Abstract
Vitamin K epoxide reductase (VKOR) sustains blood coagulation by reducing vitamin K epoxide to the hydroquinone, an essential cofactor for the γ-glutamyl carboxylation of many clotting factors. The physiological redox partner of VKOR remains uncertain, but is likely a thioredoxin-like protein. Here, we demonstrate that human VKOR has the same membrane topology as the enzyme from Synechococcus sp., whose crystal structure was recently determined. Our results suggest that, during the redox reaction, Cys43 in a luminal loop of human VKOR forms a transient disulfide bond with a thioredoxin (Trx)-like protein located in the lumen of the endoplasmic reticulum (ER). We screened for redox partners of VKOR among the large number of mammalian Trx-like ER proteins by testing a panel of these candidates for their ability to form this specific disulfide bond with human VKOR. Our results show that VKOR interacts strongly with TMX, an ER membrane-anchored Trx-like protein with a unique CPAC active site. Weaker interactions were observed with TMX4, a close relative of TMX, and ERp18, the smallest Trx-like protein of the ER. We performed a similar screen with Ero1-α, an ER-luminal protein that oxidizes the Trx-like protein disulfide isomerase. We found that Ero1-α interacts with most of the tested Trx-like proteins, although only poorly with the membrane-anchored members of the family. Taken together, our results demonstrate that human VKOR employs the same electron transfer pathway as its bacterial homologs and that VKORs generally prefer membrane-bound Trx-like redox partners.
Keywords: disulfide bond formation, warfarin, quinone, blood coagulation, electron transfer
Vitamin K epoxide reductase (VKOR) is a crucial enzyme in blood coagulation and the target of the most commonly used oral anticoagulant warfarin. VKOR catalyzes the reductive branch of the vitamin K cycle (for review, see refs. 1 and 2). The cycle begins with the vitamin K-dependent γ-glutamyl carboxylase modifying select glutamate residues in blood-clotting factors, a modification required for their activation at sites of tissue injury (for review, see ref. 3). During the carboxylation reaction, vitamin K hydroquinone is converted to the epoxide form, and VKOR is needed to regenerate the hydroquinone, thus completing the cycle. The VKOR reaction requires cooperation with a redox partner that delivers reducing equivalents. The redox partner remains uncertain, although protein disulfide isomerase (PDI), a thioredoxin (Trx)-like protein in the endoplasmic reticulum (ER) lumen, has been proposed to serve this role (4, 5).
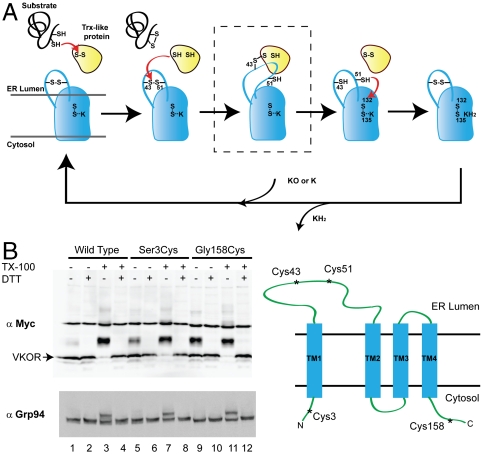
Homologs of VKOR have been identified in many organisms, even species that do not have blood (6–8). These include several classes of bacteria in which these enzymes are involved in disulfide bridge formation of newly synthesized secretory proteins (7–10). The reduced cysteines of exported proteins are initially oxidized by a Trx-like protein in the periplasm, leading to the reduction of the disulfide bridge in the active site CXXC motif of the Trx-like protein. VKOR then couples the reoxidation of the CXXC motif in the Trx-like protein with reduction of a quinone, such as vitamin K2 (menaquinone), to the hydroquinone (Fig. 1A). A recent crystal structure of the VKOR homolog from Synechococcus sp., in which the enzyme and a Trx-like domain are contained in a single polypeptide chain, revealed many details of this reaction (9). In a first step (Fig. 1A), the reduced CXXC motif of the Trx-like protein domain transfers electrons to two conserved cysteines in a periplasmic loop of VKOR (Fig. S1); this loop is located between transmembrane (TM) segments 1 and 2 of the four-TM bundle surrounding the quinone. The reaction proceeds through a mixed disulfide bridge intermediate, in which the N-terminal cysteine in the CXXC motif of the Trx-like domain is linked with the N-terminal loop cysteine in VKOR (Fig. 1A; boxed intermediate). This mixed disulfide bridge is subsequently resolved by attack of the C-terminal cysteine in the CXXC motif of the Trx-like domain, resulting in its oxidation with concomitant reduction of the VKOR loop cysteines. In the next step, the electrons are transferred from the loop cysteines to a CXXC motif at the periplasmic edge of the fourth TM of VKOR (Fig. 1A). Finally, the disulfide bridge in the CXXC motif of VKOR is regenerated by transfer of electrons to vitamin K. The hydroquinone leaves into the lipid phase and is reoxidized by the electron transfer chain.
Fig. 1.
Electron transfer pathway and membrane topology of human VKOR. (A) The scheme shows how electrons flow from reduced cysteines in newly synthesized proteins in the ER lumen through a Trx-like protein and VKOR to reduce either vitamin K epoxide (KO) to the quinone (K) or the quinone to the hydroquinone (KH2). The reaction proceeds through the boxed intermediate in which the N-terminal cysteine of the CXXC motif in the Trx-like protein forms a mixed disulfide bond with Cys43 of VKOR. Other intermediates must exist, but have not yet been demonstrated. When the reaction starts with KO, the cycle needs to be repeated to generate KH2. (B) Myc-tagged versions of either wild-type VKOR or of VKOR with cysteines introduced at either the N (Ser3Cys) or C (Gly158Cys) terminus were expressed in COS-7 cells. The cells were permeabilized with digitonin, and treated with Mal-PEG in the presence or absence of DTT or Triton X-100 (TX-100). The proteins were separated by SDS-PAGE and analyzed by immunoblotting with Myc or Grp94 antibodies. The right panel shows the derived membrane topology of human VKOR.
A similar reaction mechanism has been proposed for the mammalian VKOR, with a Trx-like protein in the ER lumen serving as its redox partner (4, 5, 9). However, conflicting topology models with three or four TM segments have been proposed for human VKOR (6, 9, 11); the loop cysteines interacting with the Trx-like protein would thus be on either the cytosolic or luminal side of the ER membrane. In addition, both the cytosolic thioredoxin protein and the luminal PDI protein have been shown to support the VKOR reaction in vitro (4, 5, 12, 13). The recent crystal structure of a bacterial VKOR homolog and multiple sequence alignments suggest that the mammalian VKORs may also conform to a four-TM model, which would necessitate an ER-luminal redox partner (6, 9). However, there are many potential redox partners, as the mammalian ER contains 20 proteins with a Trx-like fold, 12 of which also have at least one CXXC motif (for review, see ref. 14). The most abundant and best-studied member of this family is PDI, which is a major catalyst of disulfide bridge formation in newly synthesized secretory and membrane proteins. After donating its disulfide bridge to a substrate, PDI is reoxidized by Ero1-α, an ER-luminal protein that uses FAD and molecular oxygen in the reaction (15–17). Another member of the ER family of Trx-like proteins, ERp57, has been implicated in the oxidation of glycosylated substrates maintained in the calnexin/calreticulin folding cycle (18). Conflicting results exist on whether ERp57 is reoxidized by Ero1-α as well (19–21). Although some of the other Trx-like ER proteins have been characterized with respect to redox potential and possible substrates, their biological functions and physiological oxidants remain unknown.
Here, we demonstrate that the human VKOR indeed contains four TM segments, indicating that it interacts with an ER-luminal redox partner. To identify this partner, we screened a panel of ER-luminal Trx-like proteins for their ability to form a key reaction intermediate, a mixed disulfide bridge between a cysteine in the CXXC motif of the Trx-like protein and a cysteine in the luminal loop of VKOR. Our results identify TMX, an ER membrane-anchored Trx-like protein with a unique CPAC active site, as a major interaction partner of VKOR. In contrast, Ero1-α prefers soluble over membrane-bound Trx-like proteins. These data indicate that human VKOR employs the same electron transfer pathway as its bacterial homologs, consistent with a general role in oxidative protein folding, and that VKORs generally prefer membrane-bound Trx-like redox partners.
Results
Given the conflicting reports in the literature, we first sought to clarify the membrane topology of human VKOR. The proposed three- and four-TM models differ in the location of the N terminus of the protein; it would be in the ER lumen in the three-TM model and in the cytosol in the four-TM model. We determined the membrane topology of human VKOR with a membrane-impermeable cysteine modification reagent. To this end, Myc-tagged versions of either wild-type VKOR or of mutant VKOR with cysteines introduced at either the N or C terminus were expressed in COS-7 cells. The cells were permeabilized with digitonin, which generates holes in the plasma membrane, but leaves the ER membrane intact. The modification reagent maleimide-polyethyleneglycol (Mal-PEG) was then added and, after quenching the reaction, the proteins were separated by SDS-PAGE and analyzed by immunoblotting with Myc antibodies. Each cysteine modified by Mal-PEG is expected to increase the molecular weight of VKOR by ∼5 kDa; modification in the presence of the quencher DTT should prevent the molecular weight shift. With wild-type VKOR, little modification was observed (Fig. 1B, lane 1). However, extensive modification was observed when the ER membrane was solubilized with Triton X-100 (lane 3 versus lane 4), as expected from the presence of multiple endogenous cysteines located inside the ER membrane and in the ER lumen. By contrast, when a cysteine was introduced at the N terminus (Ser3Cys), it was readily modified by Mal-PEG (lane 5), indicating that the N terminus is located in the cytosol. Similar results were obtained with a cysteine introduced at the C terminus (Gly158Cys), consistent with the expectation that it is also located in the cytosol. Control experiments with the luminal ER protein Grp94 showed that its single cysteine can only be modified when the membrane is solubilized by TX-100 (lanes 3, 7, and 11). These data demonstrate that both termini of VKOR are located in the cytosol, supporting a four-TM model for human VKOR and suggesting that the two loop cysteines engage a redox partner located in the ER lumen (Fig. 1B, Right).
To identify potential redox partners of human VKOR among the Trx-like proteins of the ER, we exploited a strategy that is based on the established electron transfer pathway in bacteria (9). The correct interaction partners are expected to form a transient mixed disulfide bridge, in which the N-terminal cysteine in the CXXC motif of the Trx-like protein and the N-terminal loop cysteine of VKOR (Cys43) are covalently linked (boxed intermediate in Fig. 1A). This intermediate can be stabilized by mutating either the C-terminal loop cysteine of VKOR (Cys51Ala), the C-terminal cysteine in the CXXC motif of the Trx-like protein, or both. These mutations slow the resolution of the mixed disulfide bridge and help to maintain the complex between the redox partners.
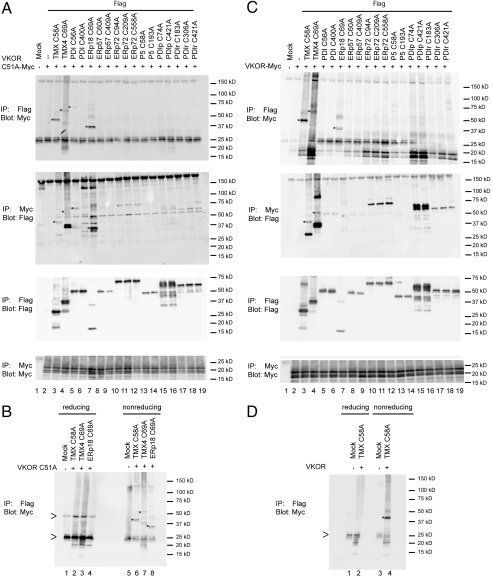
We cloned nine CXXC-containing mammalian Trx-like ER proteins, including two membrane-anchored proteins (TMX and TMX4) and the soluble PDI, ERp18, ERp57, ERp72, P5, PDIp, and PDIr proteins (see Table S1 for summary). The remaining three Trx-like proteins with CXXC motifs were excluded because they did not express well. As many of these proteins have multiple CXXC motifs that could potentially interact with VKOR, each of them was individually mutated to CXXA, resulting in a total of 17 constructs. FLAG-tagged versions of these proteins were coexpressed in COS-7 cells with Myc-tagged VKOR carrying the Cys51Ala mutation. The cells were treated with N-ethylmaleimide (NEM) to prevent disulfide rearrangements, and a cell lysate was subjected to denaturing immunoprecipitation with either FLAG or Myc antibodies. Following nonreducing SDS-PAGE, the samples were analyzed by immunoblotting with FLAG or Myc antibodies. When the Trx-like proteins were pulled down with FLAG antibodies and analyzed for interaction with VKOR (Cys51Ala)-Myc, prominent adducts of the expected molecular weights were seen for TMX and ERp18 (Fig. 2A, first panel, lanes 3 and 7). For TMX4 and the first CXXC motif of PDI, weak cross-links with VKOR (Cys51Ala) were observed (lanes 4 and 5). None of the other Trx-like proteins showed significant interactions (Fig. 2A, first panel). In the reciprocal pull-down experiment, the background was generally higher, but the same adducts were detected (Fig. 2A, second panel). The strongest three adducts, observed with TMX, TMX4, and ERp18, were sensitive to treatment with a reducing agent, as expected for a mixed disulfide bridge between a Trx-like protein and VKOR (Fig. 2B). Immunoblotting with FLAG antibodies after immunoprecipitation with FLAG antibodies showed that the Trx-like proteins were expressed and pulled down at similar levels (Fig. 2A, third panel). An equivalent conclusion could be derived for the VKOR proteins on the basis of experiments with Myc antibodies (Fig. 2A, fourth panel).
Fig. 2.
Screen for disulfide bridge formation between VKOR and Trx-like ER proteins. (A) FLAG-tagged Trx-like ER proteins bearing single CXXA mutations were coexpressed in COS-7 cells with Myc-tagged human VKOR carrying the Cys51Ala mutation. The cells were treated with NEM and lysates subjected to denaturing immunoprecipitation (IP) with either FLAG or Myc antibodies. Following nonreducing SDS-PAGE, the samples were analyzed by immunoblotting with FLAG or Myc antibodies. The stars indicate the positions of adducts between a Trx-like protein and VKOR (Cys51Ala). Note that non-cross-linked TMX4 precipitated by FLAG antibodies was detected nonspecifically by Myc antibodies (lane 4). However, because this band was seen even in the absence of VKOR (Fig. S2), this does not affect our conclusions concerning an interaction of TMX4 with VKOR. (B) Samples showing the strongest cross-links in the upper panel of A were reanalyzed by reducing and nonreducing SDS-PAGE, followed by immunoblotting with Myc antibodies. The arrows indicate the positions of the heavy and light chains of immunoglobulin used in IP. (C) As in A, but with wild-type VKOR. (D) The sample showing the strongest cross-link in the upper panel of C was reanalyzed by reducing and nonreducing SDS-PAGE, followed by immunoblotting with Myc antibodies.
Because it seemed possible that the double-mutant combination would force nonphysiological disulfide bridge formation between a Trx-like protein and VKOR, we performed more stringent experiments with wild-type VKOR. When the Trx-like proteins carrying CXXA mutations were pulled down with FLAG antibodies and analyzed for interaction with VKOR-Myc, a prominent adduct was seen with TMX (Fig. 2C, first panel, lane 3). This band disappeared upon reduction of the disulfide bond (Fig. 2D). Weaker adducts were seen with TMX4 and ERp18 (lanes 4 and 7). In the reciprocal pull-down experiments, cross-links to TMX and TMX4 were visible (second panel, lanes 3 and 4), but the ones to ERp18 were barely detectable (lane 7). All constructs were expressed and immunoprecipitated at about the same levels (third and fourth panels). Taken together, these experiments indicate that VKOR has a preference for the membrane-anchored Trx-like ER proteins, particularly for TMX.
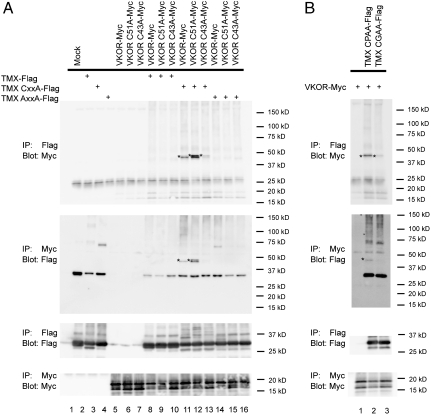
Next we characterized the formation of a mixed disulfide bridge between VKOR and TMX in more detail. As observed before, TMX with a CXXA mutation formed a covalent complex with either wild-type VKOR or a VKOR mutant in which loop Cys51 was mutated to Ala (Fig. 3A, first and second panels, lanes 11 and 12). When the other loop cysteine (Cys43) of VKOR was mutated, only weak cross-links to TMX (CXXA) were observed (lane 13). These data support the proposed electron transfer pathway, in which Cys43 would engage the CXXC motif of the Trx-like protein (Fig. 1A). As expected, when both cysteines in the CXXC motif of TMX were mutated, no cross-links were seen to any of the VKOR constructs (lanes 14–16). Likewise, for TMX with a wild-type CXXC motif, no adducts with the VKOR constructs could be detected (Fig. 3A, first and second panels, lanes 8–10), indicating that mutation of the second cysteine in the motif is required to capture the disulfide-bonded intermediate.
Fig. 3.
Characterization of the interaction between VKOR and TMX. (A) FLAG-tagged wild-type TMX or TMX mutants that carry mutations in its CXXC motif (CXXA or AXXA) were coexpressed with Myc-tagged wild-type VKOR or VKOR mutants in which the loop cysteines were changed to alanines (C43A or C51A). Controls were performed with expression of only one or neither (mock) of the two partners. The cells were treated with NEM and lysates subjected to denaturing immunoprecipitation (IP) with either FLAG or Myc antibodies. Following nonreducing SDS-PAGE, the samples were analyzed by immunoblotting with FLAG or Myc antibodies. The stars indicate the positions of adducts between TMX and VKOR. The plus sign indicates a dimer of the TMX AXXA mutant, which is seen even when it is expressed on its own. (B) The interaction of wild-type VKOR with a TMX mutant, in which the proline at the second position of the CXXC motif was altered (CPXA to CGXA), was analyzed as in A.
TMX and its paralog TMX4 are unusual among mammalian Trx-like proteins of the ER in that they have a proline residue at the second position of a CXXC motif. We therefore tested whether the proline is required for complex formation with VKOR. Indeed, its mutation to glycine greatly reduced the interaction of TMX with wild-type VKOR (Fig. 3B).
All the Trx-like proteins interacting with VKOR (TMX, TMX4, and ERp18) contained only one Trx domain, whereas the other candidate proteins had multiple Trx-like domains. As only one of the CXXC motifs in the multidomain Trx-like proteins was mutated to CXXA, it seemed possible that a transient cross-link with VKOR was missed because the disulfide bond was readily resolved by a neighboring wild-type CXXC motif. We therefore mutated both CXXC motifs to CXXA in each of the candidate redox partners containing two catalytic Trx folds. As before, no complex formation was observed (Fig. S3), indicating that these proteins indeed do not interact with VKOR.
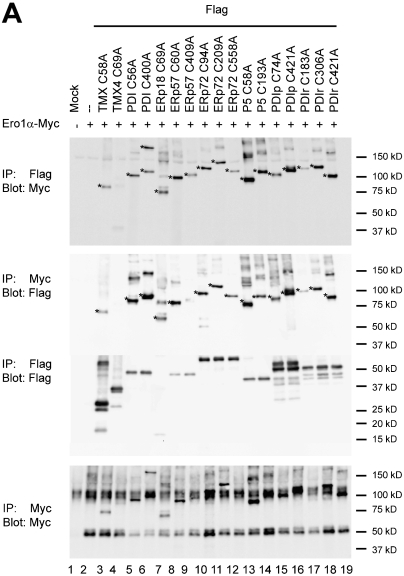
Having observed considerable specificity of VKOR for a Trx-like ER protein, we next tested the major oxidase of the ER lumen, Ero1-α, in a similar screen for redox partners. We coexpressed a Myc-tagged version of wild-type Ero1-α with FLAG-tagged Trx-like proteins carrying a CXXA mutation. Complexes of the expected molecular weight were observed with all tested constructs (Fig. 4, first and second panels), with the exception of TMX4 (lane 4). The cross-links with the membrane-anchored TMX protein were weak (lane 3). In all cases where Trx-like proteins contained multiple CXXC motifs, one of them interacted significantly stronger than TMX. These data suggest that Ero1-α interacts rather promiscuously with Trx-like ER proteins, with a bias against membrane-anchored members of the family.
Fig. 4.
Screen for disulfide bridge formation between Ero1-α and Trx-like ER proteins. FLAG-tagged Trx-like ER proteins bearing single CXXA mutations were coexpressed in COS-7 cells with Myc-tagged Ero1-α. The cells were treated with NEM and lysates subjected to denaturing immunoprecipitation (IP) with either FLAG or Myc antibodies. Following nonreducing SDS-PAGE, the samples were analyzed by immunoblotting with FLAG or Myc antibodies. The stars indicate the positions of adducts between a Trx-like protein and Ero1-α.
Discussion
Our results indicate that human VKOR has a four-TM topology, consistent with multiple sequence alignments (6) and the structure of the catalytic core of a bacterial homolog (9). In contrast to the previous three-TM model (11), this topology places most warfarin-resistance mutations close to the ER-luminal side of the vitamin K binding site and to the ER-luminal loop between TM segments 1 and 2. A four-TM topology also means that the two loop cysteines must receive electrons from an ER-luminal redox partner.
Our screen among potential Trx-like partners of the ER showed that VKOR preferentially forms a covalent complex with the membrane-anchored TMX protein, although weaker interactions were seen with its paralog TMX4 and with the soluble ERp18 protein. No interactions were seen between wild-type VKOR and the other tested Trx-like proteins, even though these proteins were highly expressed. These results attest to the specificity of the observed interactions and are in contrast to the results with the Ero1-α oxidase, which reacted more indiscriminately with Trx-like proteins in the ER. The cysteines forming the mixed disulfide bridge between VKOR and the Trx-like proteins (Cys43 in VKOR and the first cysteine in the CXXC motif of the Trx-like proteins) are equivalent to those linking the redox partners in bacteria (9, 10). Indeed, when the critical loop cysteine Cys43 of VKOR was mutated, the identified redox partners TMX, TMX4, and ERp18 no longer formed cross-links with VKOR (Fig. 3 and Fig. S4). By contrast, strong cross-links were not only observed with wild-type VKOR, but also when Cys51 was mutated. Although it is expected that this mutation would hinder the resolution of the intermolecular disulfide bridge once it is formed, the generation of such a cross-link to a CXXA-containing Trx-like protein would require an oxidizing agent, likely molecular oxygen or glutathione. A similar observation was made in Escherichia coli, where the analogous mutations lead to in vivo cross-linking between the oxidoreductase DsbB and its Trx-like redox partner DsbA (22). Both the cross-linking of VKOR to a limited set of Trx-like proteins and the specific cysteines involved argue against a random encounter of VKOR with the identified redox partners and suggest that the observed complexes are physiological intermediates in the electron transfer pathway, a pathway that seems to be conserved for all VKORs. Our experiments indicate that the two loop cysteines (Cys43 and Cys51) play an important role in electron transfer. Previous studies in which mutation of these cysteines had no effect on vitamin K epoxide reduction (23, 24) can be explained by the use of DTT as the source of redox equivalents, which bypassed the luminal redox partner and directly reduced the VKOR active site.
VKOR’s redox partners TMX and TMX4 span the ER membrane once and have a single CXXC motif in the ER lumen (25–28). It seems likely that VKOR interacts with the TM segment of these proteins because a similar interaction is seen in the structure of the Synecochoccus sp. homolog (9). In this case, the four-TM bundle of the bacterial VKOR is followed by an additional TM segment that links the enzyme to a periplasmic Trx-like domain. This fifth TM segment does not directly contribute to the enzymatic mechanism, but interacts with the core of VKOR and positions the Trx-like domain for catalysis. Several other bacterial and plant species also have VKORs that are fused through an additional TM segment with a periplasmic/ER-luminal Trx-like domain (6–8). However, there are many species in which VKORs must cooperate with a separately encoded redox partner. As for human VKOR, the preferred Trx-like protein might be expected to have a TM segment. The interaction of VKOR with a membrane-anchored Trx-like protein could accelerate the redox reaction and increase specificity. An association between the redox partners is also consistent with previous proposals that VKOR is contained in a complex (hence the gene name “VKORC1,” which stands for VKOR complex subunit 1) (29).
Another interesting general feature of the VKOR-interacting Trx-like proteins is the presence of a proline at the second position of the CXXC motif. TMX and TMX4 are unusual among mammalian Trx-like ER proteins in sharing this feature, and most bacterial Trx-like proteins fused to VKORs also have a CPXC motif. Our data show that the proline is in fact important for the formation of a mixed disulfide intermediate with VKOR. An important role for the proline at the second position of the CXXC motif has been demonstrated for DsbA, the major oxidizing Trx-like protein in E. coli, and a CPXC motif is a factor contributing to the oxidizing power of thioredoxin that is targeted to the periplasmic space of bacteria (30, 31). Its presence in VKOR-interacting redox partners is consistent with a role in oxidative protein folding.
In fact, VKOR may have a more general role in disulfide bridge formation of newly synthesized proteins than hitherto appreciated. Both mice and flies lacking Ero1 are viable with only subtle phenotypes (32, 33), indicating that ER-localized Trx-like proteins can be oxidized by alternative enzymes (34). The only other known oxidases besides VKOR and Ero1 are the QSOX proteins. However, these Erv2p-related flavoproteins have their own Trx-like redox partners (35–37), whose substrate range may not be broad enough to encompass all proteins undergoing oxidative folding. Although most highly expressed in the liver, VKOR is found in many different tissues. In addition, the closely related VKOR-like protein is also ubiquitously expressed. It therefore seems possible that there are several parallel pathways of disulfide bridge formation, each involving an oxidase (Ero1, QSOX, or VKOR) and specific Trx-like redox partners.
Our results show that human VKOR can also interact with one of the soluble Trx-like ER proteins, ERp18. This protein is the simplest of all Trx-like ER proteins, consisting of a Trx-like fold with a small insertion (38–40). It is not predicted to have extensive peptide-binding domains, which may result in a more promiscuous interaction with proteins containing a free thiol group. In fact, it also reacted efficiently with the luminal ER sulfhydryl oxidase Ero1-α, in agreement with a previous report (20). Our results do not support a major role for PDI in the VKOR reaction (4, 5). The reported inhibitory effects of bacitracin and PDI depletion could be due to pleiotropic effects (41), and the observation of a complex between PDI and VKOR under native conditions may result from chaperone activity of PDI and disulfide rearrangement.
The identification of redox partners of VKOR may also have implications for blood coagulation in humans and for recombinant production of human clotting factors. Although no mutations in TMX or TMX4 have been associated with bleeding disorders, this may be because both enzymes are expressed in liver and are at least partly redundant. It is interesting that several of the warfarin-resistance mutations map to the ER-luminal loop, relatively far away from the quinone-binding site, and likely affect electron transfer from the Trx-like protein to vitamin K. Future work will be required to fully characterize the function of TMX and TMX4 in the vitamin K cycle.
Materials and Methods
Plasmid Construction and Mutagenesis.
Full-length human cDNA clones for PDI, PDIp, P5, ERp18, ERp57, ERp72, TMX, and TMX4 and mouse PDIr were obtained from Open Biosystems. Each gene was amplified using primers designed to incorporate a C-terminal FLAG tag and to facilitate directional topoisomerase cloning into the pcDNA3.1/V5-His TOPO TA expression vector (Invitrogen). Primer sequences for subcloning are provided in Table S2. The C-terminal cysteine of each CXXC motif was mutated to alanine by site-directed mutagenesis. For TMX, an AXXA mutant was similarly generated. All mutagenesis primers are provided in Table S3.
Ero1α-myc in the pcDNA3.1 vector has been previously described (42) and was kindly provided by Roberto Sitia. Human VKORC1-MYC-FLAG construct was obtained in the pCMV6 entry expression vector (OriGene); the VKORC1-MYC construct was generated by inserting a stop codon prior to the FLAG tag via site-directed mutagenesis. Single cysteine residues were added or mutated by site-directed mutagenesis. The primer sequences are given in Table S3. All clones were verified by restriction digestion followed by direct sequencing of the entire ORF.
Cell Culture and Transfection.
COS-7 cells were cultured in DMEM (Gibco) containing 10% defined FBS (HyClone) and 100 U/mL of both penicillin and streptomycin. All cells were grown at 37 °C in a humidified incubator under 5% CO2. COS-7 cells were transfected at 70% confluence using Lipofectamine 2000 (Invitrogen) and Opti-MEM (Invitrogen) according to the manufacturer’s specifications.
Denaturing Immunoprecipitation and Immunoblotting.
COS-7 cells were washed in situ three times in freshly prepared phosphate buffered saline (pH 7.4) containing 40 mM NEM (Sigma) and then incubated in 5 mL of identical buffer for 30 min on ice. Cells were then scraped, gently pelleted, and lysed in 10 cell volumes of denaturing lysis buffer [50 mM Tris·HCl (pH 6.8), 2% sodium dodecyl sulfate, 40 mM NEM, and Complete protease inhibitor cocktail (Roche)]. Samples were denatured at 60 °C for 10 min, and then sonicated with three brief pulses at 40% amplitude (Branson). After an additional 10 min incubation on ice, insoluble material was pelleted at 10,000 × g for 10 min at 4 °C. The resulting supernatant was diluted 20-fold into IP buffer [50 mM Tris·HCl (pH 6.8), 150 mM NaCl, 1% Nonidet P-40, 40 mM NEM], allowed to stand 10 min on ice, and then centrifuged at 10,000 g at 4 °C. The lysate was then precleared with 80 μL of prewashed protein G-Sepharose beads (GE Healthcare) by rotating 1 h at 4 °C and centrifugation at 10,000 × g. One half of the clarified lysate was incubated with monoclonal anti-Myc (9E10, Roche) antibodies, the other with anti-Flag (M2, Sigma) antibodies (1 mg/mL stock), both at a 1∶100 dilution. Samples were gently rotated 16 h at 4 °C, after which 40 μL of prewashed protein G-Sepharose beads were added to the samples for a final hour of rotation. Beads were pelleted by spinning at 2,500 × g for 3 min at 4 °C and washed three times with 1 mL of cold IP buffer. After the final wash, beads were resuspended in 50 μL of 2x nonreducing SDS sample buffer, incubated 10 min at 60 °C, and removed by pelleting at 10,000 × g for 3 min. Samples were separated by SDS-PAGE, transferred to PVDF membranes, and immunoblotted using either anti-Flag M2-peroxidase (Sigma, 1∶2,000) or a combination of rabbit anti-Myc (Sigma, 1∶2,000) and goat anti-rabbit IgG-peroxidase (Sigma, 1∶2,000). Blots were visualized by chemiluminescence on a Fujifilm LAS-3000 imager using Western Lightning Plus-ECL (Perkin Elmer).
VKOR Membrane Topology Determination.
Topology determination by Mal-PEG accessibility was carried out essentially as previously described (43). In brief, COS-7 cells were transfected with wild-type or mutant (Ser3Cys or Gly158Cys) VKOR-Myc. After 48 h, cells were rinsed in situ with DMEM, trypsinized, and washed twice with HCN buffer (50 mM Hepes, pH 7.5, 150 mM NacCl, 2 mM CaCl2), and pelleted at 1,000 × g for 2 min. Cells were then semipermeabilized in 0.02% digitonin or solubilized in 1% Triton X-100 on ice for 20 min (in HCN buffer supplemented with a protease inhibitor cocktail). One millimolar Mal-PEG 5 kDa (Aektar) was then added to each sample with or without 20 mM DTT for 30 min on ice, after which all samples were brought to a final concentration of 1% TX-100/20 mM DTT and incubated an additional 15 min on ice. Lysates were clarified by spinning at 12,000 × g for 10 min and the proteins in the supernatants were separated by SDS-PAGE. Samples were analyzed by immunoblotting with anti-Myc 9E10 monoclonal (Roche) or anti-Grp94 (Santa Cruz) antibodies.
Supplementary Material
Acknowledgments.
We thank J. Beckwith, R. J. Dutton, and D. Boyd for insightful discussion and critical reading of the manuscript, Y. Shibata for invaluable technical advice, and R. Sitia for kindly providing the Ero1a plasmid. S.S. is supported by Medical Scientist Training Program fellowship Award T32GM07753 from the National Institute of General Medical Sciences. W.L. is supported by a Charles King Trust fellowship and K99 Grant 1K99HL097083 from the National Heart, Lung, and Blood Institute (NIH). T.A.R. is an Howard Hughes Medical Institute investigator.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009972107/-/DCSupplemental.
References
- 1.Oldenburg J, et al. VKORC1: Molecular target of coumarins. J Thromb Haemost. 2007;1(Suppl 5):1–6. doi: 10.1111/j.1538-7836.2007.02549.x. [DOI] [PubMed] [Google Scholar]
- 2.Tie JK, Stafford DW. Structure and function of vitamin K epoxide reductase. Vitam Horm. 2008;78:103–130. doi: 10.1016/S0083-6729(07)00006-4. [DOI] [PubMed] [Google Scholar]
- 3.Furie B, Bouchard BA, Furie BC. Vitamin K-dependent biosynthesis of gamma-carboxyglutamic acid. Blood. 1999;93:1798–1808. [PubMed] [Google Scholar]
- 4.Soute BA, et al. Stimulation of the dithiol-dependent reductases in the vitamin K cycle by the thioredoxin system. Strong synergistic effects with protein disulphide-isomerase. Biochem J. 1992;281(Pt 1):255–259. doi: 10.1042/bj2810255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wajih N, Hutson SM, Wallin R. Disulfide-dependent protein folding is linked to operation of the vitamin K cycle in the endoplasmic reticulum. A protein disulfide isomerase-VKORC1 redox enzyme complex appears to be responsible for vitamin K1 2,3-epoxide reduction. J Biol Chem. 2007;282:2626–2635. doi: 10.1074/jbc.M608954200. [DOI] [PubMed] [Google Scholar]
- 6.Goodstadt L, Ponting CP. Vitamin K epoxide reductase: Homology, active site and catalytic mechanism. Trends Biochem Sci. 2004;29:289–292. doi: 10.1016/j.tibs.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Dutton RJ, et al. Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation. Proc Natl Acad Sci USA. 2008;105:11933–11938. doi: 10.1073/pnas.0804621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh AK, Bhattacharyya-Pakrasi M, Pakrasi HB. Identification of an atypical membrane protein involved in the formation of protein disulfide bonds in oxygenic photosynthetic organisms. J Biol Chem. 2008;283:15762–15770. doi: 10.1074/jbc.M800982200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li W, et al. Structure of a bacterial homologue of vitamin K epoxide reductase. Nature. 2010;463:507–512. doi: 10.1038/nature08720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dutton RJ, et al. Inhibition of bacterial disulfide bond formation by the anticoagulant warfarin. Proc Natl Acad Sci USA. 2010;107:297–301. doi: 10.1073/pnas.0912952107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tie JK, et al. Membrane topology mapping of vitamin K epoxide reductase by in vitro translation/cotranslocation. J Biol Chem. 2005;280:16410–16416. doi: 10.1074/jbc.M500765200. [DOI] [PubMed] [Google Scholar]
- 12.Johan L, et al. Vitamin K-dependent carboxylase. Possible role for thioredoxin in the reduction of vitamin K metabolites in liver. FEBS Lett. 1987;222:353–357. doi: 10.1016/0014-5793(87)80401-5. [DOI] [PubMed] [Google Scholar]
- 13.Silverman RB, Nandi DL. Reduced thioredoxin: A possible physiological cofactor for vitamin K epoxide reductase. Further support for an active site disulfide. Biochem Biophys Res Commun. 1988;155:1248–1254. doi: 10.1016/s0006-291x(88)81274-9. [DOI] [PubMed] [Google Scholar]
- 14.Hatahet F, Ruddock LW. Protein disulfide isomerase: A critical evaluation of its function in disulfide bond formation. Antioxid Redox Sign. 2009;11:2807–2850. doi: 10.1089/ars.2009.2466. [DOI] [PubMed] [Google Scholar]
- 15.Tu BP, et al. Biochemical basis of oxidative protein folding in the endoplasmic reticulum. Science. 2000;290:1571–1574. doi: 10.1126/science.290.5496.1571. [DOI] [PubMed] [Google Scholar]
- 16.Frand AR, Kaiser CA. Ero1p oxidizes protein disulfide isomerase in a pathway for disulfide bond formation in the endoplasmic reticulum. Mol Cell. 1999;4:469–477. doi: 10.1016/s1097-2765(00)80198-7. [DOI] [PubMed] [Google Scholar]
- 17.Benham AM, et al. The CXXCXXC motif determines the folding, structure and stability of human Ero1-Lalpha. EMBO J. 2000;19:4493–4502. doi: 10.1093/emboj/19.17.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliver JD, et al. Interaction of the thiol-dependent reductase ERp57 with nascent glycoproteins. Science. 1997;275:86–88. doi: 10.1126/science.275.5296.86. [DOI] [PubMed] [Google Scholar]
- 19.Mezghrani A, et al. Manipulation of oxidative protein folding and PDI redox state in mammalian cells. EMBO J. 2001;20:6288–6296. doi: 10.1093/emboj/20.22.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jessop CE, et al. Protein disulphide isomerase family members show distinct substrate specificity: P5 is targeted to BiP client proteins. J Cell Sci. 2009;122(Pt 23):4287–4295. doi: 10.1242/jcs.059154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulp MS, et al. Domain architecture of protein-disulfide isomerase facilitates its dual role as an oxidase and an isomerase in Ero1p-mediated disulfide formation. J Biol Chem. 2006;281:876–884. doi: 10.1074/jbc.M511764200. [DOI] [PubMed] [Google Scholar]
- 22.Guilhot C, et al. Evidence that the pathway of disulfide bond formation in Escherichia coli involves interactions between the cysteines of DsbB and DsbA. Proc Natl Acad Sci USA. 1995;92:9895–9899. doi: 10.1073/pnas.92.21.9895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin DY, Tie JK, Stafford DW. The conversion of vitamin K epoxide to vitamin K quinone and vitamin K quinone to vitamin K hydroquinone uses the same active site cysteines. Biochemistry. 2007;46:7279–7283. doi: 10.1021/bi700527j. [DOI] [PubMed] [Google Scholar]
- 24.Rost S, et al. Site-directed mutagenesis of coumarin-type anticoagulant-sensitive VKORC1: Evidence that highly conserved amino acids define structural requirements for enzymatic activity and inhibition by warfarin. Thromb Haemostasis. 2005;94:780–786. doi: 10.1160/TH05-02-0082. [DOI] [PubMed] [Google Scholar]
- 25.Matsuo Y, et al. Identification of a novel thioredoxin-related transmembrane protein. J Biol Chem. 2001;276(13):10032–10038. doi: 10.1074/jbc.M011037200. [DOI] [PubMed] [Google Scholar]
- 26.Matsuo Y, et al. TMX, a human transmembrane oxidoreductase of the thioredoxin family: The possible role in disulfide-linked protein folding in the endoplasmic reticulum. Arch Biochem Biophys. 2004;423:81–87. doi: 10.1016/j.abb.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Roth D, et al. A di-arginine motif contributes to the ER localization of the type I transmembrane ER oxidoreductase TMX4. Biochem J. 2010;425:195–205. doi: 10.1042/BJ20091064. [DOI] [PubMed] [Google Scholar]
- 28.Sugiura Y, et al. Novel thioredoxin-related transmembrane protein TMX4 has reductase activity. J Biol Chem. 2010;285:7135–7142. doi: 10.1074/jbc.M109.082545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rost S, et al. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature. 2004;427:537–541. doi: 10.1038/nature02214. [DOI] [PubMed] [Google Scholar]
- 30.Grauschopf U, et al. Why is DsbA such an oxidizing disulfide catalyst? Cell. 1995;83:947–955. doi: 10.1016/0092-8674(95)90210-4. [DOI] [PubMed] [Google Scholar]
- 31.Quan S, et al. The CXXC motif is more than a redox rheostat. J Biol Chem. 2007;282:28823–28833. doi: 10.1074/jbc.M705291200. [DOI] [PubMed] [Google Scholar]
- 32.Tien AC, et al. Ero1L, a thiol oxidase, is required for Notch signaling through cysteine bridge formation of the Lin12-Notch repeats in Drosophila melanogaster. J Cell Biol. 2008;182:1113–1125. doi: 10.1083/jcb.200805001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zito E, et al. ERO1-beta, a pancreas-specific disulfide oxidase, promotes insulin biogenesis and glucose homeostasis. J Cell Biol. 2010;188:821–832. doi: 10.1083/jcb.200911086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sevier CS. New insights into oxidative folding. J Cell Biol. 2010;188:757–758. doi: 10.1083/jcb.201002114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raje S, Thorpe C. Inter-domain redox communication in flavoenzymes of the quiescin/sulfhydryl oxidase family: Role of a thioredoxin domain in disulfide bond formation. Biochemistry. 2003;42:4560–4568. doi: 10.1021/bi030003z. [DOI] [PubMed] [Google Scholar]
- 36.Sevier CS, et al. A flavoprotein oxidase defines a new endoplasmic reticulum pathway for biosynthetic disulphide bond formation. Nat Cell Biol. 2001;3:874–882. doi: 10.1038/ncb1001-874. [DOI] [PubMed] [Google Scholar]
- 37.Thorpe C, et al. Sulfhydryl oxidases: Emerging catalysts of protein disulfide bond formation in eukaryotes. Arch Biochem Biophys. 2002;405(1):1–12. doi: 10.1016/s0003-9861(02)00337-5. [DOI] [PubMed] [Google Scholar]
- 38.Rowe ML, et al. Solution structure and dynamics of ERp18, a small endoplasmic reticulum resident oxidoreductase. Biochemistry. 2009;48(21):4596–4606. doi: 10.1021/bi9003342. [DOI] [PubMed] [Google Scholar]
- 39.Alanen HI, et al. Functional characterization of ERp18, a new endoplasmic reticulum-located thioredoxin superfamily member. J Biol Chem. 2003;278:28912–28920. doi: 10.1074/jbc.M304598200. [DOI] [PubMed] [Google Scholar]
- 40.Jeong W, et al. ERp16, an endoplasmic reticulum-resident thiol-disulfide oxidoreductase: Biochemical properties and role in apoptosis induced by endoplasmic reticulum stress. J Biol Chem. 2008;283:25557–25566. doi: 10.1074/jbc.M803804200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park B, et al. Redox regulation facilitates optimal peptide selection by MHC class I during antigen processing. Cell. 2006;127:369–382. doi: 10.1016/j.cell.2006.08.041. [DOI] [PubMed] [Google Scholar]
- 42.Cabibbo A, et al. ERO1-L, a human protein that favors disulfide bond formation in the endoplasmic reticulum. J Biol Chem. 2000;275:4827–4833. doi: 10.1074/jbc.275.7.4827. [DOI] [PubMed] [Google Scholar]
- 43.Voeltz GK, et al. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.