaks in endogenous genes, without the requirement to introduce artificial nuclease recognition sites. Following this principle, ZFNs have been used to achieve efficient homologous recombination of gene targeting vectors with various endogenous loci in cultured and primary mammalian cells (10–13).
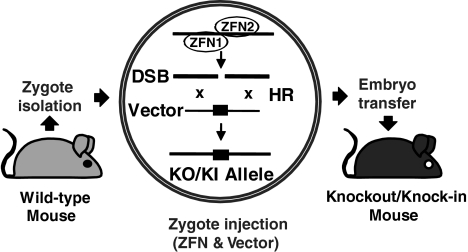
Zygotes provide a logical substrate for the single-step modification of the mammalian germ line. Whereas pronuclear DNA injection is routinely used to generate transgenic mice, rats, and other mammals by random genomic integration of transgenes (14–16), gene targeting in zygotes is not established due to the low rate of spontaneous HR. Because the mouse is the best established mammalian genetic model system, we explored whether targeted mutants can be directly obtained through ZFN-assisted homologous recombination in zygotes, upon the coinjection of ZFN mRNAs and gene targeting vectors (Fig. 1).
Fig. 1.
Principle of ZFN-assisted gene targeting in zygotes. Zygotes collected from wild-type mice are coinjected into the pronucleus and cytoplasm with DNA of a gene targeting vector and mRNAs for the expression of a pair of gene specific zinc-finger nucleases (ZFN1/2). HR of the targeting vector with the target site results in a knockout (KO) or knock-in (KI) allele. Manipulated zygotes are subsequently transferred into pseudopregnant females to recover mutant mice.
Results
We used a ZFN pair (ZFNRosa) that recognizes 18 nucleotides upstream and 12 nucleotides downstream of the XbaI site in the first intron of Rosa26 (Fig. 2A), a locus that enables widespread transgene expression in vivo (17). To confirm the stimulatory power of ZFNRosa-induced homologous recombination, we first transfected murine ES cells with a hygromycin selectable gene targeting vector (pRosa26.11; Fig. 2A) with or without ZFNRosa expression plasmids. Hygromycin-resistant ES cell clones were expanded and analyzed by Southern blotting for their Rosa26 genotype. Thirty-two ES cell clones transfected with pRosa26.11 alone exhibited only the 11.5-kb EcoRV wild-type band (Fig. 2B). The cotransfection of pRosa26.11 with ZFNRosa plasmids strongly enhanced recombination at Rosa26, such that 11 of 34 analyzed clones exhibited the predicted 4.5-kb EcoRV fragment derived from a targeted allele (Fig. 2C). One ES cell clone exhibited only the targeted band and likely represents the targeting of both Rosa26 alleles.
Fig. 2.
Zinc-finger nuclease-assisted targeting of the Rosa26 locus in ES cells. (A) Targeting vector pRosa26.11 for insertion of a hygromycin/puromycin resistance cassette, into the Rosa26 locus. The structure of the wild-type locus, including the ZFNRosa recognition sites that overlap with an intronic XbaI site (X), and of the recombined Rosa26 allele are shown. The location of the Rosa26 promoter (Pr.), first exon, of the 5′-Rosa Southern blot probe and of EcoRV (E) sites and fragments are indicated. (B) Southern blot analysis of EcoRV digested genomic DNA of 32 hygromycin resistant ES cell clones transfected with pRosa26.11 using the Rosa26 5′ probe. The wild-type Rosa26 locus exhibits a 11.5-kb EcoRV fragment (WT). (C) Southern blot analysis of EcoRV digested genomic DNA of 34 hygromycin resistant ES cell clones transfected with pRosa26.11 and ZFNRosa expression vectors using the Rosa26 5′ probe. Targeted integration of the resistance gene cassette is indicated by the presence of an additional 4.5-kb EcoRV fragment.
Targeted Integration of a β-Galactosidase Reporter Gene into the Rosa26 Locus.
To insert a β-Galactosidase gene into Rosa26 of one-cell embryos, we linked a 4.2-kb reporter cassette to homology regions flanking the target XbaI site (Fig. 3A). This targeting vector was coinjected with ZFNRosa mRNAs in a two-step procedure into the male pronucleus and the cytoplasm of mouse zygotes (see Materials and Methods). Injected zygotes were transferred into pseudopregnant females, and the resulting pups were analyzed at E18 for their Rosa26 genotype. Southern blot analysis revealed the frequent loss of the XbaI site within the ZFNRosa target sequence in one or both Rosa26 alleles, as indicated by the presence of a predicted 9-kb fragment in addition to the 4.7-kb wild-type band (Fig. 3B). Among 58 pups derived from these injections, we found 13 XbaI negative genotypes (Fig. 3B and Fig. S1A), indicating that mutagenic NHEJ events were induced by ZFNRosa in 22% of one-cell embryos. Some of these pups (Fig. 3B and Fig. S1A) had an intermediate pattern showing a majority of XbaI negative and a minor proportion of wild-type alleles, indicating that the ZFNs’ action occurred in these embryos after the first cell division. Sequence analysis of PCR-amplified target regions confirmed that the loss of the XbaI site is caused by the deletion of multiple nucleotides within the ZFN target region (Fig. 3C). The DNA of pup #22 (Fig. 3E) showing a 8.5-kb XbaI band (Fig. 3B), predicted for homologous recombination (Fig. 3A), was further characterized by Southern blotting. This analysis proved to be a bona fide homologous recombination event, as indicated by the presence of the predicted 3.6-kb EcoRV and 9.4-kb EcoRV/SwaI fragments for pup #22 (Fig. 3 B and D), detected with external 5′- and 3′-hybridization probes, respectively. The complete integration of the reporter gene in pup #22 was confirmed with an internal β-Galactosidase-specific probe that detected the predicted 8.5-kb XbaI band (Fig. 3D). The analysis of liver sections for β-Galactosidase activity proved the functionality and heterozygous state of the targeted allele in pup #22 (Fig. 3E). In addition, we verified the integrity of the recombined locus by sequence analysis of the Rosa26/reporter junction regions amplified by PCR from pup #22 (Fig. S2 A and B). Among the 58 analyzed pups, we found two, #14 and #21, that exhibit unexpected XbaI fragments of 4.7–8.5 kb (Fig. 3B) and were shown by Southern blot (Fig. 3D, #14) and PCR analysis (Fig. S2C) to contain 3′ sequences, but not the coding region of the reporter cassette (Fig. 3D), and represent incomplete or rearranged recombination products.
Fig. 3.
Targeted Integration of a β-Galactosidase reporter gene into the Rosa26 locus. (A) Targeting vector pRosa26.8 for insertion of a 4.2-kb β-Galactosidase gene, including a splice acceptor (SA) and polyA site, into the Rosa26 locus. The structure of the wild-type locus, including the ZFNRosa recognition sites that overlap with an intronic XbaI site (X), and of the recombined Rosa26 allele are shown. The location of the Rosa26 promoter (Pr.), first exon, of Southern blot probes and EcoRV (E), XbaI (X), and SwaI (S) sites and fragments are indicated. (B) Genomic DNA of E18 fetuses derived from zygote coinjections of ZFNRosa and pRosa26.8 was digested with XbaI or EcoRV and analyzed by Southern blotting using the Rosa26 5′ probe. The wild-type Rosa26 locus exhibits a 4.7-kb XbaI and a 11.5-kb EcoRV fragment (WT). The loss of the ZFNRosa target XbaI site by NHEJ results in a 9.0-kb fragment (fetus #14, #15, and #21). Targeted integration of the reporter gene is detected with the Rosa26 5′ probe by the presence of a 8.5-kb XbaI and 3.6-kb EcoRV fragment (fetus 22). (C) Sequence comparison of cloned PCR products (primers Rosa 5HA/Rosa 3HA), covering the ZFNRosa target region, from genomic DNA of fetus #9, #14, and #21, with the respective Rosa26 wild-type sequence. The location of the XbaI site within the ZFNRosa target region is indicated. In PCR amplified Rosa26 alleles from pups #9, #14, and #21, the XbaI site is lost by the deletion of multiple nucleotides (dashes). Sequence identity of all compared sequences is marked in yellow and partial sequence marked in blue; deleted nucleotides, as compared to the Rosa26 wild-type sequence, are indicated by a dash (-). Fetus #9 exhibited by Southern blot analysis an almost homozygous loss of the ZFN target XbaI site (Fig. S1A). (D) Southern blot analysis of EcoRV + SwaI digested genomic DNA with the Rosa26 3′ probe reveals a 11.5-kb band derived from the wild-type locus and a 9.4-kb fragment from the recombined allele (fetus #22 and #14). Exposure times for samples 21–55 were 2 days (2 d) and 3 days (3 d) for sample 14. Southern blot analysis of XbaI digested DNA of fetus #22 with an internal β-Galactosidase probe shows the predicted 8.5-kb band representing the recombined allele. (E) Habitus of fetus 22 (recombined) and 24 (wild type) and X-Gal staining of liver sections.
Targeted Integration of a Venus Reporter Gene into the Rosa26 Locus.
In a second experiment we used a targeting vector that inserts a 1.1-kb Venus reporter gene into the Rosa26 locus (Fig. 4A). DNA analysis of 22 pups derived from zygote coinjections of this vector with ZFNRosa mRNAs revealed the presence of one recombined allele, as shown by a 3.1-kb BamHI and a 5.6-kb XbaI fragment (Fig. 4B, fetus 6, Fig. 4D) detected with the 5′-Rosa probe and by a 3.9-kb BamHI band detected with a Venus probe (Fig 4C). The Venus reporter cassette was amplified from the genomic DNA of pup #6 and was found by sequence analysis to be entirely intact (Fig. S3). The second Rosa26 allele of pup #6 had lost the XbaI site within the ZFNRosa target sequence, as indicated by the presence of a 9-kb XbaI fragment and the absence of the wild-type 4.7-kb band (Fig. 4B), detected with the Rosa26 5′ probe.
Fig. 4.
Targeted integration of a Venus reporter gene into the Rosa26 locus. (A) Targeting vector pRosa26.3 for insertion of a 1.1-kb Venus gene, including a splice acceptor (SA) and polyA site, into the Rosa26 locus. The structure of the wild-type locus, including the ZFNRosa recognition sites that overlap with an intronic XbaI site (X), and of the recombined Rosa26 allele are shown. The location of the Rosa26 promoter (Pr.), first exon, of Southern blot probes and XbaI (X) and BamHI (B) sites and fragments are indicated. (B) Genomic DNA of E18 fetuses derived from zygotes coinjections of ZFNRosa and pRosa26.3 was digested with BamHI or XbaI and analyzed by Southern blotting using the Rosa26 5′ probe. The wild-type Rosa26 locus exhibits a 5.8-kb BamHI and a 4.7-kb XbaI band. Targeted integration of the reporter gene is indicated by the presence of a predicted 3.1-kb BamHI and a 5.6-kb XbaI fragment detected with the Rosa26 5′ probe (fetus #6). The loss of the ZFNRosa target XbaI site by NHEJ in nonrecombined alleles results in a 9.0-kb band (fetus #6). (C) Analysis of BamHI digested DNA with the internal Venus probe shows a predicted 3.9-kb band from the recombined allele of fetus 6. (D) Habitus of fetus #6 (recombined) and #7 (wild type).
Analysis of Random Vector Integrations.
Pronuclear DNA injection is routinely used for the production of transgenic mice harboring random vector integrations (14). Unexpectedly, using Southern blotting and hybridization probes for the β-Galactosidase and Venus reporter gene coding regions, we did not observe any random integration of targeting vectors among the 80 pups derived from both ZFN coinjection experiments (Figs. 3D and 4C and Fig. S1 B and C).
Discussion
Compared to a previous report on homologous recombination in mouse zygotes that described a spontaneous recombination rate below 0.1% (18), our results demonstrate that gene targeting can be achieved in pronuclei of one-cell embryos at a higher frequency (1.7–4.5%) when assisted by ZFNs. By analysis of the XbaI site located within the ZFNRosa recognition sequence, we determined that DSBs, which lead to NHEJ-induced loss of nucleotides, occurred in injected zygotes at an even higher rate of 22%. It has been demonstrated in Drosophila embryos that the suppression of NHEJ DNA repair leads to the preferential resolution of ZFN-induced DSBs by HR with a gene targeting vector, resulting in the increased recovery of targeted alleles (19). It is therefore possible that also in mammalian zygotes ZFN-assisted gene targeting can be further improved by the transient inhibition of the NHEJ pathway.
In the only reported case of HR in zygotes, the targeted allele exhibited numerous point mutations that interfered with gene expression, possibly as a result of an error-prone HR process in the pronuclei (18). Here we did not observe mutations in the analyzed regions of both targeted alleles, indicating that ZFN-assisted HR can occur with high fidelity as known for gene targeting in ES cells. As judged by the comparison of Southern blot signals derived from wild-type and mutant Rosa26 loci, both correctly targeted alleles appeared in the heterozygous state indicating that ZFN expression, DSB induction, and HR can occur in injected pronuclei within a few hours, before the completion of DNA replication and the fusion of both pronuclei. Among the 80 pups analyzed, we also observed three genotypes (#14, #15, and #21, Fig. 3B) that exhibit a contribution of mutant Rosa26 alleles below 50%, indicating that a part of the HR and NHEJ repair events occur late, only in a single cell of a 2-cell or 4-cell embryo. Homozygously modified genotypes, which exhibit the loss of the ZFN target XbaI site at both Rosa26 alleles or that exhibit one XbaI negative and one targeted allele (Fig. 4B, #6), can be explained by the import of cytoplasmic ZFN proteins into both pronuclei.
To allow an unbiased evaluation of targeting frequencies in male pronuclei, we focused here on the genetic analysis of fetuses (Figs. 3E and 4D). Because ZFN expression and NHEJ deletions were shown not to interfere with the viability and fertility of rats and zebrafish (5–8), we suggest that animals produced from ZFN-assisted HR will be similarly competent for germ-line transmission as targeted mutations generated by homologous recombination in ES cells (1).
Because ZFN-stimulated gene targeting has been demonstrated for more than 10 loci in mammalian cell lines (4), we expect that also the ZFN/zygote approach is widely applicable to other genes and species. Using ZFNs to directly manipulate the mouse genome reduces the time to obtain targeted mutants and avoids the use of selection markers, as compared to ES cell technology. Moreover, ZFN-assisted gene targeting in zygotes provides an ES cell-independent paradigm to manipulate the genome of mammals and other vertebrates with unprecedented freedom.
Materials and Methods
Injection of Zygotes.
The Fok-I based, heterodimeric ZFNRosa pair targets sequences near the XbaI site (underlined) within the first intron of the mouse Rosa26 gene; the spacer region is shown in bold: 5′-TGC-AAC-TCC-AGT-CTT-TCT-AGAAGA-TGG-GCG-GGA-GTC-3′. ZFN expression plasmids were obtained from Sigma. To prepare ZFNRosa mRNA, plasmid DNAs were linearized downstream of the ZFN coding region and precipitated. Messenger RNA was transcribed in vitro using the mMESSAGE mMACHINE® T7 Kit (Ambion) and polyadenylated using the Poly(A) Tailing Kit (Ambion). The resulting mRNA was purified using the MEGAclear™ Kit (Ambion) before resuspension in injection buffer (10 mM Tris, 0.1 mM EDTA, pH 7.2). The mRNA concentrations were determined by absorption and gel analysis, and each ZFN mRNA was diluted in injection buffer to a working concentration of 2.5 ng/μL together with targeting vector and stored at -80 °C.
Targeting vector pRosa26.8 (a derivative of pRosa26.5) (20) consists of the 1-kb 5′ Rosa26 homology arm and the 4-kb 3′ homology arm (17), flanking an adenoviral splice acceptor sequence, followed by the 3.2-kb β-Galactosidase coding region and the 220-bp SV40 polyadenylation site from plasmid pCMVβ (Fig. 2A). Targeting vector pROSA26.3 is equal to pRosa26.8, except that it contains a 1.1-kb reporter cassette for expression of the Venus GFP protein. The homology arms of both targeting vectors were sequenced and found identical to the respective genomic sequence of the strain C57BL/6J. pRosa26.8 was linearized at the 5′ end of the short homology arm with I-SceI, whereas pRosa26.3 was used circular. The DNAs were purified with the Qiaquick PCR Purification kit (Quiagen), precipitated, and dissolved in injection buffer at a working concentration of 5 ng/μL (pRosa26.8-2) or 15 ng/μL (pRosa26.3-3) together with ZFNRosa mRNA (2.5 ng/μL each) and stored at -80 °C.
Mouse zygotes were obtained by superovulation of Friend virus B (FVB) females and matings to C57BL/6N males (Charles River). The next day zygotes were collected from oviducts and microinjected in M2 embryo medium following standard procedures (21) with a mixture of targeting vector and ZFNRosa mRNAs (2.5 ng/μL each) loaded into a single microinjection needle. For microinjection a two-step procedure was applied: A first aliquot of the DNA/RNA mixture was injected into the male pronucleus (to deliver the DNA vector, as used for the production of transgenic mice). Upon the withdrawal of the injection needle from the pronucleus, a second aliquot of the DNA/RNA mixture was injected into the cytoplasm to deliver the ZFN mRNA directly to the translation machinery. Injections were performed using a Leica micromanipulator and microscope and an Eppendorf FemtoJet injection device. Injected zygotes were transferred into pseudopregnant CD1 female mice and fetuses recovered at day E18 for further analysis. From the transfer of 469 zygotes injected with targeting vector pRosa26.8 we obtained 64 fetuses (14% recovery), from which 58 were further analyzed by Southern blotting. For pRosa26.3 we obtained 22 fetuses from 120 transferred zygotes (18% recovery). Recovered fetuses showed normal development and were viable (Figs. 3E and 4D). Mice were handled according to institutional guidelines and housed in standard cages in a specific pathogen-free facility on a 12 h light/dark cycle with ad libitum access to food and water.
X-Gal Staining.
One longitudinal half of E18 fetuses were fixed and stained with X-Gal as described in ref. 21.
Preparation of Genomic DNA.
A segment of the other embryo half was used for the isolation of genomic DNA. For this purpose frozen tissue segments were homogenized in lysis buffer (50 mM Tris-HCl, pH 8.0; 100 mM EDTA, pH 8,0; 1% SDS; 100 mM NaCl; 350 μg/μL Proteinase K) and incubated at 55 °C for 2 h under vigorous shaking. Upon tissue lysis phenol/chloroform (1∶1) was added, and the mixture was vortexed and centrifugated for 10 min at 16.000 × g. The supernatant was transferred into new tubes and extracted with chloroform. The supernatant was mixed with 0.7 volumes isopropanol to precipitate the genomic DNA. Precipitated DNA was collected by centrifugation, and the pellet was washed with 70% ethanol. The pellet was air-dried, resuspended in 10 mM Tris/1 mM EDTA buffer (pH 7.5), and incubated overnight at room temperature.
Southern Blot Analysis.
Genomic DNA (6 μg) was digested overnight with 30 units restriction enzyme in a volume of 30 μL and then redigested with 10 units enzyme for 2–3 h. Samples were loaded on 0.8% agarose gels in Tris (89 mM)/borate (89 mM)/EDTA (2 mM), pH 8.0 buffer and run at 55 V overnight. The gels were then denaturated for 1 h in 1.5 M NaCl; 0.5 M NaOH, neutralized for 1 h in 0.1 M Tris-HCl pH 7.5; 0.5 M NaCl, washed with 2× SSC, and blotted for 48 h with 20× SSC on Hybond N+ membranes (GE Healthcare). The membranes were then washed with 2× SSC, UV-cross-linked, and stored at -20 °C. For hybridization the membranes were preincubated in Church buffer (1% BSA, 1 mM EDTA, 0.5 M phosphate buffer, 7% SDS) for 1 h at 65 °C under rotation. The Rosa26 5′ probe was isolated as a 460-bp EcoRI fragment from plasmid pCRII-Rosa5′ probe; the Rosa26 3′ probe was isolated as 660-bp EcoRI fragment from the plasmid pCRII-Rosa3′ probe, as described (17, 20). As β-Galactosidase probe we used the 1,250-bp SacII/EcoRV coding fragment from pCMVβ and as Venus probe the venus coding region, isolated as a 730-bp BamHI/EcoRI fragment from pCS2-venus, was used. DNA fragments used as hybridization probes were heat denatured and labeled with P32 marked dCTP (Perkin Elmer) using the high-prime DNA labeling kit (Roche). Labeled probe DNA was purified on MicroSpin™ S-200 HR columns (GE Healthcare), heat denatured, added to the hybridization buffer and membranes were rotated overnight at 65 °C. The washing buffer (2× SSC, 0,5% SDS) was prewarmed to 65 °C, and the membranes were washed three times (5 min, 30 min, and 15 min) at 65 °C under shaking. Next, the membranes were exposed at -80 °C to Biomax MS1 films and enhancing screens (Kodak) for 1–5 days until development. Photos of autoradiographs were taken with a digital camera on a transmitting light table, and segments were excised with the Adobe Photoshop software.
PCR and Sequence Analysis.
To analyze the Rosa26 alleles recombined with pRosa26.8 we amplified DNA from fetuses 22, 21, and 14 (coinjected with ZFNRosa and pRosa26.8) using the primer pairs Rosa 5HA (5′-aaagtcgctctgagttgttat-3′) and SA1 (5′-cgatccccagtactggaaag-3′) or SA2 (5′-ttgctttagcaggctctttc-3′) to amplify the 5′-homology arm/splice acceptor junction and the primer pairs Rosa 3HA (5′-cacaccaggttagcctttaagcc-3′) and pA1 (5′-tcggatcctctagagtcgagg-3′) or pA2 (5′-aaaccacaactagaatgcagtg-3′) to amplify the junction of the SV40 polyA/3′-homology arm. Amplification was performed using Herculase polymerase (Invitrogen) in 100-μL reactions with 38 cycles of 95 °C -30 s; 59 °C -1 min; 72 °C -1 min. In embryos with recombination events PCR products have the expected sizes: Rosa 5HA/SA1: 346 bp; Rosa 5HA/SA2: 365 bp; Rosa 3HA/pA1: 467 bp; Rosa 3HA/pA2: 625 bp. PCR products from embryos exhibiting homologous recombination events were directly sequenced (Sequiserve) and compared to the pRosa26.8 vector sequence using the Vector NTI software (Invitrogen).
Genomic DNA from fetuses 9, 14, and 21, which lost the XbaI site in the ZFN target region as found by Southern blotting, and of wild-type controls were further analyzed for NHEJ-induced mutations by PCR amplification using the primer pair Rosa 5HA and Rosa 3HA (wild-type PCR product: 252 bp). These PCR products were cloned using the StrataClone Blunt PCR Cloning Kit (Stratagene). Plasmid DNA was isolated from with the Qiaprep Spin Miniprep Kit (Qiagen) and analyzed by digestion with EcoRI to confirm the presence of vector inserts and digested with XbaI to analyze for the loss of the ZFNRosa target XbaI site. Selected clones were sequenced using the T3 forward primer (Sequiserve) and analyzed for mutations in comparison to the Rosa26 wild-type sequence using the Vector NTI software.
To analyze Rosa26 homologous recombined alleles with pRosa26.3, we amplified DNA from fetus 6 (coinjected with ZFNRosa and pRosa26.3) using the primer pair Rosa 5HA (5′-aaagtcgctctgagttgttat-3′) and Rosa 3HA (5′-cacaccaggttagcctttaagcc-3′). The PCR product (5HA/3HA: 1,402 bp) was directly sequenced with a single 950-bp read (Sequiserve) and compared to the pRosa26.3 vector sequence using the Vector NTI software.
Culture and Transfection of ES Cells.
JM8.A3 (22) ES cells were cultured and transfected as described for IDG3.2 ES cells (20) with 20-μg linearized targeting vector pRosa26.11 [derived from pRosa26.10 (20)] and 5 μg of a Ku70 subfragment expression vector, together with or without 15 μg of circular ZFNRosa expression plasmids. Resistant colonies were selected with 125 U Hygromycin/mL culture medium, picked, and further expanded for DNA analysis. Recombinant ES clones were identified by Southern blotting, using EcoRV digestion and the Rosa26 5′ probe, by the presence of a predicted 4.5-kb fragment in addition to the 11.5-kb wild-type band.
Supplementary Material
Acknowledgments.
We thank C. Arndt, R. Kneuttinger, A. Krause, A. Tasdemir, and S. Weidemann for excellent technical assistance, Sigma for ZFN plasmids, and J. Schick and R. Köster for reading the manuscript. This work was supported by the European Union within the European Conditional Mouse Mutagenesis Program (LSHG-CT-2005-018931, to W.W.) and by the German Ministry of Education and Research within the Disease genes to Protein pathways German genome research network (NGFN Plus) project (01GS0858, to W.W. and R.K.) of the NGFN-Plus program.
Note Added in Proof.
While this article was in print we obtained also live adult heterozygous targeted mice from coinjections of ZFNRosa and pRosa26.3.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009424107/-/DCSupplemental.
References
- 1.Capecchi MR. Gene targeting in mice: Functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet. 2005;6:507–512. doi: 10.1038/nrg1619. [DOI] [PubMed] [Google Scholar]
- 2.Lai L, Prather RS. Creating genetically modified pigs by using nuclear transfer. Reprod Biol Endocrin. 2003;1:82. doi: 10.1186/1477-7827-1-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCreath KJ, et al. Production of gene-targeted sheep by nuclear transfer from cultured somatic cells. Nature. 2000;405:1066–1069. doi: 10.1038/35016604. [DOI] [PubMed] [Google Scholar]
- 4.Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nat Biotechnol. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- 5.Doyon Y, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geurts AM, et al. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325:433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mashimo T, et al. Generation of knockout rats with X-linked severe combined immunodeficiency (X-SCID) using zinc-finger nucleases. PLoS One. 2010;5:e8870. doi: 10.1371/journal.pone.0008870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rouet P, Smih F, Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc Natl Acad Sci USA. 1994;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hockemeyer D, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 12.Santiago Y, et al. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci USA. 2008;105:5809–5814. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Urnov FD, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 14.Palmiter RD, Brinster RL. Transgenic mice. Cell. 1985;41:343–345. doi: 10.1016/s0092-8674(85)80004-0. [DOI] [PubMed] [Google Scholar]
- 15.Pursel VG, Hammer RE, Bolt DJ, Palmiter RD, Brinster RL. Integration, expression and germ-line transmission of growth-related genes in pigs. J Rep Fer S. 1990;41:77–87. [PubMed] [Google Scholar]
- 16.Rexroad CE, Jr, Hammer RE, Behringer RR, Palmiter RD, Brinster RL. Insertion, expression and physiology of growth-regulating genes in ruminants. J Rep Fer S. 1990;41:119–124. [PubMed] [Google Scholar]
- 17.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 18.Brinster RL, et al. Targeted correction of a major histocompatibility class II E alpha gene by DNA microinjected into mouse eggs. Proc Natl Acad Sci USA. 1989;86:7087–7091. doi: 10.1073/pnas.86.18.7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bozas A, Beumer KJ, Trautman JK, Carroll D. Genetic analysis of zinc-finger nuclease-induced gene targeting in Drosophila. Genetics. 2009;182:641–651. doi: 10.1534/genetics.109.101329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hitz C, Wurst W, Kuhn R. Conditional brain-specific knockdown of MAPK using Cre/loxP regulated RNA interference. Nucleic Acids Res. 2007;35:e90. doi: 10.1093/nar/gkm475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating the Mouse Embryo. 3rd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2003. [Google Scholar]
- 22.Pettitt SJ, et al. Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nat Methods. 2009;6:493–495. doi: 10.1038/nmeth.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.