Abstract
The Trf4p/Air2p/Mtr4p polyadenylation (TRAMP) complex recognizes aberrant RNAs in Saccharomyces cerevisiae and targets them for degradation. A TRAMP subcomplex consisting of a noncanonical poly(A) RNA polymerase in the Pol ß superfamily of nucleotidyl transferases, Trf4p, and a zinc knuckle protein, Air2p, mediates initial substrate recognition. Trf4p and related eukaryotic poly(A) and poly(U) polymerases differ from other characterized enzymes in the Pol ß superfamily both in sequence and in the lack of recognizable nucleic acid binding motifs. Here we report, at 2.7-Å resolution, the structure of Trf4p in complex with a fragment of Air2p comprising two zinc knuckle motifs. Trf4p consists of a catalytic and central domain similar in fold to those of other noncanonical Pol β RNA polymerases, and the two zinc knuckle motifs of Air2p interact with the Trf4p central domain. The interaction surface on Trf4p is highly conserved across eukaryotes, providing evidence that the Trf4p/Air2p complex is conserved in higher eukaryotes as well as in yeast and that the TRAMP complex may also function in RNA surveillance in higher eukaryotes. We show that Air2p, and in particular sequences encompassing a zinc knuckle motif near its N terminus, modulate Trf4p activity, and we present data supporting a role for this zinc knuckle in RNA binding. Finally, we show that the RNA 3′ end plays a role in substrate recognition.
Keywords: protein-RNA interactions, RNA quality control
The vast majority of transcripts from eukaryotic genomes do not encode proteins. These transcripts include precursors to noncoding RNAs, such as transfer and ribosomal RNAs, small nuclear and nucleolar RNAs, microRNAs, siRNAs, and piRNAs. These noncoding RNAs often fold into intricate three-dimensional structures that are critical for subsequent processing and for assembling with proteins to form ribonucleoprotein complexes. In addition, genomic sequencing experiments in both yeast and mammals have revealed the existence of a large number of short-lived noncoding transcripts that initiate near RNA polymerase II promoters. Finally, cells contain a variety of long noncoding RNAs, some of which function to regulate gene expression and chromatin structure (1, 2).
Because defective RNAs can arise by synthesis from mutant genes, transcriptional errors, or aberrant processing events, and because some products of pervasive genome transcription may be harmful, cells have evolved surveillance mechanisms to recognize aberrant and unneeded noncoding RNAs and target them for degradation by exoribonucleases. Because all exoribonucleases require a single-stranded end to initiate decay, the initial recognition of target RNAs is often carried out by polymerases that, by adding extra nucleotides to the 3' ends, recruit the decay machinery. Because many exonucleases are unable to degrade structured RNAs, this machinery frequently includes RNA helicases (3).
One of the best established noncoding RNA degradation pathways in Saccharomyces cerevisiae involves the Trf4p/Air2p/Mtr4p polyadenylation (TRAMP) complex and the nuclear exosome, a major 3′ → 5′ exonuclease (3, 4). In vivo, TRAMP and the exosome are involved in the degradation of a large variety of RNA substrates, including hypomodified and unspliced pre-tRNAs, truncated 5S rRNA and signal recognition particle RNAs, aberrant rRNA processing intermediates, and a large class of bidirectional transcripts that initiate near RNA polymerase II promoters. The exosome is conserved in higher eukaryotes, and higher eukaryotes have homologs for each component of TRAMP (3, 4), suggesting that the TRAMP-exosome pathway may be widely conserved. TRAMP consists of the poly(A) polymerase Trf4p or its close homolog Trf5p (65% sequence identity), the zinc knuckle protein Air2p or its close homolog Air1p (45% sequence identity), and Mtr4p, a member of the DExH/D box RNA helicase superfamily 2. The Trf4p/Air2p subcomplex polyadenylates the 3' ends of aberrant noncoding RNAs, providing a single-stranded “landing pad” that allows the exosome to initiate RNA decay (3, 4).
A key question in noncoding RNA quality control is how RNAs are recognized as aberrant. In the TRAMP-exosome pathway, substrate recognition is mediated by the Trf4p/Air2p subcomplex (5–8), and in vitro, this subcomplex preferentially polyadenylates an unmodified form of 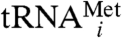 over the fully modified version (6, 9). An understanding of TRAMP substrate recognition will derive from a better understanding of Trf4p/Air2p and its interactions with RNAs.
over the fully modified version (6, 9). An understanding of TRAMP substrate recognition will derive from a better understanding of Trf4p/Air2p and its interactions with RNAs.
Trf4p/5p belongs to a family of ribonucleotide transferases in the Pol ß superfamily of nucleotidyl transferases (10, 11). It consists of N- and C-terminal sequences that have little predicted secondary structure, a catalytic domain similar in sequence to several Pol ß members with known structures, and an adjacent “central domain,” which shares a short nucleotide recognition motif hx(I/L/V)(E/Q)(E/D/N)PhxxxxNxx (h, hydrophobic; x, any residue) with so-called noncanonical Pol β RNA polymerases. Trf4p/5p differs from many structurally characterized ribonucleotidyl transferases in lacking a recognizable RNA binding domain, and it has been proposed that the Air proteins function in RNA binding (3, 4). In both Air1p and Air2p, five adjacent CCHC zinc knuckles are inserted between N- and C-terminal sequences predicted to lack secondary structure. Cx1-2Cx3-6Hx7-10C-type zinc knuckles (C, cysteine; H, histidine; x, any amino acid) and their interactions with RNAs have been studied in the context of the retroviral nucleocapsid proteins (12). In these proteins, residues x1-2 and x8-10 are typically involved in interactions with single-stranded, looped regions of RNA, and linker regions between knuckles may bind RNA duplex regions. It has not been established, however, whether the Air proteins or their zinc knuckles are involved in RNA binding, or whether these interactions resemble those of the nucleocapsid zinc knuckles.
Here we investigate the architecture and function of the Trf4p/Air2p heterodimer. We show that Air2p and its most N-terminal zinc knuckle are important for the polyadenylation activity of Trf4p, and show data suggesting that this zinc knuckle interacts with tRNA substrates. We have also determined, at a resolution of 2.7 Å, the structure of a Trf4p/Air2p subcomplex consisting of the Trf4p catalytic and central domains and a segment of Air2p that includes the fourth and fifth zinc knuckles. The fold of Trf4p is similar to that of other noncanonical Pol β family enzymes despite insignificant sequence similarity in the central domain. The structure shows that the fourth and fifth zinc knuckles of Air2p interact with the central domain of Trf4p. The Trf4p surface that they bind is highly conserved in a wide range of eukaryotes, important experimental data supporting conservation of the Trf4p/Air2p complex in higher eukaryotes.
Results and Discussion
Characterization of Trf4p/Air2p Core Complexes.
Due to significant sample degradation during purification (Fig. S1), we were not able to isolate a complex consisting of full-length forms of Trf4p and Air2p. We therefore worked with Trf4p/Air2p subcomplexes, where the proteolytically sensitive N and C termini of both proteins were removed. One subcomplex, Trf4p/Air2pZK1-5, consisted of the catalytic and central domains of Trf4p (residues 161–481) and the five zinc knuckles of Air2p (residues 58–198); a second version, Trf4p/Air2pZK4-5, included only the fourth and fifth zinc knuckles of Air2p (residues 119–198). We coexpressed Trf4p and Air2p constructs in Escherichia coli, and purified subcomplexes by affinity chromatography and gel filtration.
The truncated complexes were tested for their polyadenylation activity and also for the ability to distinguish between aberrant and correct forms of tRNAs. In these experiments, we used wild-type 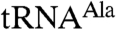 and a mutant that was identified as a preferred substrate for the intact TRAMP complex purified from yeast (6). Both truncated forms of Trf4p/Air2p are active in polyadenylation assays (Fig. 1), though less active than the full-length complexes used in other studies (6, 7, 9). The nature of the enhancement by the N and C termini is not known and will be the subject of future experiments.
and a mutant that was identified as a preferred substrate for the intact TRAMP complex purified from yeast (6). Both truncated forms of Trf4p/Air2p are active in polyadenylation assays (Fig. 1), though less active than the full-length complexes used in other studies (6, 7, 9). The nature of the enhancement by the N and C termini is not known and will be the subject of future experiments.
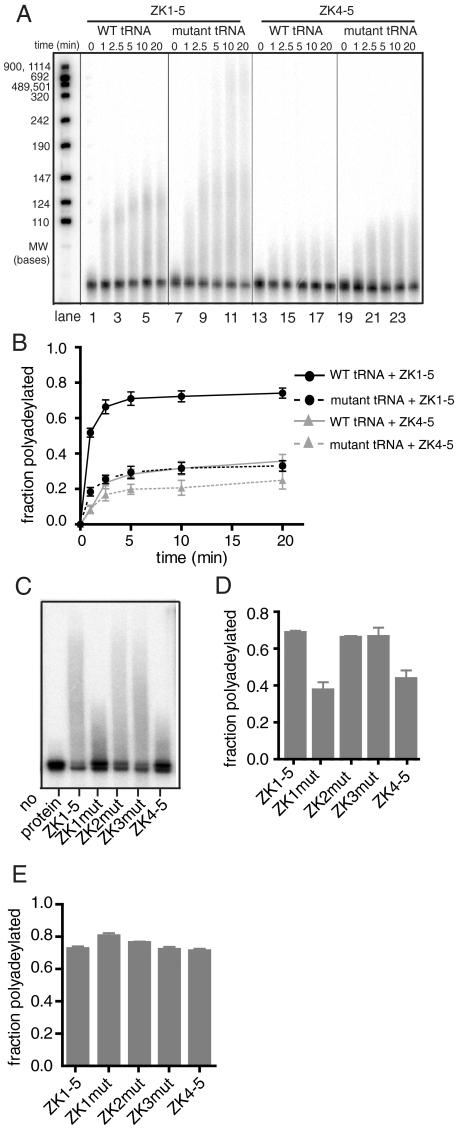
Fig. 1.
The Air2p zinc knuckles modulate the activity of Trf4p. (A) Representative gels show a time course for polyadenylation, where 2 pmol Trf4p/Air2p complex is reacted with 40 fmol of wild-type (lanes 1–6, 13–18) or mutant (lanes 7–12, 19–24) 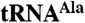 . Time courses are for Trf4p/Air2pZK1-5 (ZK1-5, lanes 1–12) and Trf4p/Air2pZK4-5 (ZK4-5, lanes 13–24). (B) Plot showing the fraction of
. Time courses are for Trf4p/Air2pZK1-5 (ZK1-5, lanes 1–12) and Trf4p/Air2pZK4-5 (ZK4-5, lanes 13–24). (B) Plot showing the fraction of 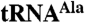 adenylated at given time points. The data are from three separate experiments. SEM is indicated. (C) Mutant
adenylated at given time points. The data are from three separate experiments. SEM is indicated. (C) Mutant 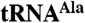 was reacted with Trf4p/Air2pZK1-5, Trf4p/Air2pZK4-5, or Trf4p/Air2pZK1-5 but with the three most N-terminal knuckles individually replaced by hexaserine linkers (ZK1mut, ZK2mut, ZK3mut). (D) Quantitation of three experiments as in C. SEM indicated. (E) A5 oligonucleotide is polyadenylated comparably by all Trf4p/Air2p complexes used in C and D.
was reacted with Trf4p/Air2pZK1-5, Trf4p/Air2pZK4-5, or Trf4p/Air2pZK1-5 but with the three most N-terminal knuckles individually replaced by hexaserine linkers (ZK1mut, ZK2mut, ZK3mut). (D) Quantitation of three experiments as in C. SEM indicated. (E) A5 oligonucleotide is polyadenylated comparably by all Trf4p/Air2p complexes used in C and D.
In examining substrate preference, we found that both truncated complexes polyadenylate the aberrant form of 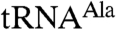 more extensively than the wild-type tRNA, as judged by both an increased poly(A) tail length and the fraction of the input substrate that underwent polyadenylation (Fig. 1 A and B). The differences are smaller for Trf4p/Air2pZK4-5, but they are reproducible and statistically significant. The Trf4p/Air2pZK1-5 complex was additionally tested with wild-type and aberrant versions of tRNAPhe, where the structure was severely disrupted by multiple mutations in the D-, T- or anticodon stems of the tRNA (Fig. S2). Structure prediction with MFOLD (13) suggests that, except in the acceptor stem, even the secondary structures of these mutant RNAs are different from the native. We found that again the mutants were more extensively polyadenylated than the wild-type RNA (Fig. S2). The finding that Trf4p/Air2pZK1-5 and Trf4p/Air2pZK4-5 differentiate between wild-type and mutant tRNAs suggests that the truncated complexes retain at least some of the sequences necessary for RNA recognition.
more extensively than the wild-type tRNA, as judged by both an increased poly(A) tail length and the fraction of the input substrate that underwent polyadenylation (Fig. 1 A and B). The differences are smaller for Trf4p/Air2pZK4-5, but they are reproducible and statistically significant. The Trf4p/Air2pZK1-5 complex was additionally tested with wild-type and aberrant versions of tRNAPhe, where the structure was severely disrupted by multiple mutations in the D-, T- or anticodon stems of the tRNA (Fig. S2). Structure prediction with MFOLD (13) suggests that, except in the acceptor stem, even the secondary structures of these mutant RNAs are different from the native. We found that again the mutants were more extensively polyadenylated than the wild-type RNA (Fig. S2). The finding that Trf4p/Air2pZK1-5 and Trf4p/Air2pZK4-5 differentiate between wild-type and mutant tRNAs suggests that the truncated complexes retain at least some of the sequences necessary for RNA recognition.
N-Terminal Zinc Knuckle of Air2p Modulates Trf4p Activity on Some Substrates.
Notably, Trf4p/Air2pZK1-5 polyadenylates both aberrant and wild-type 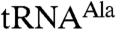 more extensively than Trf4p/Air2pZK4-5 (Fig. 1 A–D). Trf4p/Air2pZK1-5 not only polyadenylates a larger fraction of the input tRNAs than Trf4p/Air2pZK4-5, but also adds longer poly(A) tails (Fig. 1 A and C). These results demonstrate that Air2p, and specifically sequences that include its three most N-terminal zinc knuckles, enhances Trf4p activity on larger RNA substrates. They are consistent with the notion that these zinc knuckles are involved in RNA binding but do not exclude an alternative or additional role in catalysis. To further assess the possibility that the zinc knuckles are involved in RNA binding, we used identical assay conditions but with a short RNA oligonucleotide (A5) as the substrate. The finding that the oligonucleotide is polyadenylated comparably by both complexes (Fig. 1E)—and thus that the N-terminal zinc knuckle sequences in Air2p affect activity only for some substrates—argues that the N-terminal zinc knuckles of Air2p play a role in RNA binding.
more extensively than Trf4p/Air2pZK4-5 (Fig. 1 A–D). Trf4p/Air2pZK1-5 not only polyadenylates a larger fraction of the input tRNAs than Trf4p/Air2pZK4-5, but also adds longer poly(A) tails (Fig. 1 A and C). These results demonstrate that Air2p, and specifically sequences that include its three most N-terminal zinc knuckles, enhances Trf4p activity on larger RNA substrates. They are consistent with the notion that these zinc knuckles are involved in RNA binding but do not exclude an alternative or additional role in catalysis. To further assess the possibility that the zinc knuckles are involved in RNA binding, we used identical assay conditions but with a short RNA oligonucleotide (A5) as the substrate. The finding that the oligonucleotide is polyadenylated comparably by both complexes (Fig. 1E)—and thus that the N-terminal zinc knuckle sequences in Air2p affect activity only for some substrates—argues that the N-terminal zinc knuckles of Air2p play a role in RNA binding.
To delineate the relative roles of the three N-terminal-most zinc knuckles in enhancing Trf4p activity, we deleted each of the first three zinc knuckles in Trf4p/Air2pZK1-5 individually and replaced each with a hexaserine linker. The linker acts as a spacer, allowing us to delete individual zinc knuckles while not shortening the distance between the remaining ones. The mutated complexes behaved similarly to Trf4p/Air2pZK1-5 during purification. All three mutant complexes polyadenylated the short A5 oligonucleotide to similar extents as Trf4p/Air2pZK1-5 (Fig. 1E), and the complexes in which the second or third zinc knuckle was altered also polyadenylated mutant 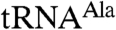 comparably (Fig. 1 C and D). The second and third zinc knuckles thus appear to be dispensable for polyadenylation of aberrant
comparably (Fig. 1 C and D). The second and third zinc knuckles thus appear to be dispensable for polyadenylation of aberrant 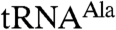 . In contrast,
. In contrast, 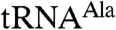 polyadenylation was reduced when the first zinc knuckle of Air2p was mutated (Fig. 1 C and D). That the first zinc knuckle enhances the polyadenylation of aberrant
polyadenylation was reduced when the first zinc knuckle of Air2p was mutated (Fig. 1 C and D). That the first zinc knuckle enhances the polyadenylation of aberrant 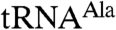 but not the A5 oligonucleotide supports a role for this zinc knuckle in binding large RNA substrates. The second and third zinc knuckles could be involved in binding other RNA substrates, or they might participate in interactions with Trf4p or the Mtr4p helicase.
but not the A5 oligonucleotide supports a role for this zinc knuckle in binding large RNA substrates. The second and third zinc knuckles could be involved in binding other RNA substrates, or they might participate in interactions with Trf4p or the Mtr4p helicase.
Similar mutagenesis experiments to assess whether the fourth and fifth zinc knuckles of Air2p also modulate Trf4p activity were not feasible because we were unable to isolate soluble Trf4p/Air2pZK1-5 when these zinc knuckles were mutated. As shown below, the fourth and fifth zinc knuckles of Air2p are involved in interactions with Trf4p, and the mutations likely disrupt complex formation between Trf4p and Air2p.
Structure of the Trf4p/Air2p Core.
Constructs of Trf4p and Air2p were coexpressed in E. coli and purified as described above. For Trf4p, we used an active-site mutant, where the third aspartate in the polymerase catalytic triad was altered to alanine (D293A), as this improved protein yields. [Trf4p activity may be harmful for E. coli, where polyadenylation also targets RNAs for degradation (14), resulting in larger yields for the inactive enzyme.] We were unable to obtain crystals with Trf4p/Air2pZK1-5, possibly because the region containing the first three zinc knuckles is conformationally heterogeneous and interferes with crystallization. Crystals of Trf4p/Air2pZK4-5 complex belong to space group P321 and diffract to 2.7-Å resolution. The structure was solved by single anomalous wavelength dispersion phasing using the anomalous signal from zinc atoms bound by Air2p. The final model includes residues 161–481 of Trf4p and residues 122–146 and 160–198 of Air2p, two zinc atoms, and 58 water molecules. Residues 147–159 of Air2p, a portion of the peptide linker between the fourth and fifth zinc knuckles, are disordered and were not modeled. Data collection and refinement statistics are in Table S1.
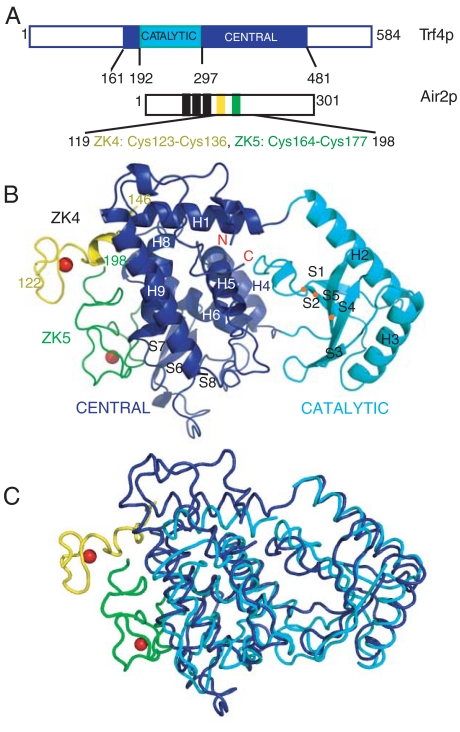
Trf4p adopts folds similar to structurally characterized Pol ß superfamily polymerases in both the catalytic and central domains, despite undetectable sequence similarity in the latter. The catalytic domain fold, common to all known Pol ß enzymes, is a five-stranded antiparallel beta sheet (strands S1–S5) flanked by two long alpha helices (H2, H3; Fig. 2) and comprises residues 190–315 in Trf4p. The central domain of Trf4p includes residues 161–189 and 316–481. It has an alpha helical core (helices H1, H4–H6, H8–H9; Fig. 2) with a three-stranded antiparallel beta sheet (S6–S8) inserted between helices H8 and H9. The nucleotide recognition motif corresponds to residues 421–433 (Fig. 2 and Fig. S3), including residues in strand S8 and C-terminal to it. The fold of the central domain is shared by other noncanonical Pol β RNA polymerases such as CCA-adding enzyme from Archaeoglobus fulgidus (15) (Fig. 2C) and the terminal uridyltransferases RET2 and TUT4 from Trypanosoma brucei (16, 17). Trf4p differs from these RNA polymerases most notably near the Air2p binding surface, in a longer helix H9 and in the length and conformation of the connector between helices H6 and H8 (residues 357–372 in Trf4p). Whereas the connector is only two residues in the archaeal CCA-adding enzyme, it consists of a short helix H7 and an adjacent loop in Trf4p. The connector in the terminal polyuridine transferases is long but differs significantly in both conformation and orientation from the Trf4p connector.
Fig. 2.
Structure of a Trf4p/Air2p subcomplex at 2.7-Å resolution. (A) Schematic of Trf4p and Air2p, indicating regions included in the crystallization construct. Air2p zinc knuckles are boxed. (B) Ribbons diagram with the catalytic and central domains of Trf4p in cyan and blue, respectively. Orange dots indicate positions of the aspartate residues in the catalytic triad. The fourth and fifth zinc knuckles of Air2p and adjacent linker regions (ZK4, ZK5) are yellow and green, respectively, and zincs are red. (C) Superposition of the Trf4p/Air2p subcomplex (blue/yellow and green) with another noncanonical RNA polymerase, CCA-adding enzyme from A. fulgidus (PDB ID 2DRA, cyan). The catalytic and central domains were superimposed separately because their relative orientation differs in the two structures.
Air2p is bound to a surface on the central domain of Trf4p formed by the connector between helices H6 and H8, the entire length of helix H8, and residues in S7 of the ß-hairpin. In Air2p, residues in the fifth zinc knuckle (residues 164–177) and in the linker sequences C-terminal to the two zinc knuckles form the interface with Trf4p (Fig. 3 A and B and Fig. S4). The fourth zinc knuckle (residues 123–136) is tethered to Trf4p via the adjacent linker region but has little direct contact with the surface of Trf4p. The fourth zinc knuckle may function in RNA binding, and residues critical for RNA binding in the nucleocapsid zinc knuckles (corresponding to residues 124–125 and 133–135 in Air2p) (12) are accessible to RNA (Fig. S5). In the fifth zinc knuckle, these same residues (165–166 and 174–176) are buried against the surface of Trf4p (Fig. S5). Thus, the fifth zinc knuckle serves as a protein–protein interaction module, and if it also interacts with RNA, its interactions are different from those of the nucleocapsid knuckles.
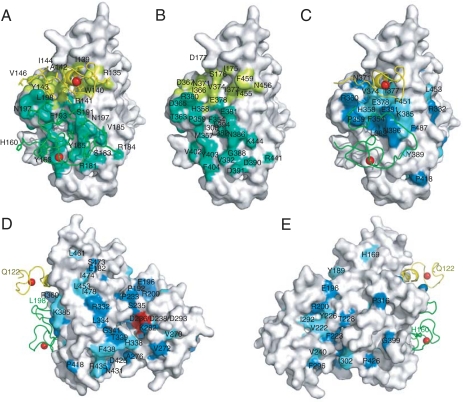
Fig. 3.
The Air2p-interaction surface of Trf4p is conserved in eukaryotes. (A) Space-filling model of Trf4p with surface residues within 4.5 Å of Air2p zinc knuckles colored lime or green. Air2p zinc knuckles are shown as worms (fourth, yellow; fifth, green), and Air2p side chains that interact with Trf4p are labeled. (B) As in A, but residues in Trf4p that are within 4.5 Å of Air2p are labeled and Air2p is not illustrated. (C) Residues that are identical or similar in Trf4p and six of seven other sequence-related polymerases are labeled and colored blue and cyan, respectively. We compared the sequence of Trf4p with sequences from S. cerevisiae, S. pombe, humans, Gallus gallus, Xenopus laevis, Drosophila melanogaster, and Caenorhabditis elegans. An alignment is in Fig. S3. Trf4p is oriented as in A and B. (D and E) Surface conservation as in C, but Trf4p is differently oriented. The conserved aspartate residues in the polymerase catalytic triad are red. Panels are enlarged in Fig. S4.
To identify functionally important surfaces, we mapped residues that are highly conserved in Trf4p and Trf5p, their Schizosaccharomyces pombe homolog Cid14, and in sequences from higher eukaryotes onto the Trf4p/Air2p structure (Fig. 3 C–E and Figs. S3 and S4). There are two highly conserved surfaces. One of these is on the central domain and corresponds to the binding site for the Air2p fragment (Fig. 3 C–E and Figs. S3 and S4). This finding is particularly significant in that it provides the only experimental evidence to date that the Trf4p/Air2p interaction is widely conserved in eukaryotes, a prerequisite for the conservation of the TRAMP-exosome surveillance pathway. A second conserved surface, as expected, is in the active-site cleft between the catalytic and central domain. The conservation also extends to surfaces adjacent to the active-site cleft, which may play roles in binding RNA or protein partners of Trf4p. These could include Mtr4p or portions of Air2p that are not in the structure.
RNA 3′ End Recognition.
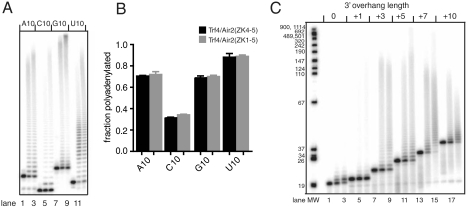
Because Trf4p must interact with the RNA 3′ end in order to polyadenylate it, we investigated whether this interaction plays a role in substrate specificity. In these experiments, we used the truncated complexes, Trf4p/Air2pZK1-5 and Trf4p/Air2pZK4-5, as well as preparations of full-length complex. (As shown in Fig. S1, the full-length complex is degraded during purification, but we would nevertheless expect similar trends as for intact sample.) To determine whether certain sequences are preferred, we performed polyadenylation assays with 10-mers consisting of A, C, G, or Us (Figs. 4 A and B and Fig. S6). We found that, for all three Trf4p/Air2p complexes, RNAs ending in cytosines are modified least. This tendency may prevent polyadenylation of deacylated nuclear tRNAs that undergo trimming of the CCA end to CC-, favoring repair by the CCA-adding enzyme (18). That all three complexes behaved similarly suggests that the Trf4p/Air2p core complex is sufficient for mediating end recognition.
Fig. 4.
RNA substrates of Trf4p/Air2p. (A) Polyadenylation of 5′ end-labeled oligomers A10, C10, G10, or U10 (30 fmol) by ∼2 pmol of Trf4p/Air2pZK4-5 (lanes 2, 5, 8, and 11), Trf4p/Air2pZK1-5 (lanes 3, 6, 9, and 12), or no protein (lanes 1, 4, 7, and 10). (B) Quantitation from three experiments in A, showing fraction of adenylated RNA. SEM is indicated. Trf4p/Air2p polyadenylates C10 less extensively than A10, G10, or U10. (C) Polyadenylation assay, where Trf4p/Air2p (2 pmol) was mixed with RNA duplexes (30 fmol) containing no overhang (lanes 1–3) or 3′- A-overhangs of length indicated (lanes 4–18). Longer extensions were added only when the 3′ overhang was three nucleotides or longer. Reactions included Trf4p/Air2pZK4-5 (lanes 2, 5, 8, 11, 14, and 17), Trf4p/Air2pZK1-5 (lanes 3, 6, 9, 12, 15, and 18), or no protein (lanes 1, 4, 7, 10, 13, and 16).
Using RNA duplexes with 3′-poly(A) overhangs of different lengths (1, 3, 5, 7, 10 nt) as substrates, we found that an overhang is required for polyadenylation by full-length Trf4p/Air2p, Trf4p/Air2pZK1-5, and Trf4p/Air2pZK4-5, but that the overhang can be as short as three nucleotides (Fig. 4C and Fig. S6). An overhang is probably required because the polymerase active-site cleft can accommodate single-stranded RNA but not a bulkier RNA duplex.
These experiments suggest that 3′ end recognition contributes to substrate specificity in Trf4p/Air2p. RNA ends have also been found to be recognition elements for other proteins involved in noncoding RNA processing and/or surveillance, including the eukaryotic proteins La and Ro (19, 20), which both bind nascent transcripts, as well as the bacterial RNase T and RNase R (21, 22). Sequences at the 3′ end may be one mechanism for recognizing such transcripts.
Summary and Model for Interactions with RNAs.
We have found that the surface on the central domain of Trf4p that interacts with Air2p is conserved in a number of other noncanonical RNA polymerases, including Trf5p in yeast as well as related polymerases in higher eukaryotes. This conservation argues that the Trf4p/Air2p subcomplex is conserved structurally and, most likely, functionally. Although our experiments have focused on the Trf4p/Air2p heterodimer, it is likely that complexes containing Trf5p and/or Air1p as well as homologous complexes in higher eukaryotes function in a similar way.
The conserved surface on Trf4p extends beyond the binding site for the fourth and the fifth zinc knuckle modules, and it is possible that additional conserved surface regions could interact with portions of Air2p not in the structure. These include the three N-terminal-most zinc knuckle modules as well as sequences at the N and C termini of Air2p. The additional conserved surface regions in Trf4p may also be important for interactions with the Mtr4p helicase or RNA substrates.
How might the Trf4p/Air2p complex interact with RNAs? In all cases, the polymerase interacts with the 3′ end of its RNA substrates, polyadenylating them while they are inserted into the polymerase active site between the central and catalytic domains, and it is likely that “misfolded” segments of RNA bound are within a certain distance of this 3′ end. We have noted that the core complexes, Trf4p/Air2pZK1-5 and Trf4p/Air2pZK4-5, are less active than the full-length complex studied by others (6), suggesting that the N and C termini are important for activity. Although the truncated ends could be important for catalysis, we favor a role in substrate binding because the crystal structure shows the catalytic regions of Trf4p appear to be structurally intact relative to other noncanonical RNA polymerases in the Pol β family. Our biochemical data show that both core complexes differentiate between correctly folded and aberrant tRNA substrates, indicating that sequences in the Trf4p/Air2p core are involved in RNA recognition. Possibly, then, the fourth and fifth zinc knuckles, included in both core complexes, could be involved in RNA binding. Additionally, we have shown that N-terminal sequences in Air2p, and in particular the first zinc knuckle, enhance the activity of Trf4p/Air2p on a longer tRNA substrate but not a short A5 oligonucleotide. That this zinc knuckle affects polyadenylation activity only on selected RNAs supports a role in RNA binding. We therefore propose a model where portions of the Trf4p/Air2p N and C termini and one (the N-terminal-most) or more of the Air2p zinc knuckles are involved in substrate binding. Different combinations of these elements could mediate interactions with a large group of structurally different RNA substrates.
Methods
Preparation, purification, and quantitation of the proteins and RNAs used in this study are described in SI Methods.
Polyadenylation Assays.
Polyadenylation assays were carried out in 10–15 μL volumes containing 50, 100, or 200 nM Trf4p/Air2p, 2.5–3.0 nM 5′ end-labeled RNA, 0.5 mM ATP, 5 mM MgCl2, 20 mM Tris·HCl, pH 7.6, and 50 mM NaCl. Reactions were incubated for 30 min at 22 °C. For the  time courses (Fig. 1A), a 15 μL reaction was set up, and 1.5 μL aliquots were taken at the times indicated. Reactions were stopped by addition of 50 mM EDTA, and for reactions involving tRNAs, the RNA was isolated by phenol-chloroform extraction. Samples were fractionated in 10% (tRNA), 12% (duplexes), or 15% (single-stranded oligomers) (wt/vol) polyacrylamide 8M urea gels. To quantitate the fraction of polyadenylated RNA, we used a PhosphorImager (Molecular Dynamics) to compare the counts in the polyadenylated portion of each lane with the total counts in the lane.
time courses (Fig. 1A), a 15 μL reaction was set up, and 1.5 μL aliquots were taken at the times indicated. Reactions were stopped by addition of 50 mM EDTA, and for reactions involving tRNAs, the RNA was isolated by phenol-chloroform extraction. Samples were fractionated in 10% (tRNA), 12% (duplexes), or 15% (single-stranded oligomers) (wt/vol) polyacrylamide 8M urea gels. To quantitate the fraction of polyadenylated RNA, we used a PhosphorImager (Molecular Dynamics) to compare the counts in the polyadenylated portion of each lane with the total counts in the lane.
Crystallization and Structure Determination.
The protein used in crystallization was concentrated to ∼8 mg/mL. Crystals were grown at 4 °C by the hanging drop vapor diffusion method. Drops consisted of 1.5 and 1.0 μL protein and reservoir solution (100 mM sodium citrate, pH 5.8, 200 mM sodium acetate, and 11% PEG 4000), respectively. Crystals appeared in 5 d and grew to a size of 200 × 150 × 50 μm over the course of 2 weeks. Crystals were transferred briefly to a solution containing the mother liquor supplemented with 25% glycerol (vol/vol) and flash-frozen in liquid nitrogen.
Data were collected at beamline ID24-C at the Advanced Photon Source in Chicago, Illinois, at a wavelength of 1.28215 Å to make use of the anomalous signal from zinc in phasing. The data were scaled and integrated using HKL2000 (23), and SHELXE (24) was used to identify zinc positions. There is one Trf4p/Air2pZK4-5 complex per asymmetric unit, and two zinc positions were located. Phases were calculated and refined with SHARP (25), and electron density maps were obtained after solvent flipping as implemented therein. The initial maps had well-defined density for the catalytic domain of Trf4p as well as for helices in the central domain, and these portions of Trf4p were modeled first. Model building was performed using the program O (26). Phases calculated from this partial model were combined with experimental phases, resulting in maps with improved density for Air2p and still unmodeled regions of Trf4p (Fig. S7). Although data from three crystals were used in calculating the initial experimental maps, regions in Air2p and the central domain of Trf4p were clearer in the phase-combined map when only data from a single crystal were used, perhaps because of slight nonisomorphism between crystals in these regions. Once model building was completed, the model was refined using the CNS software suite (27), alternating torsion angle dynamics, least-squares minimization, and individual B-factor refinement with manual rebuilding. In a final round of refinement with PHENIX (28), we also refined translation/libration/screw parameters for four domains (catalytic and central domains of Trf4p and two zinc knuckle domains) and added 58 water molecules.
Supplementary Material
Acknowledgments.
Data were collected at beamline 24-ID-C at the Advanced Photon Source, and we thank the Northeastern Collaborative Access Team staff for their support. K.M.R. thanks D. W. Rodgers for comments regarding this manuscript. Work presented here was funded by grants from the National Institute of General Medical Sciences (GM070521 and GM048410 to K.M.R. and S.L.W., respectively). S.H. is a Genentech Fellow of The Jane Coffin Childs Memorial Fund for Medical Research.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3NYB).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1003505107/-/DCSupplemental.
References
- 1.Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- 2.Brosnan CA, Voinnet O. The long and short of noncoding RNAs. Curr Opin Cell Biol. 2009;21:416–425. doi: 10.1016/j.ceb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 4.Anderson JT, Wang X. Nuclear RNA surveillance: No sign of substrates tailing off. Crit Rev Biochem Mol Biol. 2009;44:16–24. doi: 10.1080/10409230802640218. [DOI] [PubMed] [Google Scholar]
- 5.Kadaba S, et al. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vanacova S, et al. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.LaCava J, et al. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 8.Wyers F, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 9.Schneider C, Anderson JT, Tollervey D. The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Mol Cell. 2007;27:324–331. doi: 10.1016/j.molcel.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin G, Keller W. RNA-specific ribonucleotidyl transferases. RNA. 2007;13:1834–1849. doi: 10.1261/rna.652807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin G, Doublie S, Keller W. Determinants of substrate specificity in RNA-dependent nucleotidyl transferases. Biochim Biophys Acta. 2008;1779:206–216. doi: 10.1016/j.bbagrm.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D’Souza V, Summers MF. How retroviruses select their genomes. Nat Rev Microbiol. 2005;3:643–655. doi: 10.1038/nrmicro1210. [DOI] [PubMed] [Google Scholar]
- 13.Zuker M. MFOLD web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deutscher MP. Degradation of RNA in bacteria: Comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–666. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomita K, Ishitani R, Fukai S, Nureki O. Complete crystallographic analysis of CCA sequence addition. Nature. 2006;443:956–960. doi: 10.1038/nature05204. [DOI] [PubMed] [Google Scholar]
- 16.Deng J, Ernst NL, Turley S, Stuart KD, Hol WG. Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei. EMBO J. 2005;24:4007–4017. doi: 10.1038/sj.emboj.7600861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stagno J, Aphasizheva I, Rosengarth A, Luecke H, Aphasizhev R. UTP-bound and Apo bound structures of a minimal RNA uridyltransferase. J Mol Biol. 2007;366:882–899. doi: 10.1016/j.jmb.2006.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolfe CL, Hopper AK, Martin NC. Mechanism leading to and the consequences of altering the normal distribution of ATP(CTP):tRNA nucleotidyltransferase in yeast. J Biol Chem. 1996;271:4679–4686. doi: 10.1074/jbc.271.9.4679. [DOI] [PubMed] [Google Scholar]
- 19.Wolin SL, Cedervall T. The La protein. Annu Rev Biochem. 2002;71:375–403. doi: 10.1146/annurev.biochem.71.090501.150003. [DOI] [PubMed] [Google Scholar]
- 20.Fuchs G, Stein AJ, Fu C, Reinisch KM, Wolin SL. Structural and biochemical basis for misfolded RNA recognition by the Ro autoantigen. Nat Struct Mol Biol. 2006;13:1002–1009. doi: 10.1038/nsmb1156. [DOI] [PubMed] [Google Scholar]
- 21.Zuo Y, Deutscher MP. The physiological role of RNase T can be explained by its unusual substrate specificity. J Biol Chem. 2002;277:29654–29661. doi: 10.1074/jbc.M204252200. [DOI] [PubMed] [Google Scholar]
- 22.Vincent HA, Deutscher MP. Substrate recognition and catalysis by the exoribonuclease RNase R. J Biol Chem. 2006;281:29769–29775. doi: 10.1074/jbc.M606744200. [DOI] [PubMed] [Google Scholar]
- 23.Otwinowski Z, Minor W. In: Methods in Enzymology. Carter CW Jr, Sweet RM, editors. Vol. 276. New York: Academic; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 24.Sheldrick G, Schneider T. In: Methods in Enzymology. Carter CW Jr, Sweet RM, editors. Vol. 277. New York: Academic; 1997. pp. 319–343. [PubMed] [Google Scholar]
- 25.de la Fortelle E, Bricogne G. In: Methods in Enzymology. Carter CW Jr, Sweet RM, editors. Vol. 276. New York: Academic; 1997. pp. 472–494. [DOI] [PubMed] [Google Scholar]
- 26.Kleywegt GJ, Jones TA. In: Methods in Enzymology. Carter CW Jr, Sweet RM, editors. Vol. 277. New York: Academic; 1997. pp. 208–230. [DOI] [PubMed] [Google Scholar]
- 27.Brunger AT. Version 1.2 of the crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 28.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.