Abstract
Between the genetic extremes of rare monogenic and common polygenic diseases lie diverse oligogenic disorders involving mutations in more than one locus in each affected individual. Elucidating the principles of oligogenic inheritance and mechanisms of genetic interactions could help unravel the newly appreciated role of rare sequence variants in polygenic disorders. With few exceptions, however, the precise genetic architecture of oligogenic diseases remains unknown. Isolated gonadotropin-releasing hormone (GnRH) deficiency caused by defective secretion or action of hypothalamic GnRH is a rare genetic disease that manifests as sexual immaturity and infertility. Recent reports of patients who harbor pathogenic rare variants in more than one gene have challenged the long-held view that the disorder is strictly monogenic, yet the frequency and extent of oligogenicity in isolated GnRH deficiency have not been investigated. By systematically defining genetic variants in large cohorts of well-phenotyped patients (n = 397), family members, and unaffected subjects (n = 179) for the majority of known disease genes, this study suggests a significant role of oligogenicity in this disease. Remarkably, oligogenicity in isolated GnRH deficiency was as frequent as homozygosity/compound heterozygosity at a single locus (2.5%). Among the 22% of patients with detectable rare protein-altering variants, the likelihood of oligogenicity was 11.3%. No oligogenicity was detected among controls (P < 0.05), even though deleterious variants were present. Viewing isolated GnRH deficiency as an oligogenic condition has implications for understanding the pathogenesis of its reproductive and nonreproductive phenotypes; deciphering the etiology of common GnRH-related disorders; and modeling the genetic architecture of other oligogenic and multifactorial diseases.
Keywords: rare variant, idiopathic hypogonadotropic hypogonadism, Kallmann syndrome, digenic, FGFR1
Traditionally, deleterious rare variants—DNA sequence variations with minor allele frequency (MAF) <1% in the general population—have been incriminated as causes of rare genetic diseases in which each patient harbors mutations in a single gene (monogenic diseases) (1). An important paradigm shift in human genetics is the increasing recognition of rare variants as major contributors to both rare diseases and common multifactorial disorders (2, 3). In silico analyses suggest that many of the disease associations with common variants—MAF ≥5%—identified in genome-wide association studies may actually be “synthetic associations” involving nearby uncommon (1% ≤ MAF <5%) or rare variants that occur, by chance, more frequently in association with one allele at a common variant site than with the other (4). In addition, rare variants in genes linked to monogenic forms of common disease (such as familial hyperlipidemia and familial hypertension) have been shown to contribute to the population variation of the corresponding physiological parameters (plasma levels of HDL cholesterol and blood pressure, respectively) (5, 6).
According to conventional Mendelian frameworks, a monogenic disease is caused by one dominant or two recessive allelic variants in the responsible gene. However, in most monogenic disorders, the correspondence between genotype and phenotype is less than perfect. The concepts of incomplete penetrance and variable expressivity have been coined to make allowance for this discrepancy, but do not provide a molecular mechanistic explanation. In fact, monogenic diseases with limited genotype/phenotype correlations, such as Bardet–Biedl syndrome (7), retinitis pigmentosa (8), nephronopthisis (9), and several others (10), were subsequently shown to be oligogenic, i.e., caused by contributions of rare variants in a more than one gene in each patient. Elucidating the principles of oligogenic inheritance and the underlying genetic interactions should help to understand how rare variants contribute to the more complex multifactorial disorders. However, systematic investigation of oligogenic diseases is a formidable task because it requires the comprehensive sequencing of all known disease genes in sizeable cohorts of patients, which are especially challenging to assemble for such rare genetic conditions. Thus, with few exceptions, the precise genetic architecture of oligogenic diseases remains largely unknown because examples of oligogenicity have been described mostly in case reports and small studies that genotyped patient cohorts of limited sizes for subsets of disease-associated genes.
Isolated gonadotropin-releasing hormone (GnRH) deficiency caused by defects in the secretion or action of hypothalamic GnRH is one of the rare genetic diseases originally thought to be strictly monogenic (11, 12). It manifests as absent or incomplete puberty, sexual immaturity, and infertility, and is clinically diagnosed as idiopathic hypogonadotropic hypogonadism (IHH). The disease can be associated with either a normal sense of smell (normosmic IHH, nIHH) or with anosmia/hyposmia [Kallmann syndrome (KS)], and variably also with additional nonreproductive phenotypes such as unilateral renal agenesis, skeletal abnormalities, midline malformations, or hearing loss. Isolated GnRH deficiency can be sporadic or inherited as an autosomal-dominant, autosomal-recessive, or X-linked trait (13). Genetic dissection of this disorder has led to the discovery of several new loci (>12) with critical roles for the developmental and neuroendocrine control of mammalian reproduction (12, 14, 15). Genes underlying isolated GnRH deficiency have been shown to be important for the specification and proliferation of GnRH neurons (16, 17), their migration to the hypothalamus during embryonic development (18–22), the regulation of GnRH secretion (23–25), and the response of pituitary gonadotropes to GnRH stimulation (26). Although multiple mutations have been identified in each gene, no mutations have been identified in the majority of patients (>60%), signifying that yet more disease loci remain to be unearthed. In addition to this substantial locus and allelic heterogeneity, isolated GnRH deficiency is characterized by variable expressivity of the reproductive and nonreproductive phenotypes both within and across families segregating identical single-gene mutations (26–31). Pathogenic rare variants are occasionally present in family members who display only nonreproductive-associated phenotypes suffer from milder forms of GnRH deficiency, such as delayed puberty, or are asymptomatic (incomplete penetrance). Recently, a few patients were reported who harbor mutations in two genes rather than in a single locus (16, 21, 32–36). In some of these cases, the segregation pattern of these digenic mutations in the pedigree can partially account for the phenotypic variability among individual family members. Thus, the clinical heterogeneity of isolated GnRH deficiency might be explained to some degree by oligogenicity.
If isolated GnRH deficiency is oligogenic, then the availability of large cohorts of well-phenotyped patients, family members, and appropriate control subjects represents an opportunity to improve our understanding of oligogenicity in human disease. By systematically searching such cohorts for mutations in the majority of genes underlying isolated GnRH deficiency, the present study tested the following hypotheses: (i) oligogenicity is a common feature of the disease; (ii) rare variants that contribute to oligogenic GnRH deficiency when cooccurring can also be found individually in the normal population; (iii) the segregation patterns of rare variants in oligogenic pedigrees correlate with the varying phenotypes of family members; and (iv) isolated GnRH deficiency has a diverse oligogenic architecture corresponding to its complex developmental pathophysiology.
Results and Discussion
Oligogenicity in Isolated GnRH Deficiency Is as Frequent as Homozygosity/Compound Heterozygosity.
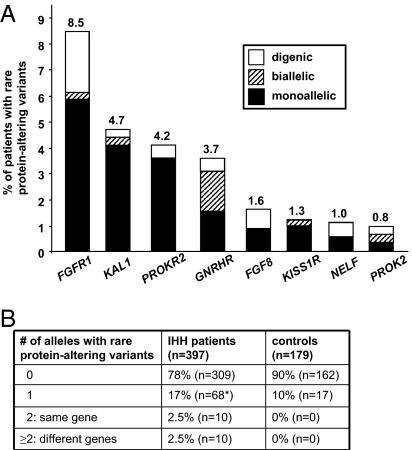
Of the 147 DNA sequence variants identified among the 397 patients and/or 179 controls in the 8 genes sequenced, 8 variants occurred commonly and 2 uncommonly, whereas the large majority (137) were rare (Table S1 and Fig. S1A). Thirty-eight of the 137 rare variants (28%) were synonymous (Table S1 and Fig. S1B). Though some of these synonymous variants might affect mRNA splicing or translation, most are presumably innocuous. Similar percentages of patients (9%) and controls (10%) harbored rare variants that were exclusively synonymous (Fig. S1C). Importantly, however, patients were significantly more likely than controls to harbor rare protein-altering variants (22% vs. 10%, P = 0.001; Fig. S1C). Indeed, nonsense, frameshift, and splice-site variants were found almost exclusively (19 of 20) in the GnRH-deficient cohort, and only 14 of 79 rare missense variants were seen even once in controls (Table S1). In addition, 66 of 79 missense variants had been previously shown to be loss-of-function in in vitro studies (n = 32) and/or were predicted in silico to be damaging by the software programs PolyPhen-2 (37), SIFT (38), or both (n = 62; Table S1). Furthermore, all four identified splice-site variants were predicted to affect splicing, and in one case this had been previously confirmed experimentally (Table S1). For these reasons, we considered only the 99 rare protein-altering variants. Each of the screened genes harbored such variants in 0.8–8.5% of patients (Fig. 1A). To our knowledge, this is the largest patient cohort screened for all eight genes so far, thus providing the best direct estimate to date of the relative frequency of variants in each gene (Fig. 1A). Half of the genes harbor rare variants at very low frequencies (≤1.6%), suggesting that discovery of all remaining genes for isolated GnRH deficiency will require very large numbers of patients, which can be achieved only by an international collaborative consortium (39).
Fig. 1.
(A) Percentages of patients with monoallelic, biallelic, and digenic rare protein-altering variants in each gene. (B) Number of alleles with rare protein-altering variants in patients and controls.
A single affected allele was found in 17% of patients (Fig. 1B). Some of these were men with KS who had KAL1 variants that were mostly frameshift or nonsense and thus in principle sufficient to account for the disease (Table S2). Others, however, had single-allele variants in either KISS1R (9%) or GNRHR (6%) (Table S2), which are genes traditionally thought to underlie recessive isolated GnRH deficiency (23, 24, 26). Because intragenic deletions in GNRHR and KISS1R are rare in GnRH-deficient patients (40, 41), it is likely that additional variants in other genes contribute to disease pathogenesis in these patients. Indeed, the fact that no variants were found in the majority of patients (78%; Fig. 1B) indicates that the eight genes sequenced account for only a fraction of the disease's genetic etiology.
Of the 397 patients, 10 had rare protein-altering variants in two or more alleles of different genes (i.e., digenicity), and 10 patients had such variants in both alleles of a single gene (i.e., homozygosity/compound heterozygosity; Fig. 1B and Table S2). Almost all of the variants involved were either known or predicted to be deleterious (18 of 21 in oligogenicity and 14 of 14 in homozygosity/compound heterozygosity; Table S1). In the present cohort, oligogenicity was as frequent as homozygosity/compound heterozygosity (2.5% of patients). Even if none of the disease genes remaining to be discovered and analyzed participates in an oligogenic mechanism of pathogenesis, this 2.5% would provide a minimum estimate for the frequency of oligogenicity in IHH (95% confidence interval: 1.2–4.6%). Among the 88 patients who had at least one rare protein-altering variant, a second affected allele was found in 10 (Fig. 1B). Thus, the likelihood of oligogenicity among the analyzed genes is 11.3% (10/88; 95% confidence interval: 5.6–19.9%). These estimates are based on the identification of rare protein-altering variants in a mere 22% of patients (Fig. 1B). Because every patient presumably harbors at least one causal variant, we expect the true frequency of oligogenicity in isolated GnRH deficiency to be much higher. We anticipate that as the broader availability and reduced cost of whole-exome sequencing facilitate the discovery of numerous genes underlying the disease over the next few years, most patients will ultimately prove to be oligogenic.
Unaffected Control Subjects Harbor Monoallelic but Not Biallelic or Digenic Variants.
The frequency of rare variants in genes underlying isolated GnRH deficiency among control subjects has not been previously determined. Ten percent of control subjects harbored a single, rare protein-altering variant, which was missense with one exception (nonsense; Fig. 1B and Table S3). Theoretically, such variants could be neutral, could contribute to isolated GnRH deficiency only in combination with other variants (oligogenicity), or could even be protective. Importantly, however, among the seven variants also seen in patients either in the present cohort (n = 5; Tables S2 and S3) or in an independent study (n = 2, PROKR2 P268C and P290S) (21), each of the five variants that have been functionally characterized in vitro (GNRHR Q106R, FGF8 P26L, and PROKR2 L173R, R268C, and P290S) has been shown to cause loss of function (16, 28, 33, 42). This indicates that isolated rare variants present in unaffected controls are not sufficient to cause GnRH deficiency on their own even when they are deleterious to protein function. Notably, the PROKR2 L173R and GNRHR Q106R mutations have been found in patients in heterozygous, compound heterozygous, homozygous, and digenic states (21, 26, 32–34, 43, 44). Thus, rare protein-altering variants that are individually present in the heterozygous state in unaffected subjects can contribute to isolated GnRH deficiency, including its oligogenic form, when cooccurring with additional deleterious alleles.
In further support of the notion that genes contributing alleles to oligogenic-isolated GnRH deficiency can individually harbor deleterious variants in the normal population, a nonsense NELF variant was present in a female control (Table S3). This indicates that monoallelic deleterious NELF variants are not sufficient to cause isolated GnRH deficiency, consistent with the fact that they have been found to underlie the disease only in the digenic state (32). Importantly, no control subject harbored two or more variants (Fig. 1B and Table S3), and the observed difference in the frequency of oligogenicity between patients (10 of 397) and controls (0 of 179) was significant (P = 0.035; Fig. 1B). These findings suggest that compound heterozygosity, homozygosity, and oligogenicity involving deleterious variants are incompatible with normal neuroendocrine control of human reproduction.
Oligogenicity Partially Accounts for the Phenotypic Variability of Isolated GnRH Deficiency.
In 7 of the 10 new (Fig. S2A) and previously identified (Fig. S2B) (16, 32, 33, 35, 36) digenic pedigrees in the present cohort, the higher the number of affected genes and alleles that an individual harbored, the more likely he or she was to have isolated GnRH deficiency manifested as IHH as opposed to a milder or partial phenotype, such as delayed puberty, anosmia, or cleft lip/palate (Fig. S2A, pedigrees I and II; Fig. S2B, pedigrees IV–VIII). For example, in pedigree IV, nIHH was present only in the two digenic subjects with one affected FGFR1 allele and two affected GNRHR alleles (Fig. S2B) (32). All three subjects with monoallelic GNRHR variants and wild-type FGFR1 were unaffected. Interestingly, of the two digenic subjects with monoallelic FGFR1 and GNRHR variants, one had delayed puberty and the other was unaffected. This discordance may be due to the different functional properties of each individual's GNRHR allele (Q106R vs. R262Q) (26, 45) and/or to additional genetic or nongenetic factors that differentially affect the two subjects. In pedigree IX, although the proband harbored digenic mutations, genotype-phenotype correlations could not be performed because she was adopted (Fig. S2B) (33). The correlation between digenicity and severe phenotype did not hold true for pedigrees III and X. In pedigree III, a digenic family member had a milder phenotype (delayed puberty) than the proband (Fig. S2A). In pedigree X, the same phenotype (nIHH) was observed in the proband with digenic mutations in FGFR1 and PROKR2, a family member harboring only the FGFR1 mutation, and four family members with no mutations in these genes (Fig. S2B). This pedigree has been shown to harbor an additional rare protein-altering variant in the gene encoding GnRH (GNRH1) (35). Even when the segregation pattern of this variant was taken into account, the phenotypic variability among family members could not be fully explained. Such exceptions further support the notion that oligogenicity in isolated GnRH deficiency is even more frequent and extensive than assessed here, and its full scope will be revealed only after all genes underlying the disease have been identified and sequenced. A recent study reached similar conclusions regarding digenicity that involves monoallelic PROKR2 variants, whose interacting partners are yet largely unknown (46). Nevertheless, it is already evident that mutations in genes thought to underlie autosomal-dominant (FGFR1), autosomal-recessive (GNRHR), or X-linked (KAL1) forms of isolated GnRH deficiency can actually underlie oligogenic disease (Fig. S2). Thus, as in other oligogenic diseases (10), the mode of inheritance of isolated GnRH deficiency in families (i.e., the Mendelian segregation of the trait) should be conceptually dissociated from the inheritance pattern of specific disease alleles.
In oligogenic-isolated GnRH deficiency, the clinical presentation of the disease is likely determined by the number and nature of affected genes and alleles. This concept is supported by a recent study showing larger phenotypic variability and reduced disease severity in KS patients harboring monoallelic vs. biallelic variants in PROK2 or PROKR2, presumably due to variants in undiscovered genes with pleiotropic roles that interact genetically with the monoallelic PROK2/PROKR2 variants (46). It is thus tempting to speculate that many of the nonreproductive phenotypes associated with isolated GnRH deficiency, such as anosmia/hyposmia, renal agenesis, midline defects, and hearing loss, may be in part or in whole due to contributions of rare variants in genes that act in both the neuroendocrine system and in nonreproductive tissues. Indeed, some of the known genes associated with the disease are also expressed in developing nonreproductive tissues that are affected in these patients, including KAL1 in the kidney (47, 48) and FGF8/FGFR1 in the craniofacial midline (49, 50). This observation raises the possibility that other genes known to regulate the development of nonreproductive tissues may also have unsuspected roles in the neuroendocrine control of reproduction. Alternatively, the pathogenesis of the associated nonreproductive phenotypes may entail interactions between variants in pleiotropic genes with variants in tissue-specific genes. These two mechanisms are not mutually exclusive, but may operate synergistically within an oligogenic framework.
Genetic Networks in Isolated GnRH Deficiency.
The molecular mechanisms by which the observed oligogenic interactions (Fig. 2) cause isolated GnRH deficiency are poorly understood. One possibility is that the mutations act independently in space and/or developmental time, such that their additive effects compromise the ontogeny of GnRH neurons and/or pituitary gonadotropes sufficiently to impair sexual maturation and fertility. Alternatively, the mutations may synergistically affect the same ligand/receptor pair or signaling pathway. At present, it is impossible to unequivocally distinguish between these models, because the precise roles of the known disease genes in the regulation of GnRH biology require further elucidation. As numerous additional genes underlying the disease are discovered through high-throughput sequencing in the near future, their functional categorization into signaling pathways, the characterization of their roles in GnRH ontogeny and action, and the study of their genetic interactions in model organisms will help unravel the mechanisms by which oligogenicity causes isolated GnRH deficiency.
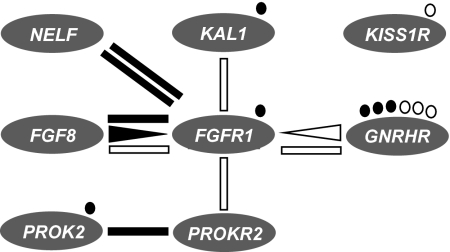
Fig. 2.
Genetic interactions in isolated GnRH deficiency. The majority of oligogenic patients (lines) showed digenic biallelic inheritance, in which two genes are implicated, each one having an affected and a wild-type allele. Two patients (triangles) showed digenic triallelic inheritance, which involves both alleles of one gene (triangle base) and one allele of a second gene (triangle apex). The same types of inheritance were observed in patients with KS (solid symbols) or nIHH (empty symbols), suggesting that oligogenicity underlies both clinical forms of isolated GnRH deficiency. Circles denote patients who had single-gene variants and were homozygous or compound heterozygous.
More recently, TAC3, TACR3, and GNRH1 have been identified as additional genes that harbor mutations at frequencies from moderate to very low in patients with the normosmic variant of isolated GnRH deficiency (25, 35, 51, 52). Even though it was not economically feasible to screen all patients in the present cohort for mutations in TAC3, TACR3, and GNRH1, the eight genes screened here showed no oligogenicity with TAC3 or TACR3, and only one case of digenicity with GNRH1 (Fig. S2B) among the partially overlapping screened cohorts (35, 51). With the possible exception of GNRHR, the genes found to interact in the present study affect the developmental biology of GnRH neurons, whereas TAC3, TACR3, GNRH1, and KISS1R (which was also not involved in digenicity, Fig. 2) affect GnRH secretion and may thus form a distinct network of oligogenic interactions in other patients with isolated GnRH deficiency.
Isolated GnRH Deficiency as a Prototypical Oligogenic Disease.
Together with similar advances in other genetic diseases demonstrating convincing oligogenicity, such as Bardet–Biedl syndrome (7, 53), the conceptual shift from monogenic to oligogenic framework for isolated GnRH deficiency can guide genetic research in disorders traditionally considered monogenic. As a disease model, isolated GnRH deficiency is especially instructive for how oligogenicity should be suspected and sought in “monogenic” diseases with marked locus heterogeneity, incomplete penetrance, and/or variable expressivity of pathogenic alleles. Among the known oligogenic disorders, isolated GnRH deficiency appears to match Bardet–Biedl syndrome in the number of disease-associated genes (53), and is among the most diverse in mechanisms of oligogenic interactions (10). This richness offers examples on which to model the architecture of other disorders suspected or shown to be oligogenic.
The fact that rare variants in genes for isolated GnRH deficiency are also present in unaffected individuals and cooperate with other variants to cause IHH raises the possibility that such variants may also underlie milder forms of GnRH deficiency when combined with certain environmental factors. In support of this idea, we have recently identified heterozygous rare variants in FGFR1, KAL1, PROKR2, and GNRHR in women with hypothalamic amenorrhea (HA), a frequent and reversible inhibition of the hypothalamic-pituitary-gonadal (HPG) axis commonly triggered by weight loss, excessive exercise, or psychological stress (54). By lowering the threshold for HPG axis inhibition, isolated rare variants in genes underlying GnRH deficiency may have conferred an evolutionary advantage in times of pandemic, famine, population migration, or climate change, which may explain why they have been retained in the human gene pool (54). We propose that when such variants form oligogenic interactions in the same individual due to consanguinity, endogamy, or chance, more severe GnRH deficiency and inhibition of the reproductive axis ensue, which manifests clinically as IHH. The pathogenesis of isolated GnRH deficiency is also very likely to entail a nongenetic component, as indicated by the occasional adult onset of the disease after normal puberty and reproductive function (55), as well as by the reversal of isolated GnRH deficiency in 10% of cases after pharmacological normalization of sex steroid levels (56). The role of nongenetic factors in isolated GnRH deficiency, its oligogenic nature, and its shared genetic basis with HA offer valuable paradigms through which gene/gene and gene/environment interactions can be modeled, and could thus help to address one of the major challenges for the elucidation of common multifactorial diseases.
Methods
Subjects.
The cohort of GnRH-deficient patients was comprised of 397 Caucasians who met the following diagnostic criteria: (i) absent/incomplete puberty by age 18 y; (ii) serum testosterone ≤100 ng/dL in men or serum estradiol ≤20 pg/mL in women in conjunction with low or normal levels of serum gonadotropins; (iii) otherwise normal pituitary function; (iv) normal serum ferritin levels; and (v) normal MRI of the hypothalamic-pituitary region. Forty-four women and 155 men had anosmia/hyposmia by history and/or olfactory testing (57) and were diagnosed with KS; 52 women and 146 men had nIHH. The cohort of control subjects was comprised of 179 unaffected Caucasians (72 women and 107 men) who had normal sexual maturation and reproductive function. The study was approved by human research committees at Massachusetts General Hospital and Newcastle-upon-Tyne Hospital. All subjects provided written informed consent before participation.
DNA Sequencing.
Genomic DNA was obtained from peripheral blood samples by standard phenol-chloroform extraction. For all patients and control subjects, the exonic and proximal intronic (at least 15 bp from splice sites) DNA sequences of the first eight genes implicated in the etiology of isolated GnRH deficiency were amplified by PCR and determined by direct sequencing. These genes are KAL1 (anosmin-1, MIM: 308700), GNRHR (GnRH receptor, MIM: 138850), KISS1R (KISS1 receptor, MIM: 604161), NELF (nasal embryonic LHRH factor, MIM: 608137), FGF8 (fibroblast growth factor 8, MIM: 600483), FGFR1 (fibroblast growth factor receptor 1, MIM: 136350), PROK2 (prokineticin 2, MIM: 607002), and PROKR2 (prokineticin receptor 2, MIM: 607212). Respective PCR primers and amplification conditions have been described previously (16, 22, 23, 26, 33, 58–60). All sequence variations were found on both DNA strands and were confirmed in a separate PCR. Genes and proteins are described using standard nomenclature (61).
Classification and Evaluation of DNA Sequence Variants.
Variants were identified by comparing the exonic and proximal intronic sequences of the screened genes in patients and control subjects to the GenBank reference sequences. Nucleotide substitutions beyond intronic position ±5 and insertions/deletions beyond intronic position ±15 were disregarded. For each variant, the frequency of the minor allele in controls or patients was calculated by dividing the number of chromosomes harboring the variant by the total number of respective chromosomes in that cohort. Based on its MAF in the control cohort, each variant was classified as common (MAF ≥5%), uncommon (1% ≤ MAF <5%), or rare (MAF <1%, which includes patient-only variants with MAF = 0% in controls). Each variant was also classified according to the nature of the DNA change as either synonymous or protein altering (frameshift, nonsense, splice site, or missense). The potential damaging effect of each missense and splice-site variant was evaluated by considering previously published modeling and/or in vitro functional studies and by using in silico prediction methods: PolyPhen-2 (37) and SIFT (38) for missense variants and Human Splicing Finder (62) for splice-site variants.
Assessment of Oligogenicity.
To assess oligogenicity, only rare protein-altering variants were considered. In subjects harboring more than one such variant in the same gene, it was determined whether the variants were on the same or different alleles by sequencing the subject's parents and/or by sequencing individual alleles after cloning PCR amplicons encompassing the variants in the pTOPO vector (Invitrogen). Each patient and control subject was scored as to the total number of alleles harboring rare protein-altering variants. In subjects with more than a single allele with such variants (biallelic or triallelic), it was further noted whether these variants were on alleles of the same or different genes (monogenic or digenic, respectively). Statistical comparisons were performed using the χ2 test. To examine whether the segregation patterns of variants identified in oligogenic pedigrees helped explain the phenotypes of individual family members, all available relatives of oligogenic GnRH-deficient patients were evaluated by history and/or physical examination for reproductive and associated nonreproductive phenotypes and tested for the respective patient's variants.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants U54 HD028138, HD015788-23, HD056264, and GM061354.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009622107/-/DCSupplemental.
References
- 1.Brinkman RR, Dubé MP, Rouleau GA, Orr AC, Samuels ME. Human monogenic disorders— a source of novel drug targets. Nat Rev Genet. 2006;7:249–260. doi: 10.1038/nrg1828. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360:1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 3.McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141:210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 4.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen JC, et al. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 6.Ji W, et al. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet. 2008;40:592–599. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katsanis N. The oligogenic properties of Bardet–Biedl syndrome. Hum Mol Genet. 2004;13(Spec No 1):R65–R71. doi: 10.1093/hmg/ddh092. [DOI] [PubMed] [Google Scholar]
- 8.Kajiwara K, Berson EL, Dryja TP. Digenic retinitis pigmentosa due to mutations at the unlinked peripherin/RDS and ROM1 loci. Science. 1994;264:1604–1608. doi: 10.1126/science.8202715. [DOI] [PubMed] [Google Scholar]
- 9.Hoefele J, et al. Evidence of oligogenic inheritance in nephronophthisis. J Am Soc Nephrol. 2007;18:2789–2795. doi: 10.1681/ASN.2007020243. [DOI] [PubMed] [Google Scholar]
- 10.Badano JL, Katsanis N. Beyond Mendel: An evolving view of human genetic disease transmission. Nat Rev Genet. 2002;3:779–789. doi: 10.1038/nrg910. [DOI] [PubMed] [Google Scholar]
- 11.Seminara SB, Hayes FJ, Crowley WF., Jr Gonadotropin-releasing hormone deficiency in the human (idiopathic hypogonadotropic hypogonadism and Kallmann's syndrome): Pathophysiological and genetic considerations. Endocr Rev. 1998;19:521–539. doi: 10.1210/edrv.19.5.0344. [DOI] [PubMed] [Google Scholar]
- 12.Bianco SD, Kaiser UB. The genetic and molecular basis of idiopathic hypogonadotropic hypogonadism. Nat Rev Endocrinol. 2009;5:569–576. doi: 10.1038/nrendo.2009.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waldstreicher J, et al. The genetic and clinical heterogeneity of gonadotropin-releasing hormone deficiency in the human. J Clin Endocrinol Metab. 1996;81:4388–4395. doi: 10.1210/jcem.81.12.8954047. [DOI] [PubMed] [Google Scholar]
- 14.Jameson JL. Rites of passage through puberty: A complex genetic ensemble. Proc Natl Acad Sci USA. 2007;104:17247–17248. doi: 10.1073/pnas.0708636104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balasubramanian R, et al. Human GnRH deficiency: A unique disease model to unravel the ontogeny of GnRH neurons. Neuroendocrinology. 2010 doi: 10.1159/000314193. 10.1159/000314193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falardeau J, et al. Decreased FGF8 signaling causes deficiency of gonadotropin-releasing hormone in humans and mice. J Clin Invest. 2008;118:2822–2831. doi: 10.1172/JCI34538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung WC, Moyle SS, Tsai PS. Fibroblast growth factor 8 signaling through fibroblast growth factor receptor 1 is required for the emergence of gonadotropin-releasing hormone neurons. Endocrinology. 2008;149:4997–5003. doi: 10.1210/en.2007-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwanzel-Fukuda M. Origin and migration of luteinizing hormone-releasing hormone neurons in mammals. Microsc Res Tech. 1999;44:2–10. doi: 10.1002/(SICI)1097-0029(19990101)44:1<2::AID-JEMT2>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 19.Franco B, et al. A gene deleted in Kallmann's syndrome shares homology with neural cell adhesion and axonal path-finding molecules. Nature. 1991;353:529–536. doi: 10.1038/353529a0. [DOI] [PubMed] [Google Scholar]
- 20.Legouis R, et al. The candidate gene for the X-linked Kallmann syndrome encodes a protein related to adhesion molecules. Cell. 1991;67:423–435. doi: 10.1016/0092-8674(91)90193-3. [DOI] [PubMed] [Google Scholar]
- 21.Dodé C, et al. Kallmann syndrome: Mutations in the genes encoding prokineticin-2 and prokineticin receptor-2. PLoS Genet. 2006;2:e175. doi: 10.1371/journal.pgen.0020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pitteloud N, et al. Loss-of-function mutation in the prokineticin 2 gene causes Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci USA. 2007;104:17447–17452. doi: 10.1073/pnas.0707173104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seminara SB, et al. The GPR54 gene as a regulator of puberty. N Engl J Med. 2003;349:1614–1627. doi: 10.1056/NEJMoa035322. [DOI] [PubMed] [Google Scholar]
- 24.de Roux N, et al. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc Natl Acad Sci USA. 2003;100:10972–10976. doi: 10.1073/pnas.1834399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Topaloglu AK, et al. TAC3 and TACR3 mutations in familial hypogonadotropic hypogonadism reveal a key role for Neurokinin B in the central control of reproduction. Nat Genet. 2009;41:354–358. doi: 10.1038/ng.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Roux N, et al. A family with hypogonadotropic hypogonadism and mutations in the gonadotropin-releasing hormone receptor. N Engl J Med. 1997;337:1597–1602. doi: 10.1056/NEJM199711273372205. [DOI] [PubMed] [Google Scholar]
- 27.Dodé C, et al. Loss-of-function mutations in FGFR1 cause autosomal dominant Kallmann syndrome. Nat Genet. 2003;33:463–465. doi: 10.1038/ng1122. [DOI] [PubMed] [Google Scholar]
- 28.de Roux N, et al. The same molecular defects of the gonadotropin-releasing hormone receptor determine a variable degree of hypogonadism in affected kindred. J Clin Endocrinol Metab. 1999;84:567–572. doi: 10.1210/jcem.84.2.5449. [DOI] [PubMed] [Google Scholar]
- 29.Massin N, et al. X chromosome-linked Kallmann syndrome: Clinical heterogeneity in three siblings carrying an intragenic deletion of the KAL-1 gene. J Clin Endocrinol Metab. 2003;88:2003–2008. doi: 10.1210/jc.2002-021981. [DOI] [PubMed] [Google Scholar]
- 30.Pitteloud N, et al. Mutations in fibroblast growth factor receptor 1 cause both Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci USA. 2006;103:6281–6286. doi: 10.1073/pnas.0600962103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seminara SB, et al. Successful use of pulsatile gonadotropin-releasing hormone (GnRH) for ovulation induction and pregnancy in a patient with GnRH receptor mutations. J Clin Endocrinol Metab. 2000;85:556–562. doi: 10.1210/jcem.85.2.6357. [DOI] [PubMed] [Google Scholar]
- 32.Pitteloud N, et al. Digenic mutations account for variable phenotypes in idiopathic hypogonadotropic hypogonadism. J Clin Invest. 2007;117:457–463. doi: 10.1172/JCI29884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cole LW, et al. Mutations in prokineticin 2 and prokineticin receptor 2 genes in human gonadotrophin-releasing hormone deficiency: Molecular genetics and clinical spectrum. J Clin Endocrinol Metab. 2008;93:3551–3559. doi: 10.1210/jc.2007-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Canto P, Munguía P, Söderlund D, Castro JJ, Méndez JP. Genetic analysis in patients with Kallmann syndrome: Coexistence of mutations in prokineticin receptor 2 and KAL1. J Androl. 2009;30:41–45. doi: 10.2164/jandrol.108.005314. [DOI] [PubMed] [Google Scholar]
- 35.Chan YM, et al. GNRH1 mutations in patients with idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci USA. 2009;106:11703–11708. doi: 10.1073/pnas.0903449106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raivio T, et al. Impaired fibroblast growth factor receptor 1 signaling as a cause of normosmic idiopathic hypogonadotropic hypogonadism. J Clin Endocrinol Metab. 2009;94:4380–4390. doi: 10.1210/jc.2009-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 39.Sykiotis GP, Pitteloud N, Seminara SB, Kaiser UB, Crowley WF., Jr Deciphering genetic disease in the genomic era: The model of GnRH deficiency. Sci Transl Med. 2010;2 doi: 10.1126/scitranslmed.3000288. 32 rv2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pedersen-White JR, Chorich LP, Bick DP, Sherins RJ, Layman LC. The prevalence of intragenic deletions in patients with idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Mol Hum Reprod. 2008;14:367–370. doi: 10.1093/molehr/gan027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trarbach EB, et al. Screening of autosomal gene deletions in patients with hypogonadotropic hypogonadism using multiplex ligation-dependent probe amplification: Detection of a hemizygosis for the fibroblast growth factor receptor 1. Clin Endocrinol (Oxf) 2010;72:371–376. doi: 10.1111/j.1365-2265.2009.03642.x. [DOI] [PubMed] [Google Scholar]
- 42.Monnier C, et al. PROKR2 missense mutations associated with Kallmann syndrome impair receptor signalling activity. Hum Mol Genet. 2009;18:75–81. doi: 10.1093/hmg/ddn318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cerrato F, et al. Coding sequence analysis of GNRHR and GPR54 in patients with congenital and adult-onset forms of hypogonadotropic hypogonadism. Eur J Endocrinol. 2006;155(Suppl 1):S3–S10. doi: 10.1530/eje.1.02235. [DOI] [PubMed] [Google Scholar]
- 44.Pitteloud N, et al. The fertile eunuch variant of idiopathic hypogonadotropic hypogonadism: Spontaneous reversal associated with a homozygous mutation in the gonadotropin-releasing hormone receptor. J Clin Endocrinol Metab. 2001;86:2470–2475. doi: 10.1210/jcem.86.6.7542. [DOI] [PubMed] [Google Scholar]
- 45.Bedecarrats GY, Linher KD, Kaiser UB. Two common naturally occurring mutations in the human gonadotropin-releasing hormone (GnRH) receptor have differential effects on gonadotropin gene expression and on GnRH-mediated signal transduction. J Clin Endocrinol Metab. 2003;88:834–843. doi: 10.1210/jc.2002-020806. [DOI] [PubMed] [Google Scholar]
- 46.Sarfati J, et al. A comparative phenotypic study of kallmann syndrome patients carrying monoallelic and biallelic mutations in the prokineticin 2 or prokineticin receptor 2 genes. J Clin Endocrinol Metab. 2010;95:659–669. doi: 10.1210/jc.2009-0843. [DOI] [PubMed] [Google Scholar]
- 47.Duke VM, et al. KAL, a gene mutated in Kallmann's syndrome, is expressed in the first trimester of human development. Mol Cell Endocrinol. 1995;110:73–79. doi: 10.1016/0303-7207(95)03518-c. [DOI] [PubMed] [Google Scholar]
- 48.Hardelin JP, et al. Anosmin-1 is a regionally restricted component of basement membranes and interstitial matrices during organogenesis: Implications for the developmental anomalies of X chromosome-linked Kallmann syndrome. Dev Dyn. 1999;215:26–44. doi: 10.1002/(SICI)1097-0177(199905)215:1<26::AID-DVDY4>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 49.Abu-Issa R, Smyth G, Smoak I, Yamamura K, Meyers EN. Fgf8 is required for pharyngeal arch and cardiovascular development in the mouse. Development. 2002;129:4613–4625. doi: 10.1242/dev.129.19.4613. [DOI] [PubMed] [Google Scholar]
- 50.Trokovic N, Trokovic R, Mai P, Partanen J. Fgfr1 regulates patterning of the pharyngeal region. Genes Dev. 2003;17:141–153. doi: 10.1101/gad.250703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gianetti E, et al. TAC3/TACR3 mutations reveal preferential activation of gonadotropin-releasing hormone release by neurokinin B in neonatal life followed by reversal in adulthood. J Clin Endocrinol Metab. 2010;95:2857–2867. doi: 10.1210/jc.2009-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bouligand J, et al. Isolated familial hypogonadotropic hypogonadism and a GNRH1 mutation. N Engl J Med. 2009;360:2742–2748. doi: 10.1056/NEJMoa0900136. [DOI] [PubMed] [Google Scholar]
- 53.Zaghloul NA, Katsanis N. Mechanistic insights into Bardet–Biedl syndrome, a model ciliopathy. J Clin Invest. 2009;119:428–437. doi: 10.1172/JCI37041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carronia L, et al. A genetic basis for functional hypothalamic ammenorrhea. N Engl J Med. doi: 10.1056/NEJMoa0911064. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nachtigall LB, Boepple PA, Pralong FP, Crowley WF., Jr Adult-onset idiopathic hypogonadotropic hypogonadism—a treatable form of male infertility. N Engl J Med. 1997;336:410–415. doi: 10.1056/NEJM199702063360604. [DOI] [PubMed] [Google Scholar]
- 56.Raivio T, et al. Reversal of idiopathic hypogonadotropic hypogonadism. N Engl J Med. 2007;357:863–873. doi: 10.1056/NEJMoa066494. [DOI] [PubMed] [Google Scholar]
- 57.Doty RL, Shaman P, Kimmelman CP, Dann MS. University of Pennsylvania Smell Identification Test: A rapid quantitative olfactory function test for the clinic. Laryngoscope. 1984;94:176–178. doi: 10.1288/00005537-198402000-00004. [DOI] [PubMed] [Google Scholar]
- 58.Oliveira LM, et al. The importance of autosomal genes in Kallmann syndrome: Genotype-phenotype correlations and neuroendocrine characteristics. J Clin Endocrinol Metab. 2001;86:1532–1538. doi: 10.1210/jcem.86.4.7420. [DOI] [PubMed] [Google Scholar]
- 59.Miura K, Acierno JS, Jr, Seminara SB. Characterization of the human nasal embryonic LHRH factor gene, NELF, and a mutation screening among 65 patients with idiopathic hypogonadotropic hypogonadism (IHH) J Hum Genet. 2004;49:265–268. doi: 10.1007/s10038-004-0137-4. [DOI] [PubMed] [Google Scholar]
- 60.Pitteloud N, et al. Reversible Kallmann syndrome, delayed puberty, and isolated anosmia occurring in a single family with a mutation in the fibroblast growth factor receptor 1 gene. J Clin Endocrinol Metab. 2005;90:1317–1322. doi: 10.1210/jc.2004-1361. [DOI] [PubMed] [Google Scholar]
- 61.Antonarakis SE Nomenclature Working Group. Recommendations for a nomenclature system for human gene mutations. Hum Mutat. 1998;11:1–3. doi: 10.1002/(SICI)1098-1004(1998)11:1<1::AID-HUMU1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 62.Desmet FO, et al. Human Splicing Finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.