Abstract
Cell wall degrading enzymes have a complex molecular architecture consisting of catalytic modules and noncatalytic carbohydrate-binding modules (CBMs). The function of CBMs in cell wall degrading processes is poorly understood. Here, we have evaluated the potential enzyme-targeting function of CBMs in the context of intact primary and secondary cell wall deconstruction. The capacity of a pectate lyase to degrade pectic homogalacturonan in primary cell walls was potentiated by cellulose-directed CBMs but not by xylan-directed CBMs. Conversely, the arabinofuranosidase-mediated removal of side chains from arabinoxylan in xylan-rich and cellulose-poor wheat grain endosperm cell walls was enhanced by a xylan-binding CBM but less so by a crystalline cellulose-specific module. The capacity of xylanases to degrade xylan in secondary cell walls was potentiated by both xylan- and cellulose-directed CBMs. These studies demonstrate that CBMs can potentiate the action of a cognate catalytic module toward polysaccharides in intact cell walls through the recognition of nonsubstrate polysaccharides. The targeting actions of CBMs therefore have strong proximity effects within cell wall structures, explaining why cellulose-directed CBMs are appended to many noncellulase cell wall hydrolases.
Keywords: cell wall degradation, fluorescence quantification, pectate lyase, xylanase, arabinofuranosidase
Polysaccharide-rich cell walls are structurally complex and metabolically dynamic cell compartments that underpin many aspects of plant growth. The major polysaccharide components of plant cell walls are currently classed as cellulose, hemicelluloses (e.g., xyloglucans, heteroxylans, heteromannans, mixed-linkage glucans), and pectins. Cell wall structures are diverse across taxa and also within a plant (1, 2). Broadly, in the primary cell walls of dicotyledons, cellulose microfibrils are cross-linked by xyloglucan, and these polymers are coextensive with, and possibly linked to, pectic polysaccharides, providing a hydrated and relatively porous matrix. The major pectic polysaccharide is homogalacturonan (HG), which can be variably methyl-esterified (3). Commelinid monocotyledons, which include grasses and cereals, have biochemically distinct cell walls in which acidic glucuronoarabinoxylans typically carry out some of the roles of pectins (1). In secondary cell walls, cellulose content can range from 30% to >90% of the polysaccharides present and there can be distinct sets of hemicelluloses, such as unsubstituted xylans, in dicotyledons.
Cell walls are the most abundant source of organic carbon in the biosphere, and the recycling of these composite structures is of considerable biological importance. Understanding how microbes deconstruct cell walls is also of growing industrial significance for the biofuel and bioprocessing sectors. Although polysaccharides present a source of carbon for numerous microorganisms, plants have evolved many traits that resist microbial and enzymatic assaults. The deconstruction of cell wall polysaccharides is mediated by an array of enzymes, including glycoside hydrolases (GHs), polysaccharide lyases, and carbohydrate esterases. These enzymes are grouped into sequence-based families in the Carbohydrate-Active EnZymes (CAZy) database (http://www.cazy.org;) (4, 5). Cell wall-directed enzymes, particularly those that degrade cellulose and hemicelluloses, are frequently modular in that they contain one or more noncatalytic carbohydrate-binding modules (CBMs) in addition to the catalytic module(s) (6–8). CBMs have also been grouped into CAZy sequence-based families; currently, around half of the 59 CBM families contain members that bind to cell wall polymers. CBMs are suggested to enhance the efficiency of enzymes by mediating prolonged and intimate contact between the respective catalytic module and its target substrate (9, 10). Recent studies have shown that CBMs, which apparently exhibit the same specificities against isolated structural polysaccharides, can display differential recognition of the equivalent polymers when they are located in cell walls (11, 12). There have been several reports proposing that CBMs may also potentiate polysaccharide deconstruction through a cell wall disruptive mechanism (13–16). However, the improvement in enzyme performance through the addition of isolated CBMs and catalytic modules has been modest (15, 17), arguing against a disruptive function for these modules. The one notable exception is CBM33 (in nature, it is not appended to enzymes), which greatly enhances the capacity of chitinases to degrade highly crystalline forms of chitin (18). In general, the specificity of CBMs reflects the substrates targeted by the associated catalytic modules (19). Cellulose-binding CBMs are exceptions to this general rule in that, in addition to cellulases, these modules are often components of enzymes that hydrolyze xylans, mannans, and pectins (20–23). Numerous studies have shown that CBMs appended to cell wall hydrolases significantly enhance the enzyme activity against insoluble substrates (9, 10, 24). Currently, however, analysis of the functional importance of CBMs in enzyme action has been limited to exploration of their role in vitro against purified substrates or simple highly processed composites, wherein the integrity of the cell wall has been disrupted (9, 24, 25). Such approaches have not dissected the specificity of CBMs or their capacity to enhance catalytic activity in the context of intact cell walls. For example, it is possible that cellulose-directed CBMs may only benefit enzymes that degrade hemicellulose or pectins when the cell structure has been disrupted, possibly subsequent to initial fungal invasion. It is also unclear when substrate-targeting CBMs benefit their appended catalytic modules. For example, xylan-specific CBMs may only benefit their cognate enzymes once the degradative process has been initiated by removal of the side chains of the hemicellulosic structures. We have recently developed methodologies to detect CBM binding to ligands within intact cell walls (11, 12) and also to quantitate enzyme action on these materials using immunofluorescence techniques (26). These studies showed that xylanases from GH families 10 and 11 displayed contrasting efficiencies in the degradation of xylan when a component of intact secondary cell walls (as quantified in terms of epitope detection) compared with their in vitro activities against isolated polymers (26). Such studies indicate that intact cell walls with varying architectures, as revealed most effectively in sections through plant organs, can provide distinct structural contexts and specific limitations to GH and CBM access.
Here, through the use of series of synthetic modular constructs, we report the impact of appended CBMs on the activity of a range of polysaccharide degrading enzymes against substrates in intact primary and secondary cell walls. To achieve this objective, the catalytic modules of a pectate lyase, an arabinofuranosidase, and two xylanases were fused to selected xylan- and cellulose-directed CBMs (shown schematically in Fig. S1). The relative activities of the enzymes were determined using quantitative immunofluorescence microscopy. The data showed that the CBMs potentiated the hydrolytic action of the appended enzymes in both primary and secondary cell walls. The functional significance of CBMs is therefore demonstrated in a setting that closely reflects an in vivo context. The results also provide an explanation for the presence of crystalline cellulose-binding CBMs in enzymes that hydrolyze pectins or xylans.
Results
Pectic HG Degradation by a Pectate Lyase Is Promoted by Appended Cellulose-Binding Modules but Not by Xylan-Binding Modules.
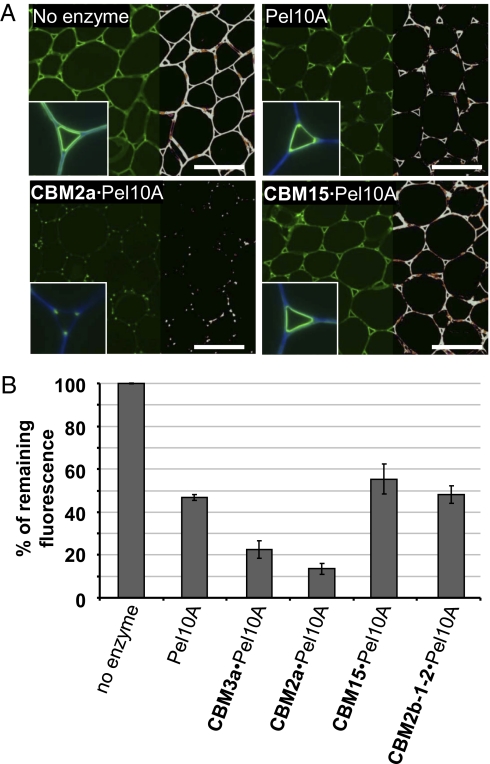
Pectic HG is abundant in the cell walls of parenchymal tissues of tobacco stems, as evidenced by JIM5 antibody binding (Fig. 1A). There is a complete loss of the JIM5 epitope after pretreatment of cell walls with an excess (300 nM) of pectate lyase. At lower concentrations of the enzyme, the degree of HG degradation could be quantified by determining the depletion of JIM5 immunofluorescence intensities captured in the micrographs (26) (Methods). After a 1-h treatment with 10 nM Cellvibrio japonicus pectate lyase Pel10A, 50 ± 3% of JIM5 binding was lost compared with the control, in which no enzyme was added (Fig. 1B). This concentration of Pel10A was then used to study the impact of appended CBMs directed to cell wall polysaccharides (Fig. 1). When crystalline cellulose-directed CBMs from families 3a and 2a were appended to the lyase, there was an increase in the depletion of JIM5 fluorescence (Fig. 1); the levels of JIM5 fluorescence were reduced to <25% and <15% of control levels, respectively. In these experiments, the remaining JIM5 fluorescence was restricted to the corners of intercellular space at the junctions of adhered and unadhered cell walls, examples of which are shown for CBM2a·Pel10A (Fig. 1A). In contrast, there was no increase in activity when xylan-binding CBMs from family 15 or 2b were appended to the pectate lyase. This is consistent with the observation that no xylan has been detected in tobacco stem pith parenchymal cell walls, and these CBMs bind strongly to the secondary cell walls of the vascular tissues in other regions of the stem (26).
Fig. 1.
Effect of pectate lyase (Pel10A) treatments on the detection of the JIM5 pectic HG epitope in primary cell walls of transverse sections of tobacco stem pith parenchyma. (A) Indirect immunofluorescence microscopy of JIM5 binding after Pel10A (no treatment, Pel10A alone, CBM2a·Pel10A, CBM15·Pel10A). JIM5-tagged FITC fluorescence is shown, as observed on the left half of each micrograph. The right half of each micrograph shows the same micrograph with overlaid false colors reflecting fluorescence intensities. (Insets) Images of JIM5 fluorescence combined with Calcofluor White fluorescence (blue) show higher magnification of cell walls in the region of intercellular spaces. (Scale bar: 200 μm.) (B) Histogram showing relative fluorescence intensities reflecting Pel10A treatments on JIM5 binding to the sections. The cellulose-binding modules CBM2a and CBM3a show a positive impact on the appended pectate lyase, whereas the xylan-binding modules CBM15 and CBM2b-1-2 show no impact. The results are expressed as percentages of the remaining fluorescence compared with a control without enzymatic treatment. The histogram shows mean ± SEM.
These studies indicate that there are distinct regions of primary cell walls that contain different amounts of HG and/or the tissue location of the pectic polysaccharide influences its susceptibility to degradation by pectate lyase. The pectic HG in adhered cell walls is most readily lost, followed by cell wall regions lining intercellular space and, finally, the corners of intercellular space at the junctions between adhered and unadhered regions of adjacent cells. The appending of a crystalline cellulose-directed CBM to the pectate lyase can promote the degradation of pectic HG in all these regions of primary cell walls.
Arabinofuranosidase Removal of Arabinosyl Residues from Arabinoxylan in Wheat Endosperm Cell Walls Is Promoted by Appended Xylan-Binding Modules.
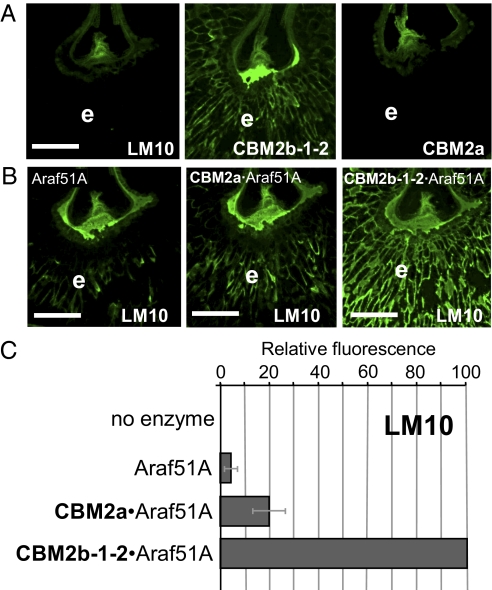
Wheat endosperm cell walls are typified by a high level of arabinoxylan and a low level of cellulose, which is in the region of 2% dry weight (27). The capacity of xylan- and cellulose-directed molecular probes to bind to cell walls of wheat grain was assessed by immunohistochemistry. Thus, the monoclonal antibody LM10 (raised against unsubstituted xylan) has been demonstrated previously to bind only to no- or low-substituted xylans and showed no recognition of the cell walls in the wheat grain endosperm (28) (Fig. 2A). The xylan-directed CBM2b-1–2 does bind to endosperm arabinoxylan in an equivalent section of wheat grain. In contrast, CBM2a, which binds to crystalline cellulose, showed no recognition of the walls of the wheat grain endosperm, reflecting its low cellulose content (Fig. 2A).
Fig. 2.
Indirect immunofluorescence microscopy of CBMs and monoclonal antibodies binding to transverse sections of wheat grain endosperm cell walls and effect of arabinofuranosidase treatments on the detection of the LM10 xylan epitope. (A) Lack of binding of LM10 indicated that the endosperm (e) cell walls are heavily arabinosylated. The xylan-binding module CBM2b-1-2 binds effectively and cellulose-directed CBM2a does not bind effectively to equivalent cell walls. (Scale bar: 500 μm.) (B) Indirect immunofluorescence detection of the LM10 epitope in wheat grain sections after treatments with the arabinofuranosidase Abf51A alone or appended with CBM2a or CBM2b-1-2. (Scale bar: 500 μm.) (C) Histogram showing relative LM10-associated fluorescence in wheat grain endosperm cell walls after no treatment and treatments with Abf51A constructs. The results are expressed as percentages of fluorescence relative to corresponding micrographs showing maximum fluorescence (i.e., LM10 epitope detection after CBM2b-1-2·Abf51A treatment). LM10 epitopes are revealed by the loss of arabinosyl residues. The histogram shows mean ± SEM.
The GH51 arabinofuranosidase, Abf51A, from C. japonicus releases O2 and O3 linked arabinofuranose side chains from monosubstituted backbone residues in xylan and arabinan (29). To study the impact of appended CBMs on arabinofuranosidase action, hybrid enzymes were generated by fusing the cellulose-binding CBM2a or the xylan-binding CBM2b-1-2 to the catalytic module of Abf51A. Although some LM10 binding to wheat grain central endosperm cell walls was observed after treatment with 100 nM Abf51A, the binding was sparse (Fig. 2B). The Abf51A derivatives containing CBM2a and CBM2b-1-2 were more active in generating the LM10 epitope than the catalytic module alone when used at equimolar concentrations. Appending the cellulose (CBM2a)- and xylan (CBM2b-1-2)-binding modules to Abf51A resulted in 4- and 20-fold increases in LM10 epitope detection, respectively (Fig. 2 B and C). These data indicate that LM10 is an effective probe for arabinofuranosidase action on endosperm cell wall arabinoxylan. The substantial potentiation of arabinofuranosidase activity by the xylan-binding module and the lesser impact of the cellulose-binding module on side chain removal reflect the relative abundance of these polysaccharide ligands in the endosperm.
Xylanase Degradation of Xylan Polysaccharides in Secondary Cell Walls Is Potentiated by Both Xylan- and Cellulose-Binding Modules.
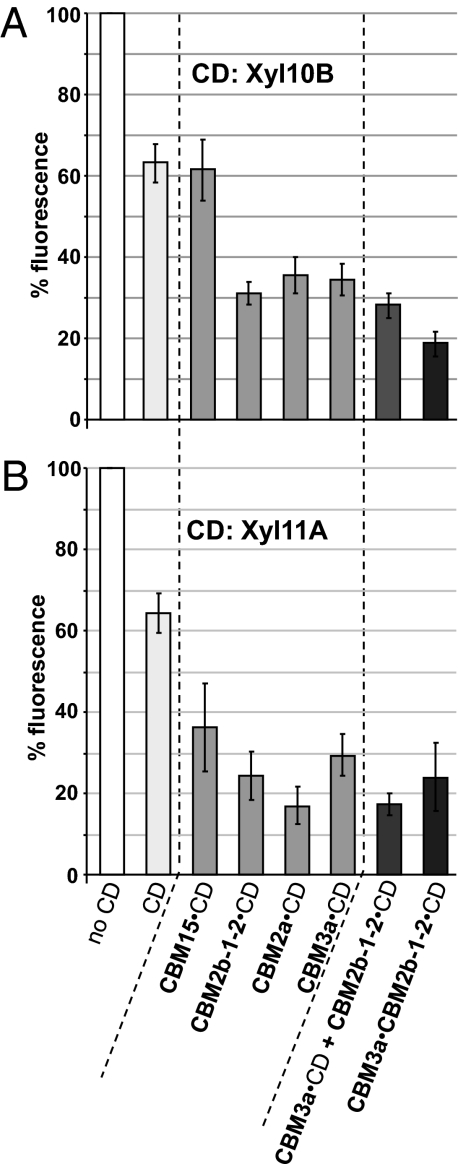
A previous study has shown that the enzymatic treatment of tobacco stem sections with xylanase Xyl10B or Xyl11A reduced but did not completely abolish xylan epitopes in secondary cell walls (26), indicative of the recalcitrant nature of these composite structures. Variants of Xyl10B and Xyl11A containing the xylan-binding modules CBM15 and CBM2b-1-2 and the crystalline cellulose-binding modules CBM2a and CBM3a were generated. Equimolar amounts of enzyme (50 nM for Xyl10B and its derivatives and 250 nM for Xyl11A and its derivatives) were incubated with a series of tobacco stem sections, and the removal of xylan was determined using CBM2b-1-2 appended to GFP. A 5-fold higher concentration of Xyl11A derivatives was used compared with the Xyl10B derivatives, because the GH11 enzyme is less active than the GH10 xylanase in degrading xylan epitopes in tobacco secondary cell walls (26). Histograms in Fig. 3 summarize the impact of the Xyl10B and Xyl11A derivatives on the capacity of CBM2b-1-2 to detect xylan within tobacco stem secondary walls. The catalytic modules of Xyl10B and Xyl11A, as discrete entities, resulted in 37 ± 5% and 36 ± 5% reductions in CBM2b1-2–mediated fluorescence, respectively. Appending the CBMs to the two xylanases generally resulted in an increase in the capacity of the enzymes to remove xylan from secondary cell walls; only Xyl10B linked to CBM15 did not display an increase in activity compared with Xyl10B. CBM2b-1-2, which binds to a broad range of xylans, potentiated the activities of both xylanases by decreasing the fluorescence intensities by ∼2-fold compared with the respective catalytic modules alone. That CBM15 only enhanced Xyl11A activity may reflect the specificity of CBM15 for highly exposed xylans (30), which are readily removed by the Xyl10B catalytic module on its own (discussed further below). Both cellulose-directed CBM2a and CBM3a increased the catalytic activities of the two enzymes to a similar extent as CBM2b-1-2 (∼2- to 4-fold decrease in fluorescence compared with the catalytic modules alone). There was a general trend that appending CBMs had an increased impact on Xyl11A activity relative to Xyl10B.
Fig. 3.
Impact of appended single CBMs, an enzyme mixture, and an appended tandem CBM on the action of Xyl10B (A) and Xyl11A (B) catalytic domains (CDs) on CBM2b-1-2 recognition of xylan in secondary cell walls of transverse sections of tobacco stem. The results are expressed as percentages of the remaining CBM2b-1-2:GFP fluorescence compared with the control without enzymatic treatment. In all cases, the same final molarities of CDs were used for the Xyl10B derivatives (50 nM) (A) and the Xyl11A derivatives (250 nM) (B). Histograms show mean ± SEM.
It is possible that the xylan-binding CBM2b-1-2 and the two cellulose-binding modules target their attached catalytic modules to distinct xylan substructures or contexts in the cell walls. If this hypothesis is correct, one might expect enhanced xylan degradation when the CBM3a- and CBM2b-1-2–linked xylanases are coincubated with cell walls. To address this question, tobacco stem sections were treated with equal quantities of CBM3a- and CBM2b-1-2–containing enzymes, either individually or in combination, while keeping the total concentration of the catalytic module constant (Fig. 3). No significant additional xylan degradation by the CBM enzyme mixtures was observed compared with incubations containing CBM3a·Xyl10B or CBM2b-1-2·Xyl10B. These data suggest that the different CBMs are not directing the enzyme to xylans that are in differing cell wall contexts. There was some impact of the enzyme combination relative to CBM3a-Xyl11A. The fluorescence level for the enzyme combination was 17% compared with the no-enzyme control, whereas the corresponding value for CBM3a·Xyl11A was 29%, which was significantly different at the 95% confidence level. There was no significant difference, however, between the enzyme combination and CBM2b-1-2·Xyl11A (Fig. 3).
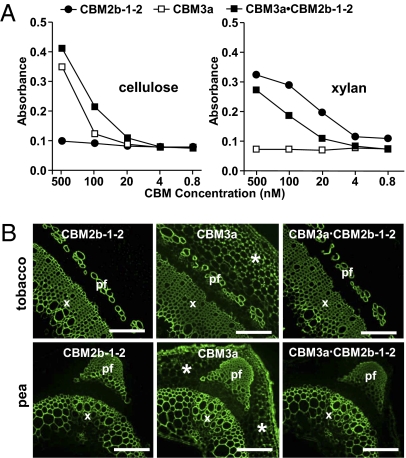
There are numerous examples of modular microbial plant cell wall GHs that contain more than one CBM. Within a single enzyme, CBMs can be members of different families that target distinct ligands (19, 22, 31, 32). A synergistic effect between CBMs with distinct specificities in targeting composite structures has been proposed (33, 34). To evaluate this possibility, CBM3a was covalently linked to CBM2b-1-2 and the ability of this tandem module to recognize isolated ligands in vitro and in intact cell walls was assessed. A microtiter plate assay demonstrated that the CBM3a·CBM2b-1-2 construct displayed the expected dual specificity, binding to both isolated xylan and cellulose in a manner similar to the individual modules alone (Fig. 4A). The capacity of the CBM3a·CBM2b-1-2 construct to bind to ligands in intact cell walls of both tobacco and pea stem sections was then assessed using immunohistochemistry. CBM2b-1-2 only bound to secondary cell walls, whereas CBM3a recognized both secondary and primary cell walls. The CBM3a·CBM2b-1-2 tandem construct only bound to secondary cell walls in stem sections, wherein cellulose and xylan are closely associated (Fig. 4B).
Fig. 4.
Binding properties of CBM3a, CBM2b-1-2, and the tandem CBM3a·CBM2b-1-2 module to isolated polymers and intact cell walls. (A) Effective binding of CBM3a, CBM2b-1-2, and CBM3a·CBM2b-1-2 to ligands in vitro using microtiter plate assays. (B) Indirect immunofluorescence analysis of CBM3a, CBM2b-1-2, and CBM3a·CBM2b-1-2 binding to transverse sections of tobacco stems (Upper) and pea stems (Lower). All probes bind effectively to the secondary cell walls of phloem fibers (pf) and xylem cells (x). In addition, CBM3a binds to cortical primary cell walls (*) in both stems. (Scale bar: 200 μm.)
The CBM3a·CBM2b-1-2 construct was appended to both Xyl10B and Xyl11A to determine its capacity to influence catalytic activity on secondary cell walls. The tandem module enhanced the xylanase activity of both enzymes against tobacco secondary cell walls relative to the catalytic domains alone (Fig. 3). For Xyl11A, the addition of the two modules was similar to constructs containing either CBM3a or CBM2b-1-2. The double-CBM construct, however, did enhance the cell wall activity of Xyl10B activity in comparison to each CBM appended individually to the catalytic domain, and this was the only Xyl10B construct that reduced mean fluorescence to a level <20% of that of the untreated control section (Fig. 3).
Discussion
This report shows that xylan- and cellulose-targeting CBMs can modulate the activity of appended catalytic modules against polysaccharides in both primary and secondary cell walls, as evidenced by the immunohistochemical detection of epitope/ligand loss or appearance. We show that appended CBMs directed to a cell wall polymer, other than the substrate of the attached catalytic domain, can promote enzyme action, although if the target ligand for the CBM is at a low level or absent, the impact of the noncatalytic module is greatly reduced or absent. These observations are significant for understanding the function of CBMs within complex modular enzymes while also providing insights into cell wall structures. These in situ studies of cell wall deconstruction demonstrate that CBMs can potentiate enzyme activity against cell walls by targeting polymers that are in close proximity to the substrate of the appended catalytic module.
Cell walls contain a range of interacting polysaccharides. With respect to the detection of polysaccharides by protein-based molecular probes, recent studies have shown that the enzymatic removal of pectic HG can increase the access of CBMs to cellulose (11) and is required to reveal xylan and xyloglucan in discrete regions of cell walls (26, 35). These observations indicate that even in cell walls exposed by sectioning, polysaccharide interactions could restrict access of enzymes to their insoluble substrates. In the case of pectate lyase degradation of HG in tobacco pith parenchymal cell walls, it is perhaps surprising that an appended cellulose-binding CBM could have any impact, because the uronic acid polymer is generally believed to be highly accessible. The capacity of cellulose-directed CBMs to potentiate the loss of HG from cell walls was particularly apparent for regions lining pith parenchymal intercellular spaces, which are known to have, in addition to cellulose, distinct components of pectin and hemicellulose (35, 36), which may reflect the structural stabilization of the parenchymal systems. Appended cellulose-directing CBMs promoted loss of HG from the lining of the space but did not remove the HG epitopes at junctions of adhered and unadhered cell walls as effectively. This indicates that the pectic HG was less tractable to pectate lyase action in these key points of cell attachment, or it may simply reflect the higher abundance of pectic HG at these regions. Together, these observations indicate that distinct cell wall structures are present in these locations and that this spatial structural heterogeneity has implications for cell wall functions and for protein/enzyme access and action. Endogenous cell wall-modifying enzymes are known to contain CBMs (37); thus, the action of these noncatalytic modules may be required for precise targeting to and modification of polymers in specific regions of plant cell walls during developmental processes.
In three structurally distinct cell walls, the appending of CBMs to enzymes can promote the activity of the respective catalytic modules. To be effective, the CBM may target the substrate hydrolyzed by the catalytic module or nonsubstrate polysaccharides in the same cell wall. It is thus apparent that appended CBMs can provide an advantage to enzyme action when cell walls present tightly packed structures with restricted access to the polysaccharides. Even the degradation of HG, generally viewed as the most accessible polymer in primary cell walls, can be potentiated by CBMs, consistent with the occurrence of these modules in a small subsection of pectate lyases (23, 38). However, a component of the pectic polymers found in intractable cell wall regions, where they are potentially attached to cellulose and/or hemicellulose polymers, is likely to be difficult to degrade. The experiments presented here are consistent with the view that CBMs, which are targeted to nonpectic polysaccharides in these regions, can lead to enhanced degradation of these recalcitrant populations of HG polysaccharides. The use of cellulose-poor wheat endosperm cell walls as the target for arabinofuranosidase action on arabinoxylan emphasizes the relevance of the appropriate CBMs for the cell wall context.
It therefore seems likely that in the context of intact plant cell walls, the possession of a cellulose-directed CBM would confer a selective advantage for many polysaccharide degrading enzymes. This advantage would occur by allowing the enzyme to remain in intimate contact with cell wall materials. In this proposed mechanism, the enzyme is bound to the cell wall through its cellulose-specific CBM, whereas the catalytic module is able to access its target substrate, which must be in close association with the cellulosic microfibrils. Thus, the CBM greatly increases the concentration of the enzyme in the vicinity of the substrate, leading to the observed increase in polysaccharide hydrolysis. It could be argued, however, that by reducing diffusion rates, CBMs may limit enzyme access, particularly when all available substrate has been hydrolyzed within the vicinity of the bound CBM. However, the crystalline cellulose-directed CBMs are able to diffuse over the surface of the cellulose microfibrils (39), enabling the enzyme to access substrate molecules rapidly within the wall. Indeed, the capacity of the xylan-specific CBMs used in this study to diffuse over cell walls reflects the relatively weak affinity these modules display for soluble ligands (Kd was typically 10−4 to 10−5 M at ambient temperature) (9, 40), enabling rapid dissociation and reassociation, particularly within the context of complete cell walls, where entropic factors, particularly within specific microenvironments, may greatly increase affinity. It should be recognized, however, that CBMs may also enhance activity by disrupting the interface between the substrate and other polysaccharides within the wall, although no compelling in vitro data indicate that cellulose- or hemicellulose-targeting CBMs display such a function.
The differential impact of appended CBMs on the action of the catalytic modules of Xyl10B and Xyl11A is intriguing. The most obvious differential effect is mediated by CBM15, which can potentiate Xyl11A activity but not that of Xyl10B. This may be related to the observation that 5-fold more Xyl11A than Xyl10B (when they lack CBMs) was required to provide an equivalent loss of xylan, whereas against soluble pure forms of substrate, the GH11 enzyme is considerably more active than the GH10 xylanase (Table S1). This is consistent with the topology of the substrate-binding cleft of Xyl11A and Xyl10B. In Xyl11A, the binding cleft is narrow and deep, and thus adapted to bind single xylan chains. In Xyl10B, substrate is accommodated in a more open cleft; thus, the enzyme is able to bind xylan molecules that are in relatively close proximity to other components of cell walls. Therefore, it is likely that Xyl10B does not require a CBM to hydrolyze xylans that are in sparse association with other cell wall components, whereas Xyl11A will still benefit from the targeting function of these modules to deconstruct such structures. CBM15 is particularly well adapted to direct Xyl11A to highly exposed regions of xylan because its major role in nature is to bind soluble xylans and xylooligosaccharides on the surface of the endogenous bacterium C. japonicus (40). It is likely that the beneficial effects of CBM2b-1-2 and CBM2a on Xyl11A, as opposed to Xyl10A, may also reflect the capacity of these modules to direct the GH11 xylanase to regions of the substrate that are in an appropriate context for the enzyme.
Based on the argument above, it is interesting that fusing the CBM3a·CBM2b-1-2 double module to the two xylanases appeared to confer more benefit to the GH10 xylanase compared with Xyl11A. It is possible that when bound to just a single CBM, the enzymes have more freedom to explore cell wall structure; when bound to CBM3a, the enzyme can slide along cellulose microfibrils, whereas the CBM2b-1-2 can freely associate and dissociate from its ligand. By contrast, when both modules are appended to a xylanase, the enzyme may become locked into specific regions of the wall in which xylan and cellulose are in very close association. It is possible that xylan is accessible to GH11 xylanases in only a proportion of these cell wall contexts, whereas such substructures are more suited to the topology of the GH10 substrate-binding clefts.
In conclusion, this report demonstrates that CBMs can be effective in potentiating the activity of pectic and hemicellulosic polysaccharide-active enzymes in both primary and secondary cell wall contexts. Moreover, the data indicate that CBMs binding cellulose, which is abundant in most cell types, confer a significant benefit to enzymes that cleave matrix polysaccharides.
Methods
Monoclonal Antibodies and Recombinant Proteins.
Xylan monoclonal antibody LM10 (28) and the pectic HG monoclonal antibody JIM5 (35) were used as unpurified hybridoma cell culture supernatants. CBM2b-1-2, CBM15, CBM2a, and CBM3a were derived from Cellulomonas fimi xylanase Xyl11A (9), C. japonicus xylanase Xyl10C (40), C. japonicus xylanase Xyl10A (21), and Clostridium thermocellum cellulosome-integrating protein CipA (41), respectively. The enzymes used to explore the functional importance of CBMs in cell wall deconstruction were the C. japonicus pectate lyase Pel10A (38), C. japonicus arabinofuranosidase Abf51A (29), Cellvibrio mixtus xylanase Xyl10B (42), and Neocallimastix patricarium xylanase Xyl11A (43). Schematics of the catalytic modules and CBM constructs are shown in Fig. S1, the specific activities of all constructs against soluble substrates are shown in Table S1, and the construction of plasmids encoding these proteins is detailed in SI Text and Table S2.
Preparation of Plant Materials, Enzymatic Treatment of Cell Walls in Organ Sections, and Polysaccharide Detection Procedures.
Tobacco (Nicotiana tabacum L.) and pea (Pisum sativum L.) plants were grown as described (35). Excised stem regions and wheat (Triticum aestivum L.) grains were fixed in PEM buffer (50 mM Pipes, 5 mM EGTA, 5 mM MgSO4, pH 6.9) containing 4% (vol/vol) paraformaldehyde. After fixation, all plant materials were wax-embedded and sectioned as described previously (12).
Pectate lyase treatments were carried out as described previously (11) using an enzyme concentration of 10 nM for all constructs for 1 h at 21 °C. Treatments with arabinofuranosidase and associated constructs were carried out overnight at an enzyme concentration of 100 nM in 50 mM sodium phosphate buffer (pH 7.0). Xylanase treatments were carried out as described previously (26) at enzyme concentrations of 50 and 250 nM for Xyl10B derivatives and Xyl11A derivatives, respectively. Sections not treated with the enzymes were incubated for an equivalent time with the corresponding buffers. All sections were subsequently treated for 20 min with 5 μg·mL−1 proteinase K (Sigma–Aldrich) in PBS before substrate detection to remove any enzymes still attached to cell walls through their CBMs.
Detection of pectic HG with JIM5 and of xylans with LM10 antibodies was carried out as described (12). The binding of His-tagged CBMs to plant sections was assessed by a three-stage immunolabeling technique described previously (26), except for the CBM2b-1-2:GFP construct, which was used to label sections directly by incubating with 50 μg·mL−1 for 1 h in 3% (wt/vol) milk protein/PBS at room temperature before washing with PBS and mounting for microscopy. Microtiter plate assays of CBM binding to isolated cellulose and xylan polymers was carried out using ELISA procedures as described previously (35). Birchwood xylan and hydroxylethylcellulose were used as polysaccharide substrates and coated on to microtiter plates at 50 μg·mL−1.
Immunofluorescence Microscopy and Quantification of Enzyme Impacts on Cell Wall Polymers.
Immunofluorescence was observed with an Olympus BX-61 microscope equipped with epifluorescence irradiation, and all micrographs were captured with an ORCA 285 camera (Hamamatsu) using the Volocity software (Improvision). The relative capacities of the enzymes to degrade/modify their substrates directly within the cell wall of sections were determined by quantitative assessments of the immunofluorescence intensities captured in equivalent micrographs using a protocol that has been described (26). Briefly, using Improvision Volocity quantitation software, the absolute level of fluorescence contained in the micrographs was determined. The spatial mapping of the pixels was performed using a color-coding method. For cell wall deconstruction, the expected modulation is the disappearance of epitopes following polysaccharide degradation. Control micrographs obtained without enzymatic treatment were designated as 100% of initial fluorescence, and fluorescence levels in micrographs of treated sections were determined accordingly. Some cell wall polysaccharide epitopes are only fully revealed after a corresponding enzymatic treatment. This is the case for the LM10 xylan epitope in wheat grain endosperm cell walls, which is more abundant after arabinofuranosidase treatments. In this particular instance, the micrograph featuring the most intense signal after enzymatic treatment was designated to contain a 100% fluorescence signal and other treatments were quantified accordingly, as indicated in Fig. S2. For quantification of arabinofuranosidase action, equivalent regions of the micrographs containing only endosperm cells were selected for quantification. In all cases, the fluorescence quantification reflects the analysis of micrographs obtained from a minimum of three separate sections that were prepared from at least three separate plants. The fluorescence quantification values shown are means of a minimum of four assessments.
Supplementary Material
Acknowledgments
This work was supported by funding from the UK Biotechnology and Biological Sciences Research Council (Grant BB/E014364/1).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1005732107/-/DCSupplemental.
References
- 1.Harris PJ. Diversity in plant cell walls. In: Henry RJ, editor. Plant Diversity and Evolution: Genotypic and Phenotypic Variation in Higher Plants. Wallington, Oxfordshire, UK: CAB International Publishing; 2005. pp. 201–227. [Google Scholar]
- 2.Knox JP. Revealing the structural and functional diversity of plant cell walls. Curr Opin Plant Biol. 2008;11:308–313. doi: 10.1016/j.pbi.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Mohnen D. Pectin structure and biosynthesis. Curr Opin Plant Biol. 2008;11:266–277. doi: 10.1016/j.pbi.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cantarel BL, et al. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009;37(Database issue):D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. Carbohydrate-binding modules: Fine-tuning polysaccharide recognition. Biochem J. 2004;382:769–781. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hashimoto H. Recent structural studies of carbohydrate-binding modules. Cell Mol Life Sci. 2006;63:2954–2967. doi: 10.1007/s00018-006-6195-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guillén D, Sánchez S, Rodríguez-Sanoja R. Carbohydrate-binding domains: Multiplicity of biological roles. Appl Microbiol Biotechnol. 2010;85:1241–1249. doi: 10.1007/s00253-009-2331-y. [DOI] [PubMed] [Google Scholar]
- 9.Bolam DN, et al. Pseudomonas cellulose-binding domains mediate their effects by increasing enzyme substrate proximity. Biochem J. 1998;331:775–781. doi: 10.1042/bj3310775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carrard G, Koivula A, Söderlund H, Béguin P. Cellulose-binding domains promote hydrolysis of different sites on crystalline cellulose. Proc Natl Acad Sci USA. 2000;97:10342–10347. doi: 10.1073/pnas.160216697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blake AW, et al. Understanding the biological rationale for the diversity of cellulose-directed carbohydrate-binding modules in prokaryotic enzymes. J Biol Chem. 2006;281:29321–29329. doi: 10.1074/jbc.M605903200. [DOI] [PubMed] [Google Scholar]
- 12.McCartney L, et al. Differential recognition of plant cell walls by microbial xylan-specific carbohydrate-binding modules. Proc Natl Acad Sci USA. 2006;103:4765–4770. doi: 10.1073/pnas.0508887103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knowles J, Lehtovaara P, Teeri T. Cellulase families and their genes. Trends in Biotechnology. 1987;5:255–261. [Google Scholar]
- 14.Teeri TT. Crystalline cellulose degradation: New insight into the function of cellobiohydrolases. Trends in Biotechnology. 1997;15:160–167. [Google Scholar]
- 15.Din N, et al. C1-Cx revisited: Intramolecular synergism in a cellulase. Proc Natl Acad Sci USA. 1994;91:11383–11387. doi: 10.1073/pnas.91.24.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L, Zhang Y, Gao P. A novel function for the cellulose binding module of cellobiohydrolase I. Sci China C Life Sci. 2008;51:620–629. doi: 10.1007/s11427-008-0088-3. [DOI] [PubMed] [Google Scholar]
- 17.Moser F, Irwin D, Chen SL, Wilson DB. Regulation and characterization of Thermobifida fusca carbohydrate-binding module proteins E7 and E8. Biotechnol Bioeng. 2008;100:1066–1077. doi: 10.1002/bit.21856. [DOI] [PubMed] [Google Scholar]
- 18.Vaaje-Kolstad G, Horn SJ, van Aalten DM, Synstad B, Eijsink VG. The non-catalytic chitin-binding protein CBP21 from Serratia marcescens is essential for chitin degradation. J Biol Chem. 2005;280:28492–28497. doi: 10.1074/jbc.M504468200. [DOI] [PubMed] [Google Scholar]
- 19.Montanier C, et al. Evidence that family 35 carbohydrate binding modules display conserved specificity but divergent function. Proc Natl Acad Sci USA. 2009;106:3065–3070. doi: 10.1073/pnas.0808972106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kellett LE, et al. Xylanase B and an arabinofuranosidase from Pseudomonas fluorescens subsp. cellulosa contain identical cellulose-binding domains and are encoded by adjacent genes. Biochem J. 1990;272:369–376. doi: 10.1042/bj2720369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferreira LM, Durrant AJ, Hall J, Hazlewood GP, Gilbert HJ. Spatial separation of protein domains is not necessary for catalytic activity or substrate binding in a xylanase. Biochem J. 1990;269:261–264. doi: 10.1042/bj2690261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hogg D, et al. The modular architecture of Cellvibrio japonicus mannanases in glycoside hydrolase families 5 and 26 points to differences in their role in mannan degradation. Biochem J. 2003;371:1027–1043. doi: 10.1042/BJ20021860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKie VA, et al. A new family of rhamnogalacturonan lyases contains an enzyme that binds to cellulose. Biochem J. 2001;355:167–177. doi: 10.1042/0264-6021:3550167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Black GW, et al. Evidence that linker sequences and cellulose-binding domains enhance the activity of hemicellulases against complex substrates. Biochem J. 1996;319:515–520. doi: 10.1042/bj3190515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolam DN, et al. Evidence for synergy between family 2b carbohydrate binding modules in Cellulomonas fimi xylanase 11A. Biochemistry. 2001;40:2468–2477. doi: 10.1021/bi002564l. [DOI] [PubMed] [Google Scholar]
- 26.Hervé C, Rogowski A, Gilbert HJ, Knox JP. Enzymatic treatments reveal differential capacities for xylan recognition and degradation in primary and secondary plant cell walls. Plant J. 2009;58:413–422. doi: 10.1111/j.1365-313X.2009.03785.x. [DOI] [PubMed] [Google Scholar]
- 27.Mares DJ, Stone BA. Studies on wheat endosperm. I. Chemical composition and ultrastucture of the cell walls. Aust J Biol Sci. 1973;26:793–812. [Google Scholar]
- 28.McCartney L, Marcus SE, Knox JP. Monoclonal antibodies to plant cell wall xylans and arabinoxylans. J Histochem Cytochem. 2005;53:543–546. doi: 10.1369/jhc.4B6578.2005. [DOI] [PubMed] [Google Scholar]
- 29.Beylot MH, McKie VA, Voragen AG, Doeswijk-Voragen CH, Gilbert HJ. The Pseudomonas cellulosa glycoside hydrolase family 51 arabinofuranosidase exhibits wide substrate specificity. Biochem J. 2001;358:607–614. doi: 10.1042/0264-6021:3580607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szabo L, et al. Structure of a family 15 carbohydrate-binding module in complex with xylopentaose. Evidence that xylan binds in an approximate 3-fold helical conformation. J Biol Chem. 2001;276:49061–49065. doi: 10.1074/jbc.M109558200. [DOI] [PubMed] [Google Scholar]
- 31.Araki R, et al. Essential role of the family-22 carbohydrate-binding modules for beta-1,3-1,4-glucanase activity of Clostridium stercorarium Xyn10B. FEBS Lett. 2004;561:155–158. doi: 10.1016/S0014-5793(04)00160-7. [DOI] [PubMed] [Google Scholar]
- 32.DeBoy RT, et al. Insights into plant cell wall degradation from the genome sequence of the soil bacterium Cellvibrio japonicus. J Bacteriol. 2008;190:5455–5463. doi: 10.1128/JB.01701-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomme P, Creagh AL, Kilburn DG, Haynes CA. Interaction of polysaccharides with the N-terminal cellulose-binding domain of Cellulomonas fimi CenC. 1. Binding specificity and calorimetric analysis. Biochemistry. 1996;35:13885–13894. doi: 10.1021/bi961185i. [DOI] [PubMed] [Google Scholar]
- 34.Boraston AB, et al. Co-operative binding of triplicate carbohydrate-binding modules from a thermophilic xylanase. Mol Microbiol. 2002;43:187–194. doi: 10.1046/j.1365-2958.2002.02730.x. [DOI] [PubMed] [Google Scholar]
- 35.Marcus SE, et al. Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol. 2008;8:60. doi: 10.1186/1471-2229-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ordaz-Ortiz JJ, Marcus SE, Knox JP. Cell wall microstructure analysis implicates hemicellulose polysaccharides in cell adhesion in tomato fruit pericarp parenchyma. Molecular Plant. 2009;2:910–921. doi: 10.1093/mp/ssp049. [DOI] [PubMed] [Google Scholar]
- 37.Urbanowicz BR, et al. A tomato endo-beta-1,4-glucanase, SlCel9C1, represents a distinct subclass with a new family of carbohydrate binding modules (CBM49) J Biol Chem. 2007;282:12066–12074. doi: 10.1074/jbc.M607925200. [DOI] [PubMed] [Google Scholar]
- 38.Brown IE, Mallen MH, Charnock SJ, Davies GJ, Black GW. Pectate lyase 10A from Pseudomonas cellulosa is a modular enzyme containing a family 2a carbohydrate-binding module. Biochem J. 2001;355:155–165. doi: 10.1042/0264-6021:3550155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jervis EJ, Haynes CA, Kilburn DG. Surface diffusion of cellulases and their isolated binding domains on cellulose. J Biol Chem. 1997;272:24016–24023. doi: 10.1074/jbc.272.38.24016. [DOI] [PubMed] [Google Scholar]
- 40.Pell G, et al. Structural and biochemical analysis of Cellvibrio japonicus xylanase 10C: How variation in substrate-binding cleft influences the catalytic profile of family GH-10 xylanases. J Biol Chem. 2004;279:11777–11788. doi: 10.1074/jbc.M311947200. [DOI] [PubMed] [Google Scholar]
- 41.Poole DM, et al. Identification of the cellulose-binding domain of the cellulosome subunit S1 from Clostridium thermocellum YS. FEMS Microbiol Lett. 1992;78:181–186. doi: 10.1016/0378-1097(92)90022-g. [DOI] [PubMed] [Google Scholar]
- 42.Pell G, et al. The mechanisms by which family 10 glycoside hydrolases bind decorated substrates. J Biol Chem. 2004;279:9597–9605. doi: 10.1074/jbc.M312278200. [DOI] [PubMed] [Google Scholar]
- 43.Gilbert HJ, Hazlewood GP, Laurie JI, Orpin CG, Xue GP. Homologous catalytic domains in a rumen fungal xylanase: Evidence for gene duplication and prokaryotic origin. Mol Microbiol. 1992;6:2065–2072. doi: 10.1111/j.1365-2958.1992.tb01379.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.