Abstract
Toll-like receptor 9 (TLR9) senses microbial DNA and triggers type I IFN responses in plasmacytoid dendritic cells (pDCs). Previous studies suggest the presence of myeloid differentiation primary response gene 88 (MyD88)-dependent DNA sensors other than TLR9 in pDCs. Using MS, we investigated C-phosphate-G (CpG)-binding proteins from human pDCs, pDC-cell lines, and interferon regulatory factor 7 (IRF7)-expressing B-cell lines. CpG-A selectively bound the aspartate-glutamate-any amino acid-aspartate/histidine (DExD/H)-box helicase 36 (DHX36), whereas CpG-B selectively bound DExD/H-box helicase 9 (DHX9). Although the aspartate-glutamate-alanine-histidine box motif (DEAH) domain of DHX36 was essential for CpG-A binding, the domain of unknown function 1605 (DUF1605 domain) of DHX9 was required for CpG-B binding. DHX36 is associated with IFN-α production and IRF7 nuclear translocation in response to CpG-A, but DHX9 is important for TNF-α and IL-6 production and NF-κB activation in response to CpG-B. Knocking down DHX9 or DHX36 significantly reduced the cytokine responses of pDCs to a DNA virus but had no effect on the cytokine responses to an RNA virus. We further showed that both DHX9 and DHX36 are localized within the cytosol and are directly bound to the Toll-interleukin receptor domain of MyD88 via their helicase-associated domain 2 and DUF domains. This study demonstrates that DHX9/DHX36 represent the MyD88-dependent DNA sensors in the cytosol of pDCs and suggests a much broader role for DHX helicases in viral sensing.
Keywords: cytosolic sensor, innate immunity
The innate immune response is the first line of the host defense system in response to microbial infections. Pattern recognition receptors (PRRs) are sentinels to detect pathogen-associated molecular patterns (PAMPs) and to initiate a downstream signaling cascade leading to the activation of type I IFN and inflammatory cytokines (1). PRRs have been categorized into several families, including Toll-like receptors (TLR) (2), retinoic acid inducible gene I (RIG-I)–like receptors (RLR) (3), and Nod-like receptors (4). In addition, absent in melanoma 2 (AIM2) (5) and RNA polymerase III (6, 7) recently have been identified as cytosolic DNA sensors.Plasmacytoid dendritic cells (pDCs), also known as “professional type I IFN-producing cells,” are a specialized cell type for mounting antiviral innate immune responses and are characterized by their selective expression of TLR7 and TLR9 for sensing viral RNA and DNA and constitutive expression of interferon regulatory factor 7 (IRF-7) for rapid IFN responses (2, 8, 9). Although TLR9 was shown to be critical for endowing pDCs with the ability to sense microbial DNA within the endosome compartment, there is a major gap in understanding how TLR9 binds DNA and whether TLR9 represents the only DNA sensor in pDCs. Recent studies suggested the presence of a myeloid differentiation primary response gene 88 (MyD88)-dependent viral sensor other than TLR9 in pDCs (10–12). However, the nature of this sensor is unknown.
Microbial nucleic acids, including their genomic DNA/RNA and replicating intermediates, work as strong PAMPs (13), so finding PRR-sensing pathogenic nucleic acids and investigating their signaling pathway is of general interest. Cytosolic RNA is recognized by RLRs, including RIG-I, melanoma differentiation-associated gene 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2). RIG-I senses 5′-triphosphate dsRNA and ssRNA or short dsRNA with blunt ends. MDA5 mainly senses long dsRNA. RIG-I and MDA5 have tandem caspase activation and recruitment domains (CARD) at the N-terminal region that interact with other CARD-containing proteins such as the mitochondrial antiviral signaling (MAVS) protein (14–17). MAVS transduces the signaling cascade through IκB kinase (IKK)-related kinases such as TNF receptor-associated factor family member-associated NF-κB activator (TANK)-binding kinase-1 (TBK1) and inducible IKK, culminating in the activation of IRF3 and inducing the transcription of type I IFNs such as IFN-β.
Cytosolic DNA, whether self or nonself, is a potent pathogenic stimulus of the innate immune system. When bacteria or virus infect the cells and their DNA is introduced into cytosol, the innate immune response is triggered to produce type I IFNs. DNase II-deficient mice, which can not digest DNA from engulfed apoptotic cells, produced robust amounts of type I IFN mediating IRF3 activation (18, 19). Several studies have suggested the existence of cytosolic DNA sensors within the innate immune system (20–22). Intracellular administration of the double-stranded B-form of DNA into mouse embryonic fibroblasts triggered TBK1/IRF3-dependent, TLR9/MyD88-independent, and RIG-I–independent type I IFN responses (21). An earlier study suggested that Z-DNA binding protein 1, also known as “DNA-dependent activator of IRF” (DAI), is a cytosolic DNA sensor (23). However, one study using DAI-knockout mice failed to confirm DAI as the cytosolic DNA sensor (24). AIM2 is another cytosolic DNA sensor that activates inflammasome but is not involved in the type I IFN response (5). Recently, RNA polymerase III was found to sense microbial DNA in cytosol, triggering an RNA intermediate-dependent type I IFN response (6, 7). Whether there are cytosolic sensors that bind DNA directly is not known.
Here we show that biochemical purification of C-phosphate-G (CpG)-binding proteins led to the identification of aspartate-glutamate-any amino acid-aspartate/histidine (DExD/H)-box helicase 36 (DHX36) and DExD/H-box helicase 9 (DHX9) as specific sensors for CpG-A and CpG-B, respectively, in pDC cells. DHX36 could sense CpG-A by direct binding via the aspartate-glutamate-alanine-histidine box motif (DEAH) domain, whereas DHX9 could sense CpG-B via its domain of unknown function (DUF). Both DHXs are critical for sensing viral DNA pathogens to trigger differential cytokine responses. Under CpG treatment, DHX36 and DHX9 are localized in cytosol but not in endosomal structures and bind to the Toll-IL receptor (TIR) domain of MyD88 via their helicase-associated 2 (HA2) and DUF domains, leading to the activation of IRF7 and p50 (NF-κB).
Results
DHX36 and DHX9 Associate with CpG-A and CpG-B, Respectively.
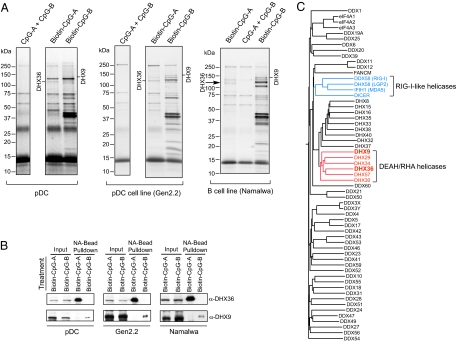
We used biochemical approaches to determine CpG-binding proteins in human primary pDCs, in a human pDC cell line (Gen2.2), and in an IRF7-transfected B-cell line (Namalwa). The Gen2.2 human pDC cell line is a human leukemia cell line that shares identical features with human primary pDCs (25). The Namalwa cell line is a TLR9-expressing human B-cell line transfected with IRF7 that has the ability to produce large amounts of type I IFN in response to CpG-oligodeoxynucleotide (ODN). We first generated biotinylated CpG-A (ODN 2216) and CpG-B (ODN 2006) and confirmed that both were as potent as nonconjugated CpG in activating human primary pDCs and Gen2.2 and Namalwa cells. Then we isolated ≈1.0 × 108 human primary pDCs with >95% purity by anti-blood dendritic cell antigen 2 (BDCA2) magnetic bead sorting from ≈100 human peripheral blood buffy coat samples. Human primary pDCs from each donor were incubated for 4 h with CpG-A?B (control) or with biotin-CpG-A or biotin-CpG-B. We chose this incubation period because CpG induced significant IFN gene transcription in pDCs at 4 h. Parallel experiments were performed using Gen2.2 and Namalwa cells.
To purify the CpG-bound protein complexes, whole-cell lysates from pDCs treated with biotin-CpG-A, biotin-CpG-B, or CpG-A?B were purified with NeutrAvidin (NA)-conjugated beads that specifically bind the biotin moiety. The polypeptides bound to biotin-CpGs were separated by gradient polyacrylamide gel electrophoresis and visualized by Coomassie staining. As shown in Fig. 1A, several polypeptide bands were observed that were unique to the biotin-CpG-A and biotin-CpG-B pulldowns and were absent from the control CpG-A?B pulldown. Moreover, these polypeptide bands, specific for either the biotin-CpG-A or biotin-CpG-B pulldown, also were observed in the pulldown experiments using Gen2.2 and Namalwa cells.
Fig. 1.
DHX36 and DHX9 associate with CpG-A and CpG-B, respectively. (A) Coomassie staining of CpG-A–associated proteins and CpG-B–associated proteins purified with NA beads from primary pDCs (Left), from a pDC cell line (Gen2.2) (Center), and from a B-cell line (Namalwa) (Right) treated with both CpG-A and CpG-B (CpG-A?B), with biotin-CpG-A, or with biotin-CpG-B. Proteins identified by LC-MS are as indicated. The sample treated with CpG-A?B served as control. (B) Purified CpG-A–bound proteins and CpG-B–-bound proteins from pDCs (Left), from Gen2.2 cells (Center), and from Namalwa cells (Right) were analyzed by immunoblotting with anti-DHX36 and anti-DHX9 antibodies, as indicated. (C) Phylogeny of 59 human DExD/H helicases was analyzed by ClustalW analysis.
The polypeptide bands specific for biotin-CpG-A and biotin-CpG-B pulldowns were excised from the gel and analyzed by liquid chromatography (LC)-MS. We found DHX36 at 110 kDa in biotin-CpG-A pulldowns and DHX9 at 140 kDa in biotin-CpG-B pulldowns. The DExD/H box family includes a large number of proteins that play important roles in RNA metabolism. Members of this family act as RNA helicases or unwindases, using the energy from ATP hydrolysis to unwind RNA structures or to dissociate RNA–protein complexes in cellular processes that require modulation of RNA structures, and are distinguished by the presence of several conserved motifs, including the characteristic DExD/H sequence (in which “x” can be any amino acid). These proteins are highly conserved from viruses and bacteria to humans (26). Although initially classified as RNA helicases, many members of the DExD/H helicase family, including DHX36 and DHX9, were found to display DNA helicase activity (27, 28). Sequence analyses revealed that DHX36 and DHX9 are in the same DEAH/RHA helicase subfamily, which is close to but is clearly separated from the RIG-I–like helicase subfamily (Fig. 1C). Because a subgroup of the DExD/H helicase family, including RIG-I, MDA5, and LGP2, was found to function as cytosolic sensors of viral RNA for mounting innate antiviral immune responses (29), we hypothesized that DHX36 and its close relative DHX9 may be the sensors for viral DNA in pDCs.
To confirm whether DHX36 specifically binds CpG-A and DHX9 specifically binds CpG-B, we performed immunoblotting analyses using antibodies specific to DHX36 or DHX9. As shown in Fig. 1B, anti-DHX36 antibody detected a 110-kDa band in the biotin-CpG-A pulldown, and anti-DHX9 antibody detected a 140-kDa band in the biotin-CpG-B pulldown.
DHX36 Binds CpG-A via the DEAH Domain, and DHX9 Binds CpG-B via the DUF Domain.
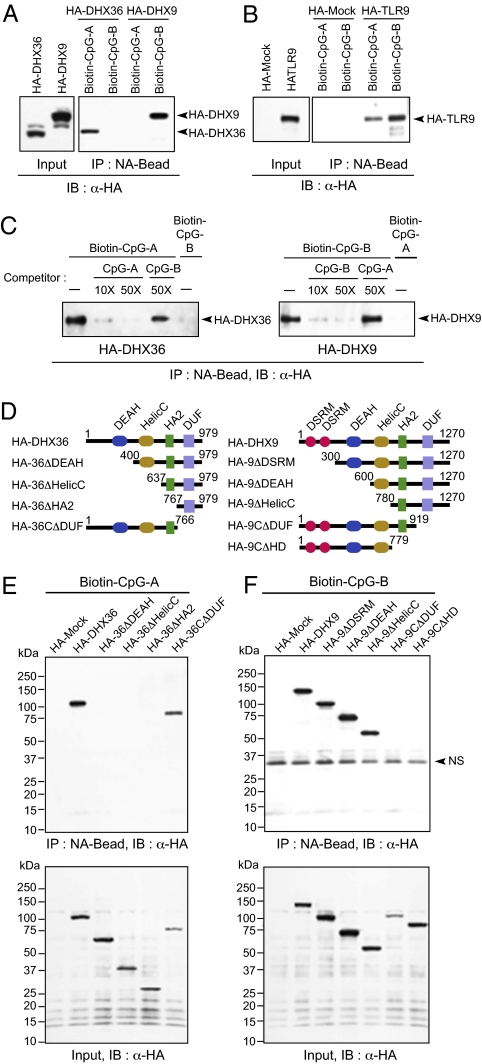
To confirm further that the recognition of DHX36 is specific to CpG-A and that the recognition of DHX9 is specific to CpG-B, competition experiments were performed. HA-tagged DHX36 and DHX9 were expressed in 293T cells. The cell lysates were incubated with biotin-CpG-A or biotin-CpG-B in the presence or absence of nonconjugated CpG-A or CpG-B as competitors. Pulldown and immunoblotting analyses showed that DHX36 bound to CpG-A but not to CpG-B (Fig. 2A). The binding of DHX36 to biotin-CpG-A was blocked by nonconjugated CpG-A in a dose-dependent fashion, but it was not blocked by nonconjugated CpG-B (Fig. 2C). Similarly, DHX9 bound to biotin-CpG-B but not to biotin-CpG-A (Fig. 2A). The binding of DHX9 to biotin-CpG-B was blocked by nonconjugated CpG-B in a dose-dependent fashion, but it was not blocked by nonconjugated CpG-A (Fig. 2C). Both biotin-CpG-A and biotin-CpG-B were shown to bind to TLR9 (Fig. 2B).
Fig. 2.
Domains responsible for specific interactions of DHX36 with CpG-A and of DHX9 with CpG-B. (A) Pulldown assays were performed by incubating whole-cell lysates from HA-DHX36– and HA-DHX9–expressing 293T cells with biotin-CpG-A or biotin-CpG-B with NA beads. Bound proteins were analyzed by immunoblotting with anti-HA antibody. (B) Pulldown and immunoblotting assays were performed as described in A but with whole-cell lysates from HA-Mock or HA-TLR9–expressing 293T cells. (C) Competitive pulldown assays. (Left) Whole-cell lysates from HA-DHX36–expressing 293T cells were preincubated in the absence or presence of nonconjugated CpG-A (10× and 50×; 1×=1 μM) or CpG-B (50×) and then were incubated with 1× of biotin-CpG-A or biotin-CpG-B along with NA beads. (Right) Whole-cell lysates from HA-DHX9–expressing 293T cells were preincubated in the absence or presence of nonconjugated CpG-B (10× and 50×) or CpG-A (50×) and then were incubated with 1× of biotin-CpG-B or biotin-CpG-A along with NA beads. Bound proteins were analyzed by immunoblotting with anti-HA antibody. (D) Schematic representations of DHX36 and DHX9 and their serial-deletion mutants. Numbers denote amino acid residues. DEAH, aspartate-glutamate-alanine-histidine box motif; DSRM, double-stranded RNA binding motif; DUF 1605, domain of unknown function 1605; HA2, helicase-associated domain 2; HelicC, helicase C-terminal domain. HA2, helicase-associated domain 2. (E and F) Pulldown assays were performed by incubating lysates from 293T cells expressing HA-tagged full-length and deletion mutants of DHX36 with biotin-CpG-A (E) or by incubating HA-tagged full-length and deletion mutants of DHX9 with biotin-CpG-B (F) along with NA beads. Bound proteins were analyzed by immunoblotting with anti-HA. NS, nonspecific band.
DHX36 and DHX9 belong to the RHA subfamily of DExD/H helicase and share the conserved domain structures DEAH, helicase C terminal domain (HelicC), HA2, and DUF1605 (Fig. 2D). To define the CpG-A–binding domain of DHX36 and the CpG-B–binding domain of DHX9, serial deletion mutants of DHX36 and DHX9 were expressed in 293T cells, and their ability to bind CpG-A or CpG-B was analyzed. We found that the DEAH domain of DHX36 was essential for binding CpG-A, and the DUF domain of DHX9 was essential for binding CpG-B (Fig. 2 E and F).
DHX36 and DHX9 Are Critical for Microbial DNA-Mediated Cytokine Responses.
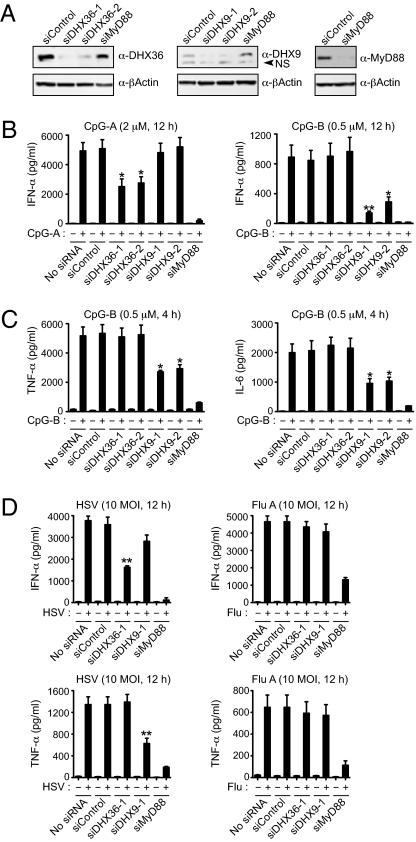
We next used siRNA technology to investigate the function of DHX36 and DHX9 in pDC responses to CpG-A and CpG-B. Because the manipulation of gene expression has been unsuccessful in human primary pDCs, we performed the experiments in Gen2.2 cells, a pDC cell line derived from human pDC leukemia that expresses TLR7/9, transcription factor 4, and IRF7 and which has all the phenotypic and functional features associated with human primary pDCs (25). Although the CpG-A response by pDCs is characterized by high IFN, the CpG-B response by pDCs is characterized by low IFN but high TNF and IL-6 synthesis (30, 31). Knocking down endogenous DHX36 by two different siRNAs (siDHX36-1 and siDHX36-2) led to >50% reduction in IFN-α production by the Gen2.2 cells in response to CpG-A, whereas knocking down DHX9 by two different siRNAs (siDHX9-1 and siDHX9-2) had no effect on CpG-A–induced IFN-α secretion (Fig. 3 A and B, Left). In contrast, DHX9 knockdown led to an approximate 50% reduction in TNF and IL-6 responses as well as a residual IFN-α response to CpG-B by Gen2.2 cells (Fig. 3 B, Right and C). To determine the function of DHX9/36 in antiviral innate immune responses, pDC cell lines were challenged with DNA virus (HSV) or RNA virus (influenza A virus, Flu A). We found that DHX9/36 knockdown inhibited pDC responses to HSV but not to Flu A virus (Fig. 3D). These results suggest that DHX36 and DHX9 are critical for sensing CpG-ODN and viral pathogens and that DHX36/DHX9-mediated sensing is dependent on MyD88.
Fig. 3.
DHX36 and DHX9 are critical for microbial DNA-mediated cytokine responses. (A) Gen2.2 cells were transfected with nonspecific siRNA (siControl), two siRNAs targeting DHX36 (siDHX36-1 and siDHX36-2), two siRNAs targeting DHX9 (siDHX9-1 and siDHX9-2), or a MyD88-targeting siRNA (siMyD88). Endogenous DHX36, DHX9, and MyD88 were monitored by immunoblotting with anti-DHX36, anti-DHX9, and anti-MyD88 antibodies, as indicated at the right. NS, nonspecific bands. (B and C) ELISAs to monitor IFN-α production from Gen2.2 cells transfected with siRNA, as indicated, upon treatment with 2 μM of CpG-A for 12 h or 0.5 μM of CpG-B for 12 h (B) or to monitor TNF-α and IL-6 production upon treatment with 0.5 μM of CpG-B for 4 h (C). (D) ELISAs to monitor IFN-α and TNF-α production from Gen2.2 cells transfected with siRNA, as indicated, upon treatment with HSV or Flu A (multiplicity of infection, 10) for 12 h. Data are mean ± SD from three independent experiments. *P < 0.05 and **P < 0.01 versus sample transfected with siControl and treated with CpG or virus.
Both DHX36 and DHX9 Bind MyD88 via the TIR Domain in the Cytosol of pDC.
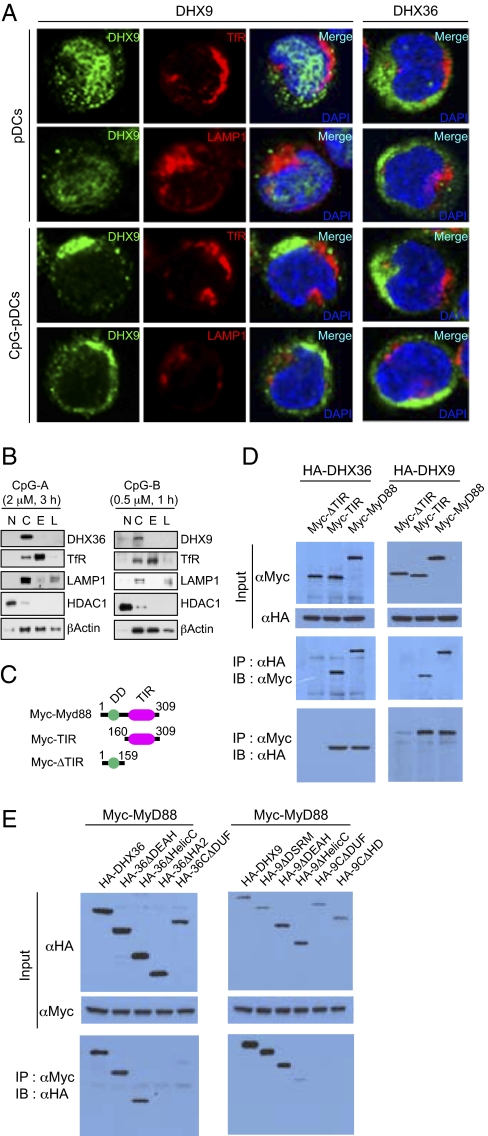
We next conducted experiments to investigate the cellular localization of DHX36 and DHX9 and whether and how they interact with MyD88. Using specific antibodies to DHX9 or DHX36, we found that both were localized within the cytosol of pDCs, and neither was localized within the early endosome marked by the transferrin receptor (TfR) or the late endosome marked by lysosomal-associated membrane protein 1 (LAMP-1) (Fig. 4A). We also fractionated endosomal structures by ultracentrifugation based on step-gradient sucrose cushion and found that DHX9 and DHX36 were not fractionated with endosomal structure (Fig. 4B). These results indicate that DHX9 and DHX36 function in the cytosol but not in endosomal structures.
Fig. 4.
DHX36 and DHX9 interact with MyD88 in the cytosol of pDCs. (A) Confocal images of pDCs untreated or treated with CpG-A or CpG-B. The three columns on the left show staining of DHX9 and TfR (an early endosome marker) or LAMP1 (a late endosome marker). The column on the right shows staining of DHX36 and TfR or LAMP1. (B) Endosomal fractionation. Fractionated early endosome and late endosome by ultracentrifugation based on step-gradient sucrose was examined by immunoblotting with anti-DHX9, anti-DHX36, anti-TfR, anti-LAMP1, anti-histone deacetylase 1 (anti-HDAC1), and anti-β-Actin antibodies. (C) Schematic representations of MyD88 and two deletion mutants. Numbers denote amino acid residues. DD, death domain; TIR, Toll-IL-1R homologous domain. (D) Lysates from 293T cells expressing Myc-tagged full-length or deletion mutants of MyD88 with HA-DHX36 or HA-DHX9 were immunoprecipitated with anti-HA antibody, followed by immunoblotting with anti-Myc antibody, or vice versa. (E) Lysates from 293T cells expressing HA-tagged full-length or deletion mutants of DHX36 or DHX9 with Myc-MyD88 were immunoprecipitated with anti-Myc antibody, followed by immunoblotting with anti-HA antibody.
A recent study showed that the tobacco plant TLR-like “R” antiviral protein, encoded by the N gene, bound directly via its TIR domain to the Tobacco mosaic virus pathogen elicitor protein, p50 helicase (32). This report prompted us to determine whether DHX9/36 binds to MyD88 directly. To do so, we expressed the DHX9 and DHX36 HA-tagged deletion mutants depicted in Fig. 2D with Myc-tagged deletion mutants of MyD88 in 293T cells (Fig. 4C) and performed coimmunoprecipitation assays. We observed that MyD88 indeed could interact with both DHX36 and DHX9 through the TIR domain of MyD88 (Fig. 4D). Moreover, the HA2 and DUF domains of DHX36 and DHX9 were critical for interaction with MyD88, and the HelicC domain of DHX9 was involved also (Fig. 4E). This result suggests that DHX9 and DHX36 indeed bind to MyD88 via interactions between the HA2 and DUF domains of DHX and the TIR domain of MyD88.
DHX36 and DHX9 Trigger Downstream Signaling to Activate IRF7 and p50 (NF-κB).
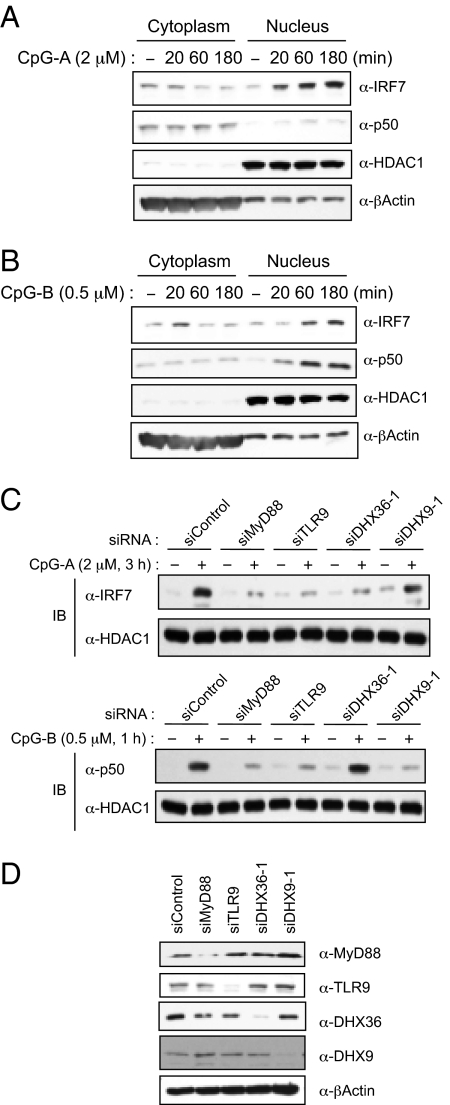
To investigate the requirement of DHX36 and DHX9 for CpG-triggered signaling culminating in the activation of IRF7 and p50 (Fig. 5 A and B), we knocked down MyD88, TLR9, DHX36, and DHX9 expression in Gen2.2 cells and monitored nuclear localization of IRF7 and p50 (Fig. 5D). As shown in Fig. 5C, when MyD88 or TLR9 expression was dampened by siRNA, nuclear localization of IRF7 by CpG-A and nuclear localization of p50 by CpG-B was diminished. Interestingly, whereas knockdown of DHX36 diminished the nuclear localization of IRF7 by CpG-A but not the nuclear localization of p50 by CpG-B, knockdown of DHX9 inhibited nuclear localization of p50 by CpG-B but not the nuclear localization of IRF7 by CpG-A. These results suggest that DHX36-mediated sensing of CpG-A and DHX9-mediated sensing of CpG-B trigger different signal pathways in pDCs.
Fig. 5.
DHX36 and DHX9 trigger downstream signaling cascades. (A and B) Nuclear fractions from Gen2.2 cells stimulated with 2 μM of CpG-A (A) or 0.5 μM of CpG-B (B) in a time-dependent manner were immunoblotted with anti-IRF7 or anti-p50 antibodies. HDAC1 and β-Actin were used as nuclear and cytosolic markers, respectively. (C) Nuclear fractions from Gen2.2 cells transfected with siRNA, as indicated, upon treatment with 2 μM of CpG-A for 3 h (Upper) or 0.5 μM of CpG-B for 1 h (Lower) were immunoblotted with anti-IRF7 or anti-p50 antibodies. HDAC1 was used as a loading control. (D) Endogenous DHX36, DHX9, TLR9, and MyD88 were monitored by immunoblotting with anti-DHX36, anti-DHX9, anti-TLR9, and anti-MyD88 antibodies, as indicated at the right.
Discussion
In this study we have demonstrated that DHX36 and DHX9, members of the RHA subfamily of DExD/H helicases, represent the CpG-A sensor and the CpG-B sensor, respectively, in human pDCs. Therefore, the DExD/H helicase family has a dedicated RIG-I–like subfamily for sensing viral RNA, as shown by previous studies (29), and a dedicated RHA subfamily for sensing viral DNA, as shown here. Interestingly, each member of the RIG-I subfamily of DExD/H helicases senses viral RNA differently. For example, RIG-I senses 5′ triphosphate ds/ssRNA, MDA5 senses cytoplasmic dsRNA (33), and LGP2 senses dsRNA independent of 5′ triphosphates (34). In parallel, we found that two members of the RHA subfamily of DExD/H helicases sense microbial DNA differently: DHX36 senses CpG-A using the DEAH domain, whereas DHX9 senses CpG-B using the DUF domain. The DEAH domain has ATP-dependent helicase activity and is known to bind RNA or DNA. As indicated by its name, the function of the DUF domain had not been identified; here we report a putative function for the DUF domain as a binding domain to unmethylated CpG ODN. Because cell lysates were used for in vitro binding assay, the possibility that indirect binding of helicases to CpGs was mediated by other proteins cannot be excluded completely. In eukaryotes, a total of 59 DExD/H helicases have been grouped into four subfamilies: RIG-1-like, DEAH/RHA, aspartate-glutamate-alanine-aspartate (DEAD)-Box, and Snf1-related kinase interacting protein 2 (Ski2)-like (26). Our study suggests that DExD/H helicases may play a much broader role in antiviral innate immune responses than previously thought. Indeed, a recent study demonstrated that Dicer-2, another RIG-I–like DExD/H helicase, senses viral nucleic acids in the Drosophila innate immune system (35). The potential roles of the other DExD/H helicase family members in sensing microbial nucleic acids remain to be explored.
Studies using Myd88-knockout mice have shown that the innate immune responses of pDCs to viral DNA, CpG-A, and CpG-B are totally dependent on MyD88 (36). We confirm these results in our current study by showing that MyD88 knock down by siRNA completely abolishes the innate immune responses of pDCs to both CpG-A and CpG-B. Previous studies have indicated the presence of TLR9-independent, MyD88-dependent viral DNA sensors in pDCs (10–12). Our study identified DHX36 and DHX9 as MyD88-dependent viral DNA sensors in pDCs that are localized within the cytosol and bind MyD88 directly via its TIR domain. These findings suggest that MyD88 serves as a key adaptor molecule for both endosomal TLRs and cytosolic helicases. Interestingly, the interaction between the TIR domain and helicases already has been documented in the plant world (32). This study suggests that the MyD88-dependent endosomal TLR9 sensor and the cytosolic DHX9/36 sensor may play complementary roles in viral DNA sensing, with TLR9 sensing viral entry and DHX9/36 sensing viral replication.
Materials and Methods
Statistical Analysis.
Statistically significant differences were determined by unpaired, two-tailed, Student's t test. P values <0.05 were considered statistically significant. Statistical analysis of data was done using GraphPad Prism version 5 for Macintosh (GraphPad Software).
Phylogenetic Analysis.
All protein sequences of listed DExD/H helicases were retrieved from the UniProt browser with the Swiss-Prot Knowledgebase, which is manually annotated and reviewed. These sequences were aligned, and all residues that contained gaps were removed from the alignment. The ClustalW multiple sequence alignment program with neighbor joining estimation was used to generate a phylogenetic tree.
Other detailed methods are provided in SI Text.
Supplementary Material
Acknowledgments
We thank Stephanie Watowich and Shao-Cong Sun for discussions and Melissa Wentz for critical reading. We thank all our colleagues in our laboratory.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006539107/-/DCSupplemental.
References
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity. 2010;32:305–315. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Yoneyama M, Fujita T. RIG-I family RNA helicases: Cytoplasmic sensor for antiviral innate immunity. Cytokine Growth Factor Rev. 2007;18:545–551. doi: 10.1016/j.cytogfr.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 4.Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- 5.Schroder K, Muruve DA, Tschopp J. Innate immunity: Cytoplasmic DNA sensing by the AIM2 inflammasome. Curr Biol. 2009;19:R262–R265. doi: 10.1016/j.cub.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 6.Ablasser A, et al. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hemmi H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 9.Colonna M, Trinchieri G, Liu YJ. Plasmacytoid dendritic cells in immunity. Nat Immunol. 2004;5:1219–1226. doi: 10.1038/ni1141. [DOI] [PubMed] [Google Scholar]
- 10.Hochrein H, et al. Herpes simplex virus type-1 induces IFN-alpha production via Toll-like receptor 9-dependent and -independent pathways. Proc Natl Acad Sci USA. 2004;101:11416–11421. doi: 10.1073/pnas.0403555101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jung A, et al. Lymphocytoid choriomeningitis virus activates plasmacytoid dendritic cells and induces a cytotoxic T-cell response via MyD88. J Virol. 2008;82:196–206. doi: 10.1128/JVI.01640-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hokeness-Antonelli KL, Crane MJ, Dragoi AM, Chu WM, Salazar-Mather TP. IFN-alphabeta-mediated inflammatory responses and antiviral defense in liver is TLR9-independent but MyD88-dependent during murine cytomegalovirus infection. J Immunol. 2007;179:6176–6183. doi: 10.4049/jimmunol.179.9.6176. [DOI] [PubMed] [Google Scholar]
- 13.Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 15.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 16.Xu LG, et al. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 17.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 18.Okabe Y, Kawane K, Akira S, Taniguchi T, Nagata S. Toll-like receptor-independent gene induction program activated by mammalian DNA escaped from apoptotic DNA degradation. J Exp Med. 2005;202:1333–1339. doi: 10.1084/jem.20051654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshida H, Okabe Y, Kawane K, Fukuyama H, Nagata S. Lethal anemia caused by interferon-beta produced in mouse embryos carrying undigested DNA. Nat Immunol. 2005;6:49–56. doi: 10.1038/ni1146. [DOI] [PubMed] [Google Scholar]
- 20.O'Riordan M, Yi CH, Gonzales R, Lee KD, Portnoy DA. Innate recognition of bacteria by a macrophage cytosolic surveillance pathway. Proc Natl Acad Sci USA. 2002;99:13861–13866. doi: 10.1073/pnas.202476699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishii KJ, et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- 22.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Takaoka A, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 24.Ishii KJ, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 25.Chaperot L, et al. Virus or TLR agonists induce TRAIL-mediated cytotoxic activity of plasmacytoid dendritic cells. J Immunol. 2006;176:248–255. doi: 10.4049/jimmunol.176.1.248. [DOI] [PubMed] [Google Scholar]
- 26.Linder P. Dead-box proteins: A family affair—active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaughn JP, et al. The DEXH protein product of the DHX36 gene is the major source of tetramolecular quadruplex G4-DNA resolving activity in HeLa cell lysates. J Biol Chem. 2005;280:38117–38120. doi: 10.1074/jbc.C500348200. [DOI] [PubMed] [Google Scholar]
- 28.Zhou K, et al. RNA helicase A interacts with dsDNA and topoisomerase IIalpha. Nucleic Acids Res. 2003;31:2253–2260. doi: 10.1093/nar/gkg328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beutler B, et al. Genetic analysis of resistance to viral infection. Nat Rev Immunol. 2007;7:753–766. doi: 10.1038/nri2174. [DOI] [PubMed] [Google Scholar]
- 30.Vollmer J, et al. Characterization of three CpG oligodeoxynucleotide classes with distinct immunostimulatory activities. Eur J Immunol. 2004;34:251–262. doi: 10.1002/eji.200324032. [DOI] [PubMed] [Google Scholar]
- 31.Verthelyi D, Ishii KJ, Gursel M, Takeshita F, Klinman DM. Human peripheral blood cells differentially recognize and respond to two distinct CPG motifs. J Immunol. 2001;166:2372–2377. doi: 10.4049/jimmunol.166.4.2372. [DOI] [PubMed] [Google Scholar]
- 32.Burch-Smith TM, et al. A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol. 2007;5:e68. doi: 10.1371/journal.pbio.0050068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 34.Pippig DA, et al. The regulatory domain of the RIG-I family ATPase LGP2 senses double-stranded RNA. Nucleic Acids Res. 2009;37:2014–2025. doi: 10.1093/nar/gkp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deddouche S, et al. The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in drosophila. Nat Immunol. 2008;9:1425–1432. doi: 10.1038/ni.1664. [DOI] [PubMed] [Google Scholar]
- 36.Adachi O, et al. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 1998;9:143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.