Abstract
Potato type I and II serine protease inhibitors are produced by solanaceous plants as a defense mechanism against insects and microbes. Nicotiana alata proteinase inhibitor (NaPI) is a multidomain potato type II inhibitor (pin II) that is produced at high levels in the female reproductive tissues of the ornamental tobacco, Nicotiana alata. The individual inhibitory domains of NaPI target the major classes of digestive enzymes, trypsin and chymotrypsin, in the gut of lepidopteran larval pests. Although consumption of NaPI dramatically reduced the growth and development of a major insect pest, Helicoverpa punctigera, we discovered that surviving larvae had high levels of chymotrypsin activity resistant to inhibition by NaPI. We found a potato type I inhibitor, Solanum tuberosum potato type I inhibitor (StPin1A), was a strong inhibitor of the NaPI-resistant chymotrypsin activity. The combined inhibitory effect of NaPI and StPin1A on H. armigera larval growth in the laboratory was reflected in the increased yield of cotton bolls in field trials of transgenic plants expressing both inhibitors. Better crop protection thus is achieved using combinations of inhibitors in which one class of proteinase inhibitor is used to match the genetic capacity of an insect to adapt to a second class of proteinase inhibitor.
Keywords: chymotrypsin, resistance, Lepidoptera
Lepidopteran insects are one of the most important groups of crop pests in the world. In Australia, two of the major lepidopteran pests of cotton are Helicoverpa punctigera and H. armigera (1). H. armigera is the dominant pest and has developed resistance to a number of chemical pesticides (2). The only commercially available transgenes for control of these insect pests encode Bacillus thuringiensis (Bt) toxins and the Vip3Aa20 toxin (3). First-generation Bt crops expressing a single Bt toxin, Cry1AC, were highly successful. However, field-evolved resistance to Cry1Ac has been reported recently for populations of H. zea (4). Second-generation Bt crops containing two different Bt toxins are considered to be more robust, because the toxins bind to different targets in the larval midgut. However, cross-resistance has been demonstrated in the laboratory where feeding Cry2Ab to Pectinophora gossypiella (pink bollworm) caused a 420-fold increase in resistance to Cry1Ac (5). Stacking of insect resistance genes probably will be the industry standard for transgenic crops, and therefore, the discovery and development of insecticidal molecules with different modes of action is critical for long-term control of insect pests. Proteinase inhibitors (PIs) are a potential component of gene stacks for the protection of important agricultural crops against insect damage.
Plants have developed both physical and molecular strategies to limit consumption by insect pests while attracting insect pollinators. A classic example of plant–insect interactions is the production of potato type I inhibitor (pin I) and type II inhibitor (pin II) serine PIs by solanaceous plants responding to damage by lepidopteran larvae (6). PIs are expressed constitutively at high levels in reproductive tissues (7), whereas expression in leaves is relatively low until the leaves are damaged by chewing insects (8, 9). Signals produced by wounded plant cells as well as by molecules in insect saliva lead to rapid accumulation of pin II transcripts (10, 11). Early observations that PI accumulation was not restricted to the wounded leaves led to the identification of mobile signals, such as the peptide hormone systemin, that activate signaling pathways and induce the transcription of the PI genes in distal leaves (12). Furthermore, wounded plants produce volatile signals that attract parasitic and predatory insects (13) and induce PI production in neighboring, nonwounded plants to arm themselves before insect invasion occurs (14).
When plant PIs bind to the digestive proteinases of insects, they block the digestion of proteins, leading to developmental delays and increased mortality. Pin I and II inhibitors target the digestive serine proteinases trypsin and chymotrypsin, the major enzymes contributing to protein digestion in the gut of lepidopteran larvae (15). Most plants produce PIs for insect protection, but insects can adapt to PI ingestion by overproducing PI-sensitive proteases (16), and/or up-regulating the expression of proteases that are insensitive to the PIs produced by that plant (17–20), or inducing the production of PI-degrading enzymes (21, 22).
In this study we investigated the effect of ingestion of a pin I and II inhibitor on the growth of Helicoverpa spp. Nicotiana alata PI (NaPI) is a pin II inhibitor from Nicotiana alata that consists of four (6-kDa) trypsin inhibitors (T1–T4) and two (6-kDa) chymotrypsin inhibitors (C1 and C2) (23, 24). Ingestion of NaPI induced an NaPI-resistant chymotrypsin that was inhibited by a pin I inhibitor (StPin1A) from wounded Solanum tuberosum leaves. In our companion paper (25) we characterize the mechanism of the resistance of this chymotrypsin to NaPI. The combination of NaPI and StPin1A in artificial diet and transgenic plants was far more effective at reducing the growth and development of Helicoverpa spp. than either inhibitor alone.
Results
H. punctigera Larvae Contain Chymotrypsin Activity Resistant to NaPI.
To test the insecticidal activity of NaPI, H. punctigera larvae were fed a cotton leaf-based artificial diet containing 0.26% (wt/vol) NaPI. At day 21, there was 80% mortality in NaPI-fed larvae compared with 40% mortality in the control-fed larvae (Fig. 1A). Larvae raised on the NaPI diet weighed about 30 mg; larvae fed control diet weighed ≈100 mg (Fig. 1B).
Fig. 1.
Survival (A) and growth (±SEM) (B) of H. punctigera larvae raised on artificial cotton leaf diets containing 0.26% (wt/vol) NaPI.
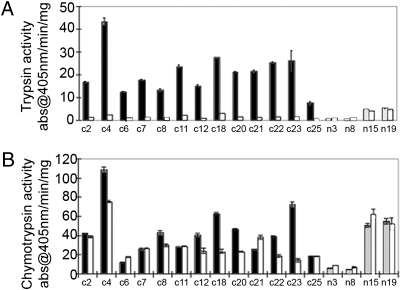
Chymotrypsin and trypsin activity was measured in unfractionated gut extracts from surviving fifth-instar larvae. The in vivo effect of NaPI substantially lowered or abolished trypsin activity (Fig. 2), but chymotrypsin activity was either unaffected or enhanced. Although subsequent in vitro inhibition of chymotrypsin activity in gut extract from control larvae by NaPI was variable, NaPI did not inhibit any of the chymotrypsin activity in gut extracts of larvae that had consumed the NaPI (Fig. 2). This result suggested that larvae produce two classes of chymotrypsins: some that are inhibited by NaPI (NaPI-susceptible) and some that are not (NaPI-resistant). In a subsequent experiment, several commercially available PIs were tested against gut extracts from H. punctigera that had been depleted of NaPI-sensitive chymotrypsins by affinity chromatography (Table S1). The pin I inhibitor completely abolished all remaining chymotrypsin activity in the gut of these H. punctigera larvae.
Fig. 2.
Trypsin and chymotrypsin activity in larvae fed artificial diet containing NaPI. H. punctigera larvae (20) were raised from neonates to fifth instar on cotton leaf-based artificial diets with (n) or without (c) 0.26% (wt/vol) NaPI. Four larvae survived to fifth instar on the NaPI diet, whereas 13 control larvae survived. The presence of NaPI-insensitive enzymes in gut extracts from individual larvae was determined by (A) trypsin and (B) chymotrypsin activity assays before (black/gray bars) and after (white bars) the addition of NaPI in vitro. Units of protease activity are expressed as change in absorbance at 405 nm/min per milligram extracted protein (±SEM).
Chymotrypsin Activity in the Gut of Two Helicoverpa Species Is Abolished by a Type I Inhibitor from Potato.
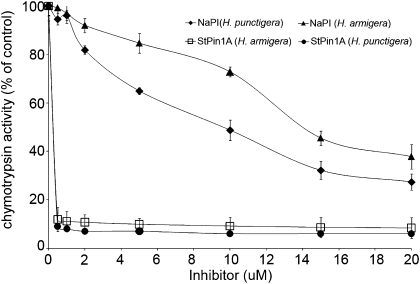
Commercial preparations of PIs often are contaminated (Fig. S1) and can contain several isoforms (26). To investigate the capacity of a pure pin I inhibitor to inhibit insect chymotrypsin activity, we obtained a pin I inhibitor cDNA using RNA from the wounded leaves of S. tuberosum. The recombinant pin I inhibitor, StPin1A, was expressed in Escherichia coli and purified to homogeneity for use in inhibition assays. We retested the gut extracts from both H. punctigera and a closely related species, H. armigera, and determined that the recombinant inhibitor StPin1A (0.2 μM) completely abolished all chymotrypsin activity in unfractionated gut extracts. In comparison, NaPI inhibited only 10% of total chymotrypsin activity at the same concentration (Fig. 3).
Fig. 3.
Inhibition of chymotrypsin activity by NaPI and StPin1A. Unfractionated gut extract (1 μg protein) from H. armigera and H. punctigera larvae was incubated with increasing concentrations of native purified NaPI and recombinant StPin1A before the addition of chymotrypsin substrate (Succ-AAPFpNA). The residual activity is expressed as percent chymotrypsin remaining compared with controls. Error bars show SEM of three independent experiments performed in duplicate.
Ingestion of NaPI and StPin1A Impedes the Development of H. armigera Larvae.
The discovery that StPin1A abolished the NaPI-resistant chymotrypsin activity led us to investigate whether the combination of StPin1A and NaPI would have a more marked effect on insect growth and development than either inhibitor on its own. To test this possibility, we placed H. armigera neonates on cotton leaf-based artificial diets with and without added PIs and recorded weight gain on days 5, 7, 9, and 11 (Fig. 4). At day 11, larvae fed diets containing NaPI or StPin1A weighed ≈50% and 40% less, respectively, than control larvae. In comparison, larvae fed an artificial diet containing both StPin1A and NaPI were on average 90% smaller than control larvae.
Fig. 4.
Growth of H. armigera larvae on cotton leaf-based diets containing various PIs. Eggs were hatched, and neonates were transferred to artificial diets containing 0.3% (wt/vol) of each PI tested. (A) Larval weight gain was recorded every second day. The average weight (±SE) of 60 larvae per treatment is shown. (B) The average size of larvae recorded on day 11. E, casein; P, StPin1A; N, NaPI; NP, NaPI/StPin1A.
Coexpression of NaPI and StPin1A Improves Cotton Production Under Insect Pressure in the Field.
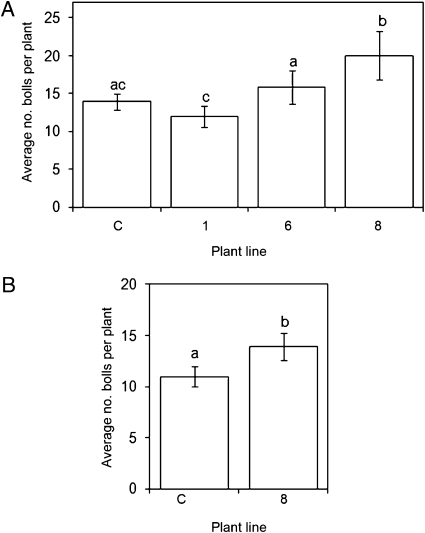
To assess the potential of NaPI and StPin1A in plant protection, genes encoding these proteins were transferred into cotton plants for field-trial assessment. Transgenic cotton plants expressing NaPI (line 1), StPin1A (line 6), or the combined NaPI-StPin1A (line 8) were tested for their performance in preventing plant damage caused by insect pests under field conditions in Queensland, Australia, during the 2004–2005 cotton-growing season. The major insect pests of cotton during the field trial period were H. armigera and H. punctigera, and the site was artificially infested with H. armigera eggs to maintain insect pressure. NaPI was highly expressed in the leaves of line 1 (1.0–1.3% of total soluble protein) and line 8 (0.4–0.6% of total soluble protein). Expression of StPin1A in line 8 was substantially lower than expression of NaPI, at 0.001–0.002% of total soluble protein. Cotton plants expressing both NaPI and StPin1A out-performed plants expressing these inhibitors alone, as measured by the number of cotton bolls per plant at maturity (Fig. 5A). Based on the strong performance of line 8 in the first field trial, homozygous line 8 cotton plants were produced for further testing in the 2006–2007 cotton-growing season in Queensland. Artificial insect infestation was not required because of the higher insect pressure; the major insect pests again were H. armigera and H. punctigera. Line 8, expressing StPin1A and NaPI, again out-performed the control plants, recording a statistically significant higher number of cotton bolls than the untransformed parental line (Figs. 5B and 6)and a higher lint weight per plant [27.8 ± 0.59 (SE) g for line 8; 22.9 ± 2.1 (SE) g for control line].
Fig. 5.
Mean number of cotton bolls per plant on transgenic cotton plant line 1 (NaPI, homozygous), line 6 (StPin1A, homozygous), line 8 (NaPI + StPin1A, hemizygous/homozygous), and untransformed parent line (C; control). (A) In field trials conducted in 2004–2005, hemizygous line 8 had a significantly higher number of bolls than all other lines (P < 0.001). (B) During the 2006–2007 field trial, homozygous line 8 also recorded significantly higher cotton boll numbers than the control line (P = 0.05). Different lowercase letters indicate significantly different values, and error bars indicate 95% confidence intervals.
Fig. 6.
Comparison of cotton bolls on field-grown transgenic cotton plants. (A) Coker 315 control plants have lower numbers of cotton bolls than (B) homozygous line 8 expressing NaPI and StPin1A.
Discussion
In this study and previously (27), we showed that ingestion of the pin II inhibitor NaPI has a much more severe impact on growth and mortality of the cotton pest H. punctigera than it does on the more serious pest H. armigera. In an attempt to improve the efficacy of NaPI against H. armigera, we tested the combination of StPin1A, a potent inhibitor of H. punctigera and H. armigera chymotrypsins, with NaPI. Artificial diet containing both inhibitors led to a marked decrease in growth rate that was not obtained with either inhibitor in isolation.
Control larvae that had not been exposed to NaPI had variable levels of NaPI-insensitive chymotrypsin activity in their gut, suggesting that the encoding gene is expressed constitutively. It is possible that the gene is also up-regulated after exposure to diets containing the inhibitor, but this possibility needs to be verified by using more larvae and examining the levels of the gene transcript. Resistance to NaPI is likely to be multitiered, as described for the cowpea bruchid and a plant cysteine PI (21), of which one component is the genetic predisposition to have high levels of the NaPI-resistant chymotrypsin.
Previous studies have demonstrated changes in chymotrypsin gene expression (17, 28) and induction of PI-resistant chymotrypsins (17, 29, 30) in response to consumption of PIs. PI-resistant chymotrypsins have been characterized only by their activity, and there are no reports of inhibitors that target the PI-resistant activity. In the current study we show that trypsin activity was severely reduced in NaPI-fed larvae, but the presence of NaPI-resistant trypsins was not investigated further. PI-resistant trypsins have been identified in numerous lepidopteran species (17, 20, 31) and have been characterized more thoroughly than PI-resistant chymotrypsins (29, 32). Volpicella and colleagues (32) discovered that PI-susceptible and PI-resistant trypsins have different substrate preferences. To determine if NaPI-resistant trypsins are present in Helicoverpa spp., we would use the substrates described by Volpicella (32) to distinguish between resistant and susceptible enzymes.
Generally, PIs are screened for their inhibitory activity against insect gut extracts before in vivo testing using artificial diets or transgenic plants. Sometimes PIs that perform well during in vitro inhibition assays do not perform well in subsequent bioassays (16). Our study highlights the importance of working with homogenous preparations of PIs that are identical to the PIs that will be used as potential transgenes.
Transgenic cotton expressing StPin1A from potato and NaPI from tobacco showed improved performance over 2 y of field trials. Pin I and pin II inhibitors accumulate naturally in the leaves of solanaceous plants in response to damage by insects or mechanical injury (33). The use of transgenes encoding PIs from structurally distinct families thus is more likely to provide better plant protection under field conditions. Although the level of StPin1A in the hemizygous cotton plants was ≈1,000-fold lower than NaPI the presence of StPin1A still improved protection against insects in field trials. Combinations of PIs that target different classes of digestive proteases have shown promising results in planta (34, 35), but individual inhibitors also have been reported to improve plant resistance to insects (36, 37).
PIs have the potential to enhance the current Bt toxin technology because they target a broader range of pests, including nematodes and fungi (38). There is a major concern that the effectiveness of Bt will be negated if field-evolved Bt resistance (39) becomes a more widespread problem. A proposed management strategy for delaying insects’ development of resistance to plant-protection transgenes, such as Bt toxins, is to deploy multiple insect-control genes (such as PIs) with different modes of action in a single plant (40). There is evidence that the combination of PIs with a sublethal-dose Bt toxin has a strong effect on the growth and development of insects (41).
Here we demonstrate the potential for using combinations of different classes of plant PIs to prevent crop damage caused by insects. The long-term aim of our approach is to select combinations of inhibitors that counter the genetic capacity of the target insect to produce various proteases under different pressures.
Materials and Methods
The synthetic substrate, N-succinyl-l-alanyl-l-alanyl-l-prolyl-phenylalanine-4-nitroanilide (succ-AAPF-pNA), α-chymotrypsin from bovine pancreas (Nα-Tosyl-l-lysine chloromethyl ketone hydrochloride treated), trypsin-agarose (N-p-Tosyl-l-phenylalanine chloromethyl ketone treated) and cyanogen bromide-activated Sepharose 4B were from Sigma-Aldrich. The NaPI series of 6-kDa chymotrypsin and trypsin inhibitors were purified from Nicotiana alata as described previously (23, 24). The Pin I inhibitor was from Calbiochem-Novabiochem. Recombinant StPin1A was supplied by Dr. Fung Lay of La Trobe University, Melbourne, Australia. All other PIs were from Sigma-Aldrich. Benzamidine agarose (35 μmol benzamidine/mL) was from MP Biomedicals.
Insect-Feeding Trials: Bioassay 1.
H. punctigera feeding trials were conducted as described previously (42), except that the artificial diet was prepared with freeze-dried cotton leaves as described for a potato leaf artificial diet (43). The gut was removed from early fifth-instar larvae and was homogenized in gut extraction buffer (500 μL ice-cold 10 mM Tris-HCl, pH 8). Insoluble material was removed by centrifugation (13,000 × g, 5 min), and the supernatant was stored at −80 °C before use in enzyme assays.
Insect-Feeding Trials: Bioassay 2.
H. armigera were raised on a cotton leaf-based artificial diet that was supplemented with NaPI (530 μM), StPin1A (530 μM), NaPI (530 μM) + StPin1A (530 μM), casein, or on an unsupplemented control diet. H. armigera neonates (60/treatment) were raised in individual microcentrifuge tubes (Sarstedt) with perforated lids. Weight gain was recorded on day 5 and every second day thereafter. Mortality was recorded for the first 5 d and then every second day thereafter. Artificial diet was replaced as required to provide a continuous supply. The larvae were kept at 25 °C with a 16-h/8-h light/dark cycle. H. armigera larvae were sourced from the Department of Primary Industries and Fisheries, Indooroopilly, Queensland, Australia. Statistical comparisons of larval weight were made by one-way ANOVA at a 99% confidence limit and Tukey–Kramer multiple-comparisons posttests, using StatPro software version 1.0 (Christopher Albright, http://www.kelley.iu.edu/albrightbooks/Free_downloads.htm).
Partial Purification of the NaPI-Insensitive Chymotrypsin for Inhibition Assays.
The midgut from 100 fourth-instar larvae were pooled and homogenized in 50 mL of ice-cold extraction buffer [10 mM Tris-HCl (pH 8.0), 5 mM EDTA, 10% (wt/vol) glycerol, 2% (wt/vol) polyvinyl pyrrolidine, 0.01% (wt/vol) NaN3] using a Sorvall Omnimixer. Insoluble material was removed by centrifugation (17,000 × g; 30 min; 4 °C) and filtration through Miracloth (Calbiochem). NaPI-sensitive proteases were removed by passage (×5) through an affinity column consisting of NaPI (C1, C2, T1–T4; 10 mg) cross-linked to cyanogen bromide-activated Sepharose 4B (1 g). The material that did not bind to the NaPI column was used to study the effect of a series of PIs on the activity of the NaPI-insensitive chymotrypsins.
Enzyme Activity and Inhibition by PIs.
Gut extracts from individual H. punctigera larvae from Bioassay 1 were assayed at pH 10 in 50 mM 3-(cyclohexylamino)-1-propanesulfonic acid using the chymotrypsin and trypsin substrates succ-AAPF-pNA and N-benzoyl-dl-arginine p-nitroanilide hydrochloride (BA-pNA), respectively. Gut extracts were preincubated with 80 nM of purified plant-derived T1 or C1 monomer for 30 min at 30 °C before the addition of substrate. The release of pNA was recorded at 405 nm on a SpectraMax 250 microtiter plate reader (Molecular Devices). Inhibition assays using StPin1A were performed using gut extracts from larvae raised on control diets.
Construction of the Binary Vectors and Transgenic Plant Lines.
DNA encoding the sequence of the NaPI gene (GenBank accession number AF105340) (44) and the StPin1A gene (GenBank accession number FJ839694) were amplified by PCR and cloned between Cauliflower Mosaic Virus (CaMV) 35S promoter and terminator sequences (45). The expression cassettes were inserted into the pBIN19 binary vector (GenBank accession number U12540) (46) and named “pHEX2” (NaPI) or “pHEX6” (StPin1A).
The binary vectors pHEX2 and pHEX6 were used to produce transgenic cotton plants by Agrobacterium-mediated transformation as described in (47) with modifications. A. tumefaciens strain LBA4404 containing the vector was used to infect hypocotyl sections of Gossypium hirsutum L. cv. Coker 315. Embryogenic callus was selected on the antibiotic kanamycin. Plantlets were transferred to soil and after acclimatization were transferred to a greenhouse. Plant lines expressing NaPI or StPin1A were identified by protein immunoblot analysis. To produce homozygous plant lines, a segregation analysis of kanamycin resistance was performed on the progeny of self-pollinated primary transformants. The hemizygous transgenic plant line 8 (NaPI-StPin1A) was produced by crossing the homozygous plant line 1 (NaPI) with the homozygous plant line 6 (StPin1A).
Field Evaluation of Transgenic Plants.
For the 2004–2005 field trial, the transgenic cotton lines 1 (homozygous), 6 (homozygous), and 8 (hemizygous) and the untransformed parent line Coker 315 were grown in the Darling Downs area of Queensland, Australia. Seed was planted by hand in three replicate plots, each plot containing 40 seed per variety. For the 2006–2007 field trial, the transgenic cotton line 8 and the untransformed parent line Coker 315 were grown in the Darling Downs region of Queensland, Australia. Seed was planted mechanically in four replicate plots per variety, each plot containing 80 seeds. Plant pests were monitored regularly throughout the growing season. In both trials, nonlepidopteran pests were controlled by the application of selective pesticides. Lepidopteran pests were controlled using a low-spray regimen consisting of three or four sprays. At weeks 6 and 12 in the 2004–2005 season, the expression of NaPI and StPin1A in the first fully expanded leaf from selected plants was determined by double-sandwich ELISA using polyclonal antibodies. At the completion of the trial, the number of mature open cotton bolls was counted on line 8, line 1, line 6 and Coker plants. Data were analyzed by ANOVA using SPSS statistics (SPSS Inc.).
Supplementary Material
Acknowledgments
We thank Prof. Adrienne Clarke, Prof. Bruce Stone, and Dr. Sonia Nikolovski for critical reading of the manuscript and Dr. Christopher Adda and Prof. Chris Lloyd for statistical analysis of the insect bioassay data. We also thank Maria Rainone and Bruce McGinness for production of transgenic plants and Dr. Jillian Hinch and Andrew Somerville for management of the field trials. K.M.D. was supported by a Cotton Research and Development Corporation Scholarship.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY618891, AY618895, and FJ839694).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009241107/-/DCSupplemental.
References
- 1.Fitt GP. The ecology of Heliothis species in relation to agroecosystems. Annu Rev Entomol. 1989;34:17–52. [Google Scholar]
- 2.Whalon M, Mota-Sanchez D, Hollingworth L, Duynslager L. Arthropod pesticide resistance database. East Lansing, MI: Michigan State University; 2004. Available at: http://www.pesticideresistance.org/search/12/0/41/0/. Accessed April 21, 2009. [Google Scholar]
- 3.United States Environmental Protection Agency 2009 Bacillus thuringiensis Cry1Ab delta-endotoxin protein and the genetic material necessary for its production (via elements of vector pZO1502) in event Bt11 Corn (OECD unique identifier: SYN-BTØ11-1)(006444) and Bacillus thuringiensis Vip3Aa20 insecticidal protein and the genetic material necessary for its production (via elements of vector pNOV1300) in event MIR162 maize (OECD unique identifier: SYN-IR162-4)(006599) and modified Cry3A protein and the genetic material necessary for its production (via elements of vector pZM26) in event MIR604 corn (OECD unique identifier: SYN-IR6Ø4-5)(006509). Fact sheet 006599-006444-006509. Available at: http://www.epa.gov/opp00001/biopesticides/ingredients/factsheets/factsheet_006599-006444.html.Accessed February 2, 2010. [Google Scholar]
- 4.Tabashnik BE, Gassmann AJ, Crowder DW, Carriére Y. Insect resistance to Bt crops: Evidence versus theory. Nat Biotechnol. 2008;26:199–202. doi: 10.1038/nbt1382. [DOI] [PubMed] [Google Scholar]
- 5.Tabashnik BE, et al. Asymmetrical cross-resistance between Bacillus thuringiensis toxins Cry1Ac and Cry2Ab in pink bollworm. Proc Natl Acad Sci USA. 2009;106:11889–11894. doi: 10.1073/pnas.0901351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green TR, Ryan CA. Wound-induced proteinase inhibitor in plant leaves: A possible defense mechanism against insects. Science. 1972;175:776–777. doi: 10.1126/science.175.4023.776. [DOI] [PubMed] [Google Scholar]
- 7.Atkinson AH, Heath RL, Simpson RJ, Clarke AE, Anderson MA. Proteinase inhibitors in Nicotiana alata stigmas are derived from a precursor protein which is processed into five homologous inhibitors. Plant Cell. 1993;5:203–213. doi: 10.1105/tpc.5.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graham JS, et al. Wound-induced proteinase inhibitors from tomato leaves. I. The cDNA-deduced primary structure of pre-inhibitor I and its post-translational processing. J Biol Chem. 1985;260:6555–6560. [PubMed] [Google Scholar]
- 9.Graham JS, et al. Wound-induced proteinase inhibitors from tomato leaves. II. The cDNA-deduced primary structure of pre-inhibitor II. J Biol Chem. 1985;260:6561–6564. [PubMed] [Google Scholar]
- 10.Bruxelles GL, Roberts MR. Signals regulating multiple responses to wounding and herbivores. Crit Rev Plant Sci. 2001;20:487–521. [Google Scholar]
- 11.Korth KL, Dixon RA. Evidence for chewing insect-specific molecular events distinct from a general wound response in leaves. Plant Physiol. 1997;115:1299–1305. doi: 10.1104/pp.115.4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryan CA, Pearce G. Systemin: A polypeptide signal for plant defensive genes. Annu Rev Cell Dev Biol. 1998;14:1–17. doi: 10.1146/annurev.cellbio.14.1.1. [DOI] [PubMed] [Google Scholar]
- 13.Kessler A, Baldwin IT. Plant responses to insect herbivory: The emerging molecular analysis. Annu Rev Plant Biol. 2002;53:299–328. doi: 10.1146/annurev.arplant.53.100301.135207. [DOI] [PubMed] [Google Scholar]
- 14.Engelberth J, Alborn HT, Schmelz EA, Tumlinson JH. Airborne signals prime plants against insect herbivore attack. Proc Natl Acad Sci USA. 2004;101:1781–1785. doi: 10.1073/pnas.0308037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Applebaum SW. Biochemistry of Digestion. In: Kerkut GA, Gilbert LI, editors. Comparative Physiology and Pharmacology of Insects. Vol. 4. Toronto: Pergamon; 1985. pp. 279–311. [Google Scholar]
- 16.Bonade-Bottino MA, Ceci LR, Gallerani R, Jouanin L, Jouanin L De Leo F. Opposite effects on Spodoptera littoralis larvae of high expression level of a trypsin proteinase inhibitor in transgenic plants. Plant Physiol. 1998;118:997–1004. doi: 10.1104/pp.118.3.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bown DP, Wilkinson HS, Gatehouse JA. Differentially regulated inhibitor-sensitive and insensitive protease genes from the phytophagous insect pest, Helicoverpa armigera, are members of complex multigene families. Insect Biochem Mol Biol. 1997;27:625–638. doi: 10.1016/s0965-1748(97)00043-x. [DOI] [PubMed] [Google Scholar]
- 18.Cloutier C, Jean C, Fournier M, Yelle S, Michaud D. Adult Colorado potato beetles, Leptinotarsa decemlineata compensate for nutritional stress on oryzacystatin I-transgenic potato plants by hypertrophic behavior and over-production of insensitive proteases. Arch Insect Biochem Physiol. 2000;44:69–81. doi: 10.1002/1520-6327(200006)44:2<69::AID-ARCH2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 19.Broadway RM. Dietary regulation of serine proteinases that are resistant to serine proteinase inhibitors. J Insect Physiol. 1997;43:855–874. doi: 10.1016/s0022-1910(97)00028-0. [DOI] [PubMed] [Google Scholar]
- 20.Bolter C, Jongsma MA. The adaptation of insects to plant protease inhibitors. J Insect Physiol. 1997;43:885–895. doi: 10.1016/s0022-1910(97)00040-1. [DOI] [PubMed] [Google Scholar]
- 21.Zhu-Salzman K, Koiwa H, Salzman RA, Shade RE, Ahn JE. Cowpea bruchid Callosobruchus maculatus uses a three-component strategy to overcome a plant defensive cysteine protease inhibitor. Insect Mol Biol. 2003;12:135–145. doi: 10.1046/j.1365-2583.2003.00395.x. [DOI] [PubMed] [Google Scholar]
- 22.Giri AP, et al. Chickpea defensive proteinase inhibitors can be inactivated by podborer gut proteinases. Plant Physiol. 1998;116:393–401. [Google Scholar]
- 23.Heath RL, et al. Characterization of the protease processing sites in a multidomain proteinase inhibitor precursor from Nicotiana alata. Eur J Biochem. 1995;230:250–257. doi: 10.1111/j.1432-1033.1995.tb20558.x. [DOI] [PubMed] [Google Scholar]
- 24.Lee MC, Scanlon MJ, Craik DJ, Anderson MA. A novel two-chain proteinase inhibitor generated by circularization of a multidomain precursor protein. Nat Struct Biol. 1999;6:526–530. doi: 10.1038/9293. [DOI] [PubMed] [Google Scholar]
- 25.Dunse KM, et al. Molecular basis for the resistance of an insect chymotrypsin to a potato type II proteinase inhibitor. Proc Natl Acad Sci USA. 2010 doi: 10.1073/pnas.1009327107. 10.1073/pnas1009327107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pouvreau L, et al. Relative abundance and inhibitory distribution of protease inhibitors in potato juice from cv. Elkana. J Agric Food Chem. 2001;49:2864–2874. doi: 10.1021/jf010126v. [DOI] [PubMed] [Google Scholar]
- 27.Anderson MA, et al. Proteinase inhibitors from Nicotiana alata enhance plant resistance to insect pests. J Insect Physiol. 1997;43:833–842. doi: 10.1016/s0022-1910(97)00026-7. [DOI] [PubMed] [Google Scholar]
- 28.Gatehouse LN, Shannon AL, Burgess EP, Christeller JT. Characterization of major midgut proteinase cDNAs from Helicoverpa armigera larvae and changes in gene expression in response to four proteinase inhibitors in the diet. Insect Biochem Mol Biol. 1997;27:929–944. doi: 10.1016/s0965-1748(97)00074-x. [DOI] [PubMed] [Google Scholar]
- 29.Volpicella M, et al. Identification and characterization of digestive serine proteases from inhibitor-resistant Helicoverpa zea larval midgut. J Chromatogr B Analyt Technol Biomed Life Sci. 2006;833:26–32. doi: 10.1016/j.jchromb.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 30.Mazumdar-Leighton S, Broadway RM. Identification of six chymotrypsin cDNAs from larval midguts of Helicoverpa zea and Agrotis ipsilon feeding on the soybean (Kunitz) trypsin inhibitor. Insect Biochem Mol Biol. 2001;31:633–644. doi: 10.1016/s0965-1748(00)00168-5. [DOI] [PubMed] [Google Scholar]
- 31.Broadway RM. Are insects resistant to plant proteinase inhibitors. J Insect Physiol. 1995;41:107–116. [Google Scholar]
- 32.Volpicella M, et al. Properties of purified gut trypsin from Helicoverpa zea, adapted to proteinase inhibitors. Eur J Biochem. 2003;270:10–19. doi: 10.1046/j.1432-1033.2003.03368.x. [DOI] [PubMed] [Google Scholar]
- 33.Gustafson G, Ryan CA. Specificity of protein turnover in tomato leaves. Accumulation of proteinase inhibitors, induced with the wound hormone, PIIF. J Biol Chem. 1976;251:7004–7010. [PubMed] [Google Scholar]
- 34.Amirhusin B, et al. Protease inhibitors from several classes work synergistically against Callosobruchus maculatus. J Insect Physiol. 2007;53:734–740. doi: 10.1016/j.jinsphys.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 35.Abdeen A, et al. Multiple insect resistance in transgenic tomato plants over-expressing two families of plant proteinase inhibitors. Plant Mol Biol. 2005;57:189–202. doi: 10.1007/s11103-004-6959-9. [DOI] [PubMed] [Google Scholar]
- 36.Duan X, et al. Transgenic rice plants harboring an introduced potato proteinase inhibitor II gene are insect resistant. Nat Biotechnol. 1996;14:494–498. doi: 10.1038/nbt0496-494. [DOI] [PubMed] [Google Scholar]
- 37.Vila L, et al. Expression of the maize proteinase inhibitor (mpi) gene in rice plants enhances resistance against the striped stem borer (Chilo suppressalis): Effects on larval growth and insect gut proteinases. Plant Biotechnol J. 2005;3:187–202. doi: 10.1111/j.1467-7652.2004.00117.x. [DOI] [PubMed] [Google Scholar]
- 38.Hilder VA, Boulter D. Genetic engineering of crop plants for insect resistance - a critical review. Crop Prot. 1999;18:177–191. [Google Scholar]
- 39.Tabashnik BE, Van Rensburg JB, Carrière Y. Field-evolved insect resistance to Bt crops: Definition, theory, and data. J Econ Entomol. 2009;102:2011–2025. doi: 10.1603/029.102.0601. [DOI] [PubMed] [Google Scholar]
- 40.Manyangarirwa W, Turnbull M, McCutcheon G, Smith J. Gene pyramiding as a Bt resistance management strategy: How sustainable is this strategy? African Journal of Biotechnology. 2006;5:781–785. [Google Scholar]
- 41.Zhu YC, Abel CA, Chen MS. Interaction of Cry1Ac toxin (Bacillus thuringiensis) and proteinase inhibitors on the growth, development, and midgut proteinase activities of the bollworm, Helicoverpa zea. Pestic Biochem Physiol. 2007;87:39–46. [Google Scholar]
- 42.Jennings C, West J, Waine C, Craik D, Anderson M. Biosynthesis and insecticidal properties of plant cyclotides: The cyclic knotted proteins from Oldenlandia affinis. Proc Natl Acad Sci USA. 2001;98:10614–10619. doi: 10.1073/pnas.191366898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gatehouse AMR, et al. Digestive proteolytic activity in larvae of tomato moth, Lacanobia oleracea; effects of plant protease inhibitors in vitro and in vivo. J Insect Physiol. 1999;45:545–558. doi: 10.1016/s0022-1910(98)00161-9. [DOI] [PubMed] [Google Scholar]
- 44.Miller EA, Lee MC, Atkinson AH, Anderson MA. Identification of a novel four-domain member of the proteinase inhibitor II family from the stigmas of Nicotiana alata. Plant Mol Biol. 2000;42:329–333. doi: 10.1023/a:1006305429013. [DOI] [PubMed] [Google Scholar]
- 45.Tabe LM, et al. A biotechnological approach to improving the nutritive value of alfalfa. J Anim Sci. 1995;73:2752–2759. doi: 10.2527/1995.7392752x. [DOI] [PubMed] [Google Scholar]
- 46.Bevan M. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 1984;12:8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Umbeck P, Johnson G, Barton K, Swain W. Genetically transformed cotton (Gossypium Hirsutum L.) plants. Nat Biotechnol. 1987;5:263–266. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.