Abstract
Monitoring changes in rhesus macaque immune cell populations during infectious disease is crucial. The aim of this work was to simultaneously analyze the phenotype of rhesus macaque lymphocyte, monocyte and dendritic cell (DC) subsets using a single 12-color flow cytometry panel. Blood from healthy non-infected rhesus macaques was labeled with a cocktail of 12 antibodies. Data were compared to three smaller lineage specific panels and absolute and relative percentages of cells were compared. Our 12-color panel allows for the identification of the following major subsets: CD4+ and CD8+ T lymphocytes, B lymphocytes, natural killer (NK) cells, natural killer T (NKT) cells, monocyte subsets and four non-overlapping HLA-DR+ Lin- cell subsets: CD34+ hematopoietic stem cells, CD11c− CD123+ plasmacytoid DC, CD11c+ CD16+ and CD11c−/dim CD1c+ myeloid DC. The development of a multiparameter flow cytometry panel will allow for simultaneous enumeration of mature lymphocyte, NK cells, monocyte and DC subsets. Studying these major players of the immune system in one panel may give us a broader view of the immune response during SIV infection and the ability to better define the role of each of these individual cell types in the pathogenesis of AIDS.
Keywords: Flow cytometry, immune cells, whole blood, rhesus monkey, dendritic cells
1. Introduction
Nonhuman primates provide essential models for studying human infectious diseases such as acquired immunodeficiency syndrome (AIDS), influenza and tuberculosis (Gardner and Luciw, 2008). The key to our understanding of the immunopathogenesis of diseases such as human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) infection is the precise identification, quantification and analysis of immune cell subsets in SIV infected rhesus macaques. Multicolor flow cytometry is a powerful tool for this (Herzenberg et al., 2002; Tung et al., 2007). Many studies using flow cytometry have underscored the role of lymphocyte, monocyte and dendritic cells (DC) subsets in SIV infection and pathogenesis of AIDS (DeMaria et al., 2000; Ibegbu et al., 2001; Pichyangkul et al., 2001; Pitcher et al., 2002; Mattapallil et al., 2004; Barratt-Boyes et al., 2006; Kim et al., 2009; Williams and Burdo, 2009).
Because of the relative genetic proximity between humans and monkeys, monoclonal anti-human antibodies (mAbs) can often recognize the simian counterpart of human antigens on monkey leukocytes (Reimann et al., 1994; Sopper et al., 1997). However, several key differences exist that can limit the use of anti-human antibodies in non-human primates (Carter et al., 1999; Webster and Johnson, 2005). These include expression of CD56 that is restricted to NK cells in humans, while it is primarily expressed on monocytes and mDC subset in monkeys (Carter et al., 1999; Brown and Barratt-Boyes, 2009). CD8 is expressed on B lymphocytes in rhesus monkeys but not in humans (Webster and Johnson, 2005).
Studies of DC in humans and monkeys are more complex not only because of issues with Abs cross-reactivity but also the nomenclature and subpopulation of these cells is evolving. DC represent less than 1% of total leukocytes and are a heterogeneous population (Palucka and Banchereau, 1999; Banchereau et al., 2000; Banchereau et al., 2003; Ju et al., 2010). Classically, human DCs have been defined as two main subsets: Lin-HLA-DR+CD11c+CD123- mDC and Lin-HLA-DR+CD11c-CD123+ pDC (Palucka and Banchereau, 1999; Banchereau et al., 2003; Steinman, 2003). Human CD11c+ mDC heterogeneity in blood was illustrated by Mac Donald et al who distinguished five non-overlapping subsets within Lin- HLA-DR+ cells: CD11c-CD123+ pDC, CD11c-CD34+ hematopoietic stem cells, and three subsets of CD11c+ mDC expressing CD16, CD1c (BDCA-1) or CD141 (BDCA-3) (MacDonald et al., 2002). We have recently described a single 12-color human flow cytometry panel that distinguished these DC subsets, in addition to major lymphocyte and monocyte subsets (Autissier et al., 2010). Like their human counterparts, rhesus monkey DC subsets are usually defined as Lin-HLA-DR +CD11c+CD123- mDC, and Lin-HLA-DR+CD11c-CD123+ pDC (Coates et al., 2003; Brown et al., 2007; Brown and Barratt-Boyes, 2009).
Based on the single 12-color panel we developed to analyze human leucocytes, we designed a single 12-color flow cytometry panel to measure in rhesus monkey major lymphocyte, monocyte and DC populations (Autissier et al., 2010). Using this panel, we characterized T and B lymphocytes, NK cells, NKT cells, monocytes and four subsets of HLA-DR+Lin- cells on normal non-infected rhesus macaques. In addition to the complete phenotypic characterization of major blood cell types, our 12-color panel pointed out phenotypic differences in DC subsets of rhesus macaques compared to humans, suggesting that more complete flow cytometry panels should be used in order to correctly study all known DC subsets in non-human primates.
2. Material and methods
2.1. Subjects
Venous blood was obtained from twelve healthy non-infected rhesus monkeys (Macaca mulatta) and collected in tubes containing anti-coagulant EDTA (Vacutainer, BD Biosciences). All animals were maintained in accordance with the guidelines of the Committee on Animals for the New England Regional Primate Research Center (NERPRC) and the “Guide for the Care and Use of Laboratory Animals” (Bayne, 1996). Blood samples were processed within 2–4 hours following collection.
2.2. Instrumentation
The optical configuration of the instrument has been previously described (Autissier et al., 2010). Briefly, a Becton Dickinson FACSAria™ cytometer with 3 lasers (BD Biosciences, San Jose, CA) was used for the study. The cytometer was optimized to measure up to 12 fluorescent parameters. The blue laser independently excites 6 fluorochromes (FITC, PE, Texas Red-PE (ECD), Cy5-PE, Cy5.5-PerCP, and Cy7-PE), the red laser can excite 3 fluorochromes (APC, Alexa Fluor 700 and Cy7-APC), and the violet laser can excite 3 fluorochromes (Pacific Blue, Aqua and QDot 655).
2.3. Antibodies used for the study
Our ultimate goal was the development of a 12-color flow cytometry panel to assess major lymphocyte, monocyte, NK cells and DC subsets. When developing a multicolor panel for monkeys, one has to test and choose the brightest antibody-fluorochrome combination available and further optimize the panel for maximum antigen detection (Mahnke and Roederer, 2007). In order to determine the accuracy of this panel, we compared it to smaller panels of select lineages already established in the laboratory, including lymphocytes (7 colors), monocytes (6 colors) and DC (9 colors). The following monoclonal antibodies were used: FITC-CD4 (clone L200), PE-CD34 (clone 563), Cy5-PE-CD16 (clone 3G8), Cy5.5-PerCP-CD123 (clone 7G3), Cy7-PE-CD20 or Cy7-APC-CD20 (clone L27), Cy7-PE-CD3 (clone SP34-2), Pacific Blue-CD14 or Cy7-PE-CD14 (clone M5E2) (all from BD Pharmingen, San Jose, CA); Alexa Fluor 700-CD11c (clone 3.9, eBiosciences, San Diego, CA), TxRed-PE-HLA-DR (clone Immu-357, Beckman Coulter, Miami, FL); APC-CD1c (clone AD5-8E7, Miltenyi Biotech, Auburn, CA); and QD655-CD8 (clone 3B5, Invitrogen, Carlsbad, CA). We also tested FITC-CD1c (clone AD5-8E7) and PE-CD141 (clone AD5-14H12) both from Miltenyi Biotec and PE-CD141 (BD, clone 1A4). Antibodies were titrated to determine optimal concentrations. Antibody-capture beads (CompBeads, BD Biosciences) were used for single-color compensation controls for each reagent used in the study. To exclude dead cells from the analysis, we used in our panel an amine reactive dye as a live/dead discrimination marker (Perfetto et al., 2006). Aqua Live/Dead kit (Invitrogen, Carlsbad, CA) was used first to gate out dead cells. The final composition of the different panels used in this study is shown in the Table. 1.
Table 1.
Flow cytometry panel description for Rhesus monkey.
| Panel | FITC | PE | TxRed-PE | Cy5-PE | Cy5.5-PerCP | Cy7-PE | APC | Alexa 700 | Cy7-APC | Pacific Blue | Aqua | QD655 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 12-Colors | CD4 | CD34 | HLA-DR | CD16 | CD123 | CD3 | CD1c | CD11c | CD20 | CD14 | Live/Dead | CD8 |
| Lymphocyte+ NK+NKT | CD4 | - | - | CD16 | - | CD3 | - | - | CD20 | CD14 | Live/Dead | CD8 |
| Monocyte | - | - | HLA-DR | CD16 | - | CD3 | - | - | CD20 | CD14 | Live/Dead | - |
| DC | - | CD34 | HLA-DR | CD16 | CD123 | CD3+CD14+ CD20 | CD1c | CD11c | - | - | Live/Dead | CD8 |
Abbreviation: FITC: fluorescein isothiocyanate, PE: phycoerythrin, Cy5: Cyanine-5, Cy5.5-PerCP: Cyanine-5.5-peridin-chlorophyll, APC: allophycocyanin, QD: quantum dot.
2.4. Blood samples and staining protocol
We routinely use two 100μl samples of whole blood in a separate tube, to ensure we have enough DC, although this can be scaled up if necessary. Erythrocytes in 100μl of whole blood were lysed using a cell lyse preparation workstation (TQ-Prep instrument, Beckman Coulter). Then, samples from two tubes, equivalent to 200 μl of whole blood, were pooled, washed with 3ml of phosphate buffered saline (PBS) containing 2% fetal bovine serum (FBS) and incubated with a pre-mixed antibody cocktail for 15 minutes at room temperature. After staining, cells were washed with 3ml of PBS containing 2% FBS, and fixed with freshly prepared 1% paraformaldehyde (PFA).
2.5. Data acquisition and sample analysis
Samples were collected on a BD FACS Aria™ flow cytometer (BD Biosciences) and analyzed using FlowJo software 8.7.1 (Treestar, Ashland, OR) on a MAC ® workstation. Instrument calibration was checked daily using rainbow fluorescent particles (BD Biosciences). After acquiring unstained and single color control samples, calculating the compensation matrix, we collected between 5×105 and 1×106 events per sample, which we determined was requested to allow for the collection of at least 200 CD34+ cells and 500 pDC. Doublets were excluded using forward scatter (FSC) area versus FSC height. Dead cells were excluded by staining with amine reactive dye. Lymphocyte, monocyte and DC subsets were identified based on FSC vs. SSC, negative selection and positive expression of their respective markers. Further gating adjustments were made based on fluorescence minus one (FMO) where all antibodies were present except the one of interest as previously described (Roederer, 2002; Autissier et al., 2010).
2.6. Percentages and absolute numbers of lymphocyte, monocyte and DC subsets
Absolute numbers of peripheral blood lymphocyte, monocyte and DC subsets were calculated by multiplying the total white blood cell count (WBC), determined with an automated hematology analyzer, with the total percentage of each cell population as determined by flow cytometry from the whole blood sample (see Table 2).
Table 2.
Percentages and absolute cell numbers of lymphocyte, monocyte and DC subsets. Data were obtained from 12 healthy adult Rhesus monkeys (Macaca Mulatta) by an automated hematology blood analyzer and FACSAria flow cytometer. Median and range are reported.
| Percentage | Absolute count (cells/μl) | |||
|---|---|---|---|---|
| Median | Range | Median | Range | |
| Total Lymphocytes | 30.09 | 8.73 – 47.44 | 1747.4 | 890.1 – 2967.4 |
| T Cells | ||||
| CD3+ | 65.80 | 52.60 – 79.50 | 1083.6 | 468.2 – 1845.5 |
| CD4+ CD8− | 43.48 | 25.51 – 47.44 | 699.3 | 365.1 –1128.5 |
| CD4− CD8+ | 18.27 | 8.04 – 28.85 | 305.9 | 71.5 – 669.7 |
| CD+ CD8+ | 1.57 | 0.45–11.54 | 29.1 | 6.6 – 274.5 |
| CD− CD8− | 3.10 | 1.27–3.93 | 50.5 | 23.2 – 116.6 |
| B Cells | ||||
| CD20+ | 25.77 | 7.78 – 41.18 | 363.7 | 185.0 – 950.1 |
| CD20+ CD8− | 22.74 | 5.06 – 36.11 | 321.5 | 118.8–945.1 |
| CD20+ CD8+ | 2.33 | 0.13–6.27 | 39.3 | 1.9–97.0 |
| CD20+ CD1c− | 4.33 | 2.60–15.17 | 44.18 | 15.48–116.2 |
| CD20+ CD1c+ | 21.44 | 10.53 – 23.29 | 125.05 | 29.28 – 305.0 |
| NK cells | ||||
| Total NK | 5.12 | 2.25 – 18.23 | 100.9 | 41.9 – 336.8 |
| CD16+CD8+CD20− | 4.10 | 1.47–16.19 | 74.4 | 31.2 – 290.9 |
| CD16-CD8+CD20− | 0.98 | 0.54 – 2.76 | 18.3 | 5.3 – 45.2 |
| CD16-CD8+CD20dim | 0.21 | 0.06 – 0.86 | 3.6 | 1.16–15.4 |
| NKT cells | ||||
| CD3+ CD16+ | 0.08 | 0.03 – 0.25 | 1.8 | 0.2 – 4.2 |
| Monocytes | ||||
| HLA-DR+ | 4.52 | 1.90 – 12.23 | 308.6 | 80.6 – 514.5 |
| CD14+ CD16− | 3.74 | 1.59–9.20 | 243.1 | 74.5 – 674.9 |
| CD14+CD16+ | 0.34 | 0.07 – 0.92 | 23.1 | 2.3 – 46.6 |
| CD14+/− CD16+ | 0.2.3 | 0.07 – 0.35 | 12.3 | 0.7–42.9 |
| Dendritic Cells | ||||
| HLA-DR+/Lin− | 1.34 | 0.30 – 2.89 | 70.8 | 17.9 – 143.6 |
| pDC CD123+ | 0.03 | 0.00 – 0.08 | 1.8 | 0.5–5.3 |
| Total mDC | 0.96 | 0.23 – 2.29 | 54.5 | 10.2–126.9 |
| mDC1 CD11c+ CD16+ | 0.61 | 0.06–2.17 | 38.1 | 4.2–108.3 |
| mDC2 CD11c− CD1c+ | 0.25 | 0.07 – 0.53 | 17.0 | 3.7 – 37.4 |
| CD34+ | 0.01 | 0.00 – 0.02 | 0.7 | 0.2–1.7 |
3. Results
3.1. Panel strategy and development
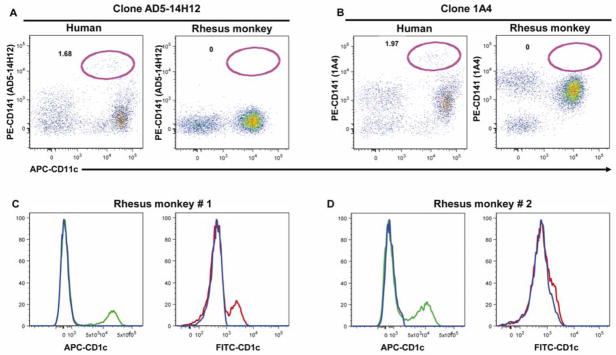
The goal of this study was to design and develop a 12-color flow cytometry panel that would allow simultaneous analysis of the lymphocyte, monocyte and DC populations in rhesus macaques. Bandpass and dichroic filters used in this study have been previously described (Autissier et al., 2010). Taking advantage that monoclonal antibodies directed against human antigens often cross-react to rhesus monkeys, our first approach was to start from a 12-color panel that we developed for human specimens (Autissier et al., 2010). Next we verified that optimal antibodies used in humans cross-reacted with rhesus macaques and their fluorescence signals were significantly robust to delineate cell types and subpopulations. The majority of antibody clones from our human panel cross-reacted to monkeys, with the exception of anti-CD141, CD11c and CD34 antibodies (Autissier et al., 2010). In humans, CD141 is expressed at a high level on a small CD11c+ mDC subset (Dzionek et al., 2000). Two different clones of the anti-CD141 antibody were tested, AD5-14H12 (Miltenyi-Biotech) and 1A4 (BD). As shown in Figure 1A, we found that the clone AD5-14H12 did not cross-react with monkeys, as previously published by others (Coates et al., 2003). The clone 1A4 cross-reacted with monkeys but did not delineate the CD141bright mDC subset described in humans (Figure 1B). Thus we removed the anti-CD141 antibody from our monkey panel. The Cy5-PE-anti-CD11c (BD, clone B-Ly6) and TxRed-PE-anti-CD34 (Beckman-Coulter, clone 581) antibodies did not cross-react with rhesus monkeys and were substituted with Alexa 700-anti-CD11c (eBiosciences, clone 3.9) and PE-anti-CD34 (BD, clone 563) antibodies, respectively. Because anti-CD56 cross-reacts to monkey monocytes and mDC, we did not include the anti-CD56 antibody in the lineage panel, which includes antibodies against CD3, CD14 and CD20 (Brown and Barratt-Boyes, 2009). Next, we tested the anti-CD1c (clone AD5-8E7) antibody, conjugated to FITC or APC. The FITC-conjugated antibody showed variable immunoreactivity giving a very weak signal in one animal (Figure 1C) or no signal in a second animal (Figure 1D). By contrast, the APC-conjugated antibody always gave a consistent signal and was therefore chosen to be included in the 12-color panel. Finally, we tested antibodies against “primary” antigens (CD3, CD4, CD8, CD14, CD16, CD20 and HLA-DR) in all possible fluorochromes available (data not shown), selecting those with the maximum signal. The final 12-color flow cytometry panel, optimized for the detection of lymphocyte, monocyte and DC in rhesus monkeys is shown in Table 1.
Figure 1. Cross reactivity assessment of anti-CD141 (A and B) and anti-CD1c (C and D) antibodies.
Two different clones of anti-CD141 antibody, AD5-14H12 (Miltenyi Biotech) (A) and 1A4 (BD Biosciences) (B) were tested on whole blood from humans and rhesus monkeys. After gating on HLA-DR+ Lin− cells [CD3 + CD14 + CD20 + CD56 (human) or CD8 (monkey)], dotplot of CD11c versus CD141 are shown. Percentages of human CD141brightCD11c+ mDC subset are comparable using either clone (A and B). In rhesus monkey, clone AD5-14H12 does not cross-react with monkey cells (A), whereas clone 1A4 cross-reacts but does not identify the CD141bright mDC subset described in human (B). Anti-CD1c antibody was tested in rhesus monkeys using either FITC- or APC- conjugate (Miltenyi Biotech). After gating on HLA-DR+Lin− cells (CD3 + CD14 + CD20 + CD8), APC-CD1c (green histogram) shows strong staining whereas FITC-CD1c (red histogram) shows either weak staining (C) or no staining at all (D). Results presented here are from 2 different monkeys.
3.2. Phenotypic analysis of lymphocyte populations
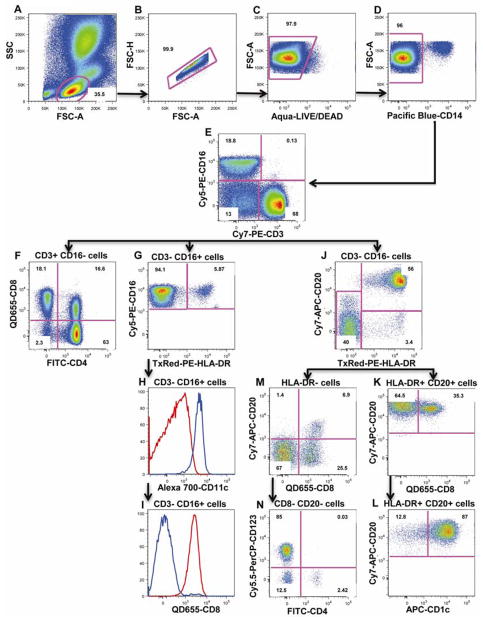
In order to identify lymphocyte populations, we first gated on cells based on forward (FSC) and side scatter (SSC) properties (Figure 2A). We excluded doublets (Figure 2B), dead cells (Figure 2C) and monocytes (Figure 2D), using FSC height, amine reactive dye and CD14 expression, respectively. From this gate, CD3+CD16− T lymphocytes (median: 65.8%, range: 52.6%–79.5%, n=12) were selected (Figure 2E) and further divided into CD4+CD8− (median: 43.48%, range: 25.51%–47.44%, n=12), CD4−CD8+ (median: 18.27%, range: 8.04%–28.85%, n=12), CD4+CD8+ (median: 1.57%, range: 0.45%–11.54%, n=12) and CD4−CD8− (likely gamma delta T cells) (median: 3.10%, range: 1.27%–3.93%, n=12) lymphocytes (Figure 2F). In addition, using an anti-CD16 antibody we identified CD3+CD16+ NKT cells (median: 0.08%, range: 0.03%–0.25%, n=12) (Figure 2E) as well as CD3−CD16+ cells (Figure 2E). Within the CD3−CD16+ cells, we identified a major HLA-DR-CD8+ NK population (median: 4.10%, range: 1.47%–16.19%, n=12; Figure 2G; 2H and 2I: red histogram), as well as a small population of myeloid DC subset expressing HLA-DR (CD3-CD16+HLA-DR+, Figure 2G), CD11c (blue histogram, Figure 2H), but not CD8 (blue histogram, Figure 2I). CD3−CD16− cells (Figure 2E) were further divided into HLA-DR+CD20+ B cells (median: 25.77%, range: 7.78%–41.18%, n=12) and HLA-DR-CD20−/dim cells (Figure 2J). HLA-DR+CD20+ B lymphocytes can be further divided into CD20+CD8− (median: 22.74%, range: 5.06%–36.11%, n=12) and CD20+CD8+ B lymphocytes (median: 2.33%, range: 0.13%–6.27%, n=12; Figure 2K). HLA-DR+CD20+ B cells can also be divided into CD1c+ resting (median: 21.44%, range: 10.53%–23.29%, n=12) and CD1c- activated (median: 4.33%, range: 2.60%–15.17%, n=12) B cells (Figure 2L). HLA-DR-CD20−/dim cells can be further divided into 2 small subsets of NK cells: CD8+CD20− cells (median: 0.98%, range: 0.54%–2.76%, n=12) and CD8+CD20dim cells (median: 0.21%, range: 0.06%–0.86%, n=12) (Figure 2M). Finally, the overall contamination of non-lymphocyte cells within the lymphocyte gate represent consistently 3% of the total lymphocyte population, and is mainly comprised of HLA-DR+CD20− cells (Figure 2J) and CD123+ basophil granulocytes (Figure 2N).
Figure 2. Flow cytometry analysis of lymphocyte populations.
First, lymphocytes were gated based on FSC and SSC (A), followed by exclusion of doublets (B), dead cells (C) and CD14+ monocytes (D). T lymphocytes (CD3+CD16−), NK (CD3−CD16+) and NKT (CD3+CD16+) populations are identified using CD3 and CD16 (E). CD4+ and CD8+ lymphocytes were distinguished within the CD3+ T lymphocytes (F). CD16+HLA-DR- NK cells and a CD16+HLA-DR+ mDC subset were distinguished within the CD3−CD16+ population (G). Both subsets can also be distinguished based on CD11c and CD8 expression (H and I). HLA-DR- cells and CD20+HLA-DR+ B cells are distinguished within CD3-CD16- cells (J). Gating on CD20+HLA-DR+ cells, 2 subsets of B cells are distinguished based on CD8 expression (K) or CD1c expression (L). From HLA-DR- cells, 2 minor subsets of NK cells can be distinguished based on CD8 and CD20 expression (M). Results presented here are from one animal and are representative of n=12. Numbers are percentages of each population within the same dotplot.
3.3. Phenotypic analysis of monocyte populations
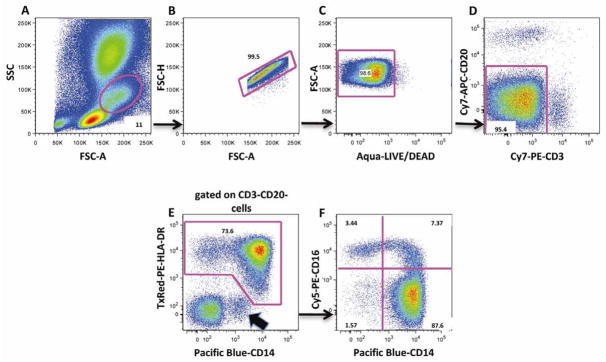
Monocyte populations were analyzed by first gating using FSC and SSC (Figure 3A). We excluded doublets (Figure 3B) and dead cells (Figure 3C) using FSC height and amine dye, respectively, and then excluded CD3+ T and CD20+ B lymphocytes (Figure 3D). From the HLA-DR+ and CD14+ cells (median: 4.52%, range: 1.90%–12.23%, n=12) (Figure 3E), three monocyte subsets were distinguished: CD14+CD16− classical monocytes (median: 3.74%, range: 1.59%–9.20%, n=12), and two subsets of activated monocytes: CD14+CD16+ (median: 0.34%, range: 0.07%–0.92%, n=12) and CD14−CD16+ (median: 0.23%, range: 0.07%–0.35%, n=10) (Figure 3F). It can be noted that potential contamination of CD14lowHLA-DR- granulocytes can be easily identified (Figure 3E).
Figure 3. Flow cytometry analysis of monocyte populations.
First, monocytes were gated based on FSC and SSC (A). Doublets (B), dead cells (C), CD3+ T and CD20+ B lymphocytes (D) were excluded. HLA-DR+ and CD14+ cells were selected (E) and three monocyte subsets were gated based on expression of CD14 and/or CD16. These include the classical monocytes (CD14+CD16−) and activated monocytes (CD14+CD16+ and CD14−CD16+) (F). Contamination from granulocytes that are HLA-DR- and CD14dim are excluded from monocyte analysis, based on HLA-DR and CD14 expression (Figure 3E, black arrow). Results presented here are from one animal and are representative of n=12. Numbers are percentages of each population within the same dotplot.
3.4. Phenotypic analysis of dendritic cell populations
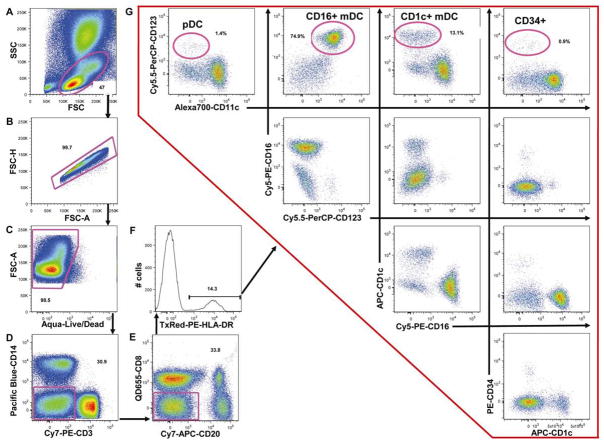
DC have an intermediate size that falls between that of monocytes and lymphocytes, when analyzed in FSC versus SSC (Figure 4A). After excluding doublets (Figure 4B) and dead cells (Figure 4C), DC were gated as HLA-DR+ cells and lineage-negative (Lin−) cells by excluding CD3+ T lymphocytes and CD14+ monocytes (Figure 4D), CD8+ NK cells and CD20+ B lymphocytes (Figure 4E) and selecting HLA-DR+ cells (Figure 4F). HLA-DR+Lin− cells (median: 1.34%, range: 0.30%–2.89%, n=12) were further divided into CD123+CD11c− pDC (median: 0.03%, range: 0%–0.08%, n=12), CD123-CD11c+CD16+ mDC (median: 0.61%, range: 0.06%–2.17%, n=12) and CD123-CD11c−/dim CD1c+ mDC (median: 0.25%, range: 0.07%–0.53%, n=12) (Figure 4G). Finally, CD34+ hematopoietic stem cells (median: 0.01%, range: 0%–0.02%, n=12) were identified within the CD123-CD11c-CD1c-CD16- cell population (Figure 4G).
Figure 4. Flow cytometry analysis of dendritic cell populations.
DC cells were gated based on FSC and SSC including both lymphocytes and monocytes (A). Doublets (B) and dead cells (C) were excluded. Lin− cells (i.e. CD3+ T lymphocytes and CD14+ monocytes (D), CD20+ B lymphocytes and CD8+ NK cells (E) were excluded and HLA-DR+ cells were gated (F). Four non-overlapping HLA-DR+Lin- cell subsets are shown in G and are distinguished based on CD123, CD1c, CD16, CD11c and CD34 expression. Results presented here are from one animal and are representative of n=12. Numbers are percentages of each population within the same dotplot.
3.5. Individual Lineage specific panels versus 12-color panel
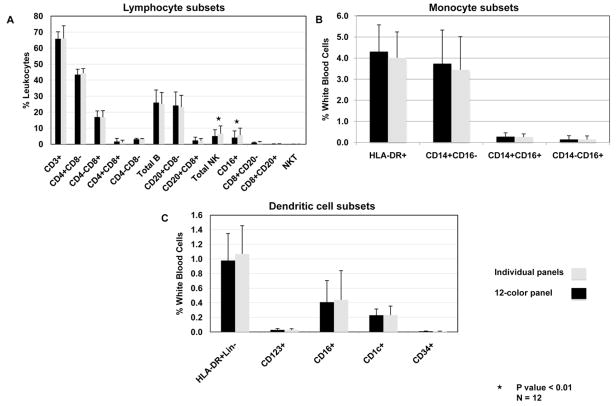
To further validate the 12-color flow cytometry panel, we compared data generated from our 12-color flow panel with data obtained using three smaller flow cytometry panels, specific for lymphocytes, monocytes and dendritic cells. The different panels are described in Table 1. Comparing the two panels (Figure 5A), the percentage of the different leukocyte subsets were very comparable with no statistical differences, except for the total NK population, where a higher percentage of CD16+ NK cells was found using the small lymphocyte panel as compared with the 12-color panel (percent change: 29.6%, P=0.0001). This difference is likely due to the inclusion of CD16+ mDC subset in the NK population using the small lymphocyte panel as opposed to exclusion of this mDC subset in the 12-color panel because of the use of HLA-DR (Figure 2G). No statistical differences were measured on the monocyte subsets and the DC subsets using either panel (Figure 5B and 5C).
Figure 5. Comparison between individual lineage specific panels and the 12-color panel.
(A) Lymphocyte subsets, (B) monocyte subsets, (C) DC subsets. Individual lineage specific panels and a 12-color panel are described in Table 1B. Percentage median and Intra Quartile Range were calculated for each subset (n=12). Statistical significance was determined by using 2-tailed paired Students’ t-test where * P< 0.01.
3.6. Absolute cell numbers of lymphocyte, monocyte and DC subsets by a single 12-color flow cytometry assay
Using the gating strategy previously described, we find a normal distribution of lymphocyte, monocyte and DC subsets in rhesus macaques that are in agreement with data obtained using three flow cytometry panels specific for each population. Percentages and absolute numbers of lymphocytes, monocytes and DC subsets are shown in Table 2.
4. Discussion
Identifying and measuring precisely immune cell subsets during longitudinal infectious studies in rhesus macaques is critical to our understanding of diseases such as SIV infection. The CD8+ lymphocyte depletion model in rhesus macaques has clearly demonstrated the importance of a strong adaptive immune response, mainly through the CD8 T lymphocyte (Schmitz et al., 1999). However, events during early acute infection where innate immune effector cells interact with the virus might be of crucial importance for the progression of the infection and ultimately the outcome of the disease (Pereira and Ansari, 2009). In addition, there is a strong B cell depletion very early after SIV infection (Mattapallil et al., 2004; Klatt et al., 2010). It suggests that a number of immune cells, from both the innate and adaptive immune response, are affected, directly or indirectly, by SIV infection. The 12-color flow cytometry panel described in this study demonstrates that we can reliably identify simultaneously major lymphocyte populations, comprising CD4+ and CD8+ T lymphocytes, CD20+ B lymphocytes, some of which express CD8, 3 subsets of NK cells (i.e. CD16+CD8+CD20−, CD16-CD8+CD20− and CD16-CD8+CD20−/dim) and NKT cells. As demonstrated in this study, identifying NK cells as CD8+CD3− or CD16+CD3− cells might overestimate the percentage of NK cells by including some CD20+ B lymphocytes that express CD8 and mDC that express CD16. An ideal phenotyping flow cytometry panel for NK cells in rhesus monkeys would minimally include at least anti-CD3, anti-CD20, anti-CD8, anti-CD16 and anti-HLA-DR antibodies.
Monocytes/macrophages are thought to be one of the main reservoirs for HIV and SIV (Collman et al., 2003; Montaner et al., 2006). They represent early targets of infection and might contribute to the central nervous system (CNS) infection by bringing the virus into the brain (Pulliam et al., 1997; Williams et al., 2001; Kim et al., 2003; Williams and Burdo, 2009). In addition, subsets of non-infected monocytes expand in bone marrow with AIDS and traffic to the CNS and accumulate with disease (Burdo et al., 2010). Monocytes are a heterogeneous population identified by the expression of CD14 and CD16 (Ellery and Crowe, 2005; Crowe and Ziegler-Heitbrock, 2010). Using our multicolor panel we can clearly identify the 3 monocyte subsets: CD14+CD16− classical monocytes, and two subsets of activated monocytes defined as CD14+CD16+ and CD14−CD16+ cells.
The study of DC in non-human primates is more complicated in part by the lack of cross-reactivity and/or poor sensitivity of some anti-human antibodies. First, because of limited channels available on flow cytometers, researchers used lineage specific cocktails as exclusion markers for DC. In humans, different lineage cocktails have been used that are usually composed of anti-CD3, anti-CD19 and/or anti-CD20, and anti-CD14 antibodies to gate out T and B lymphocytes and monocytes respectively. In addition, some groups alternatively complete this lineage cocktail with an anti-CD16 antibody (Della Bella et al., 2008), anti-CD56 antibody (Piccioli et al., 2007) or both (Ma et al., 2004; Zabel et al., 2005) to exclude NK cells. In monkeys, CD56 is mainly expressed on monocytes and not on NK cells leading to the use of CD16 instead of CD56 as a lineage marker in monkey flow cytometry analyses (Carter et al., 1999; Pichyangkul et al., 2001). However since the CD11c+ mDC in rhesus macaque express high levels of CD16 (Brown and Barratt-Boyes, 2009), using an anti-CD16 antibody in the lineage cocktail gates out a significant proportion of CD11c+ mDC. Different studies on non-human primates are using either CD11c or CD1c as markers of mDC (Malleret et al., 2008; Brown and Barratt-Boyes, 2009). Here we report that the use of these markers taken individually tends to underestimate the percentage of mDC. Using both anti-CD11c and anti-CD1c antibodies in the same panel, we found that CD11c+ mDC do not express CD1c but are CD16+ and that CD1c+ mDC are CD11c−/dimCD16-. Thus by using only CD11c as an mDC marker in rhesus macaques, the CD1c+ mDC subset is not taken into consideration in the analysis. Conversely, the use of CD1c only tends to exclude the CD11c+ CD16+ mDC subset from the analysis. In addition, by comparing the same CD1c clone (AD5-8E7) conjugated either with FITC or APC, we clearly show that the APC conjugate gives a strong signal, while the FITC gives very little or no signal suggesting that the FITC-conjugated anti-CD1c antibody is not appropriate to study mDC in non-human primates. In order to study all known DC subsets, we suggest that an optimal flow cytometry panel for DC phenotyping analyses in non-human primates should include anti-CD11c, anti-CD16, anti-CD1c and anti-CD123 antibodies in addition to anti-CD3, anti-CD14 and anti-CD19 or anti-CD20 antibodies as exclusion lineage markers.
In summary, we show that lymphocyte, monocyte and dendritic cell subsets, can be precisely measured using a 12-color multiparameter flow cytometry approach. Moreover, this assay is more precise than 4- or 5-color panels due in part to minimal contamination between separate populations. Using our 12-color flow cytometry panel, we clearly distinguished three different DC subsets in rhesus macaques based on CD123, CD11c, CD16 and CD1c expression: CD123+CD11c- pDC, and two mDC subsets (CD11c + CD16+CD1c− and CD11c−/dimCD16−CD1c+). Finally, we believe that this 12-color panel will be an important tool to study the interactions between different immune cell populations during SIV and potentially other diseases, and to better understand the key role that DC play in disease progression.
Acknowledgments
This work was supported in part by USA National Institutes of Health (R01NS37654 and R01NS40237).
The authors thank Angela Carville, veterinarian of the New England Regional Primate Center, Southborough, MA, for help with the blood draw protocol and care of the animals.
Footnotes
Abbreviations: DC, dendritic cells; NK, natural killer; NKT, natural killer T; mDC, myeloid DC; pDC, plasmacytoid DC; Lin, lineage; FMO, fluorescence minus one; CNS; central nervous system
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Autissier P, Soulas C, Burdo TH, Williams KC. Evaluation of a 12-color flow cytometry panel to study lymphocyte, monocyte, and dendritic cell subsets in humans. Cytometry A. 2010;77:410–9. doi: 10.1002/cyto.a.20859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu YJ, Pulendran B, Palucka K. Immunobiology of dendritic cells. Annu Rev Immunol. 2000;18:767–811. doi: 10.1146/annurev.immunol.18.1.767. [DOI] [PubMed] [Google Scholar]
- Banchereau J, Paczesny S, Blanco P, Bennett L, Pascual V, Fay J, Palucka AK. Dendritic cells: controllers of the immune system and a new promise for immunotherapy. Ann N Y Acad Sci. 2003;987:180–7. doi: 10.1111/j.1749-6632.2003.tb06047.x. [DOI] [PubMed] [Google Scholar]
- Barratt-Boyes SM, Brown KN, Melhem N, Soloff AC, Gleason SM. Understanding and exploiting dendritic cells in human immunodeficiency virus infection using the nonhuman primate model. Immunol Res. 2006;36:265–74. doi: 10.1385/IR:36:1:265. [DOI] [PubMed] [Google Scholar]
- Bayne K. Revised Guide for the Care and Use of Laboratory Animals available. American Physiological Society. Physiologist. 1996;39:199, 208–11. [PubMed] [Google Scholar]
- Brown KN, Barratt-Boyes SM. Surface phenotype and rapid quantification of blood dendritic cell subsets in the rhesus macaque. J Med Primatol. 2009;38:272–8. doi: 10.1111/j.1600-0684.2009.00353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KN, Trichel A, Barratt-Boyes SM. Parallel loss of myeloid and plasmacytoid dendritic cells from blood and lymphoid tissue in simian AIDS. J Immunol. 2007;178:6958–67. doi: 10.4049/jimmunol.178.11.6958. [DOI] [PubMed] [Google Scholar]
- Burdo TH, Soulas C, Orzechowski K, Button J, Krishnan A, Sugimoto C, Alvarez X, Kuroda MJ, Williams KC. Increased monocyte turnover from bone marrow correlates with severity of SIV encephalitis and CD163 levels in plasma. PLoS Pathog. 2010;6:e1000842. doi: 10.1371/journal.ppat.1000842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter DL, Shieh TM, Blosser RL, Chadwick KR, Margolick JB, Hildreth JE, Clements JE, Zink MC. CD56 identifies monocytes and not natural killer cells in rhesus macaques. Cytometry. 1999;37:41–50. [PubMed] [Google Scholar]
- Coates PT, Barratt-Boyes SM, Zhang L, Donnenberg VS, O’Connell PJ, Logar AJ, Duncan FJ, Murphey-Corb M, Donnenberg AD, Morelli AE, Maliszewski CR, Thomson AW. Dendritic cell subsets in blood and lymphoid tissue of rhesus monkeys and their mobilization with Flt3 ligand. Blood. 2003;102:2513–21. doi: 10.1182/blood-2002-09-2929. [DOI] [PubMed] [Google Scholar]
- Collman RG, Perno CF, Crowe SM, Stevenson M, Montaner LJ. HIV and cells of macrophage/dendritic lineage and other non-T cell reservoirs: new answers yield new questions. J Leukoc Biol. 2003;74:631–4. doi: 10.1189/jlb.0703357. [DOI] [PubMed] [Google Scholar]
- Crowe SM, Ziegler-Heitbrock L. Editorial: Monocyte subpopulations and lentiviral infection. J Leukoc Biol. 2010;87:541–3. doi: 10.1189/jlb.0909637. [DOI] [PubMed] [Google Scholar]
- Della Bella S, Giannelli S, Taddeo A, Presicce P, Villa ML. Application of six-color flow cytometry for the assessment of dendritic cell responses in whole blood assays. J Immunol Methods. 2008;339:153–64. doi: 10.1016/j.jim.2008.09.009. [DOI] [PubMed] [Google Scholar]
- DeMaria MA, Casto M, O’Connell M, Johnson RP, Rosenzweig M. Characterization of lymphocyte subsets in rhesus macaques during the first year of life. Eur J Haematol. 2000;65:245–57. doi: 10.1034/j.1600-0609.2000.065004245.x. [DOI] [PubMed] [Google Scholar]
- Dzionek A, Fuchs A, Schmidt P, Cremer S, Zysk M, Miltenyi S, Buck DW, Schmitz J. BDCA-2, BDCA-3, and BDCA-4: three markers for distinct subsets of dendritic cells in human peripheral blood. J Immunol. 2000;165:6037–46. doi: 10.4049/jimmunol.165.11.6037. [DOI] [PubMed] [Google Scholar]
- Ellery PJ, Crowe SM. Phenotypic characterization of blood monocytes from HIV-infected individuals. Methods Mol Biol. 2005;304:343–53. doi: 10.1385/1-59259-907-9:343. [DOI] [PubMed] [Google Scholar]
- Gardner MB, Luciw PA. Macaque models of human infectious disease. ILAR J. 2008;49:220–55. doi: 10.1093/ilar.49.2.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzenberg LA, Parks D, Sahaf B, Perez O, Roederer M. The history and future of the fluorescence activated cell sorter and flow cytometry: a view from Stanford. Clin Chem. 2002;48:1819–27. [PubMed] [Google Scholar]
- Ibegbu C, Brodie-Hill A, Kourtis AP, Carter A, McClure H, Chen ZW, Nahmias AJ. Use of human CD3 monoclonal antibody for accurate CD4+ and CD8+ lymphocyte determinations in macaques: phenotypic characterization of the CD3- CD8+ cell subset. J Med Primatol. 2001;30:291–8. doi: 10.1034/j.1600-0684.2001.300601.x. [DOI] [PubMed] [Google Scholar]
- Ju X, Clark G, Hart DN. Review of human DC subtypes. Methods Mol Biol. 2010;595:3–20. doi: 10.1007/978-1-60761-421-0_1. [DOI] [PubMed] [Google Scholar]
- Kim WK, Corey S, Alvarez X, Williams K. Monocyte/macrophage traffic in HIV and SIV encephalitis. J Leukoc Biol. 2003;74:650–6. doi: 10.1189/jlb.0503207. [DOI] [PubMed] [Google Scholar]
- Kim WK, Sun Y, Do H, Autissier P, Halpern EF, Piatak M, Jr, Lifson JD, Burdo TH, McGrath MS, Williams K. Monocyte heterogeneity underlying phenotypic changes in monocytes according to SIV disease stage. J Leukoc Biol. 2009 doi: 10.1189/jlb.0209082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatt NR, Shudo E, Ortiz AM, Engram JC, Paiardini M, Lawson B, Miller MD, Else J, Pandrea I, Estes JD, Apetrei C, Schmitz JE, Ribeiro RM, Perelson AS, Silvestri G. CD8+ lymphocytes control viral replication in SIVmac239-infected rhesus macaques without decreasing the lifespan of productively infected cells. PLoS Pathog. 2010;6:e1000747. doi: 10.1371/journal.ppat.1000747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Scheers W, Vandenberghe P. A flow cytometric method for determination of absolute counts of myeloid precursor dendritic cells in peripheral blood. J Immunol Methods. 2004;285:215–21. doi: 10.1016/j.jim.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Mahnke YD, Roederer M. Optimizing a multicolor immunophenotyping assay. Clin Lab Med. 2007;27:469–85. v. doi: 10.1016/j.cll.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malleret B, Karlsson I, Maneglier B, Brochard P, Delache B, Andrieu T, Muller-Trutwin M, Beaumont T, McCune JM, Banchereau J, Le Grand R, Vaslin B. Effect of SIVmac infection on plasmacytoid and CD1c+ myeloid dendritic cells in cynomolgus macaques. Immunology. 2008;124:223–33. doi: 10.1111/j.1365-2567.2007.02758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattapallil JJ, Letvin NL, Roederer M. T-cell dynamics during acute SIV infection. AIDS. 2004;18:13–23. doi: 10.1097/00002030-200401020-00002. [DOI] [PubMed] [Google Scholar]
- Montaner LJ, Crowe SM, Aquaro S, Perno CF, Stevenson M, Collman RG. Advances in macrophage and dendritic cell biology in HIV-1 infection stress key understudied areas in infection, pathogenesis, and analysis of viral reservoirs. J Leukoc Biol. 2006;80:961–4. doi: 10.1189/jlb.0806488. [DOI] [PubMed] [Google Scholar]
- Palucka K, Banchereau J. Dendritic cells: a link between innate and adaptive immunity. J Clin Immunol. 1999;19:12–25. doi: 10.1023/a:1020558317162. [DOI] [PubMed] [Google Scholar]
- Pereira LE, Ansari AA. A case for innate immune effector mechanisms as contributors to disease resistance in SIV-infected sooty mangabeys. Curr HIV Res. 2009;7:12–22. doi: 10.2174/157016209787048465. [DOI] [PubMed] [Google Scholar]
- Perfetto SP, Chattopadhyay PK, Lamoreaux L, Nguyen R, Ambrozak D, Koup RA, Roederer M. Amine reactive dyes: an effective tool to discriminate live and dead cells in polychromatic flow cytometry. J Immunol Methods. 2006;313:199–208. doi: 10.1016/j.jim.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Piccioli D, Tavarini S, Borgogni E, Steri V, Nuti S, Sammicheli C, Bardelli M, Montagna D, Locatelli F, Wack A. Functional specialization of human circulating CD16 and CD1c myeloid dendritic-cell subsets. Blood. 2007;109:5371–9. doi: 10.1182/blood-2006-08-038422. [DOI] [PubMed] [Google Scholar]
- Pichyangkul S, Saengkrai P, Yongvanitchit K, Limsomwong C, Gettayacamin M, Walsh DS, Stewart VA, Ballou WR, Heppner DG. Isolation and characterization of rhesus blood dendritic cells using flow cytometry. J Immunol Methods. 2001;252:15–23. doi: 10.1016/s0022-1759(01)00327-1. [DOI] [PubMed] [Google Scholar]
- Pitcher CJ, Hagen SI, Walker JM, Lum R, Mitchell BL, Maino VC, Axthelm MK, Picker LJ. Development and homeostasis of T cell memory in rhesus macaque. J Immunol. 2002;168:29–43. doi: 10.4049/jimmunol.168.1.29. [DOI] [PubMed] [Google Scholar]
- Pulliam L, Gascon R, Stubblebine M, McGuire D, McGrath MS. Unique monocyte subset in patients with AIDS dementia. Lancet. 1997;349:692–5. doi: 10.1016/S0140-6736(96)10178-1. [DOI] [PubMed] [Google Scholar]
- Reimann KA, Waite BC, Lee-Parritz DE, Lin W, Uchanska-Ziegler B, O’Connell MJ, Letvin NL. Use of human leukocyte-specific monoclonal antibodies for clinically immunophenotyping lymphocytes of rhesus monkeys. Cytometry. 1994;17:102–8. doi: 10.1002/cyto.990170113. [DOI] [PubMed] [Google Scholar]
- Roederer M. Compensation in flow cytometry. Curr Protoc Cytom. 2002;Chapter 1(Unit 1):14. doi: 10.1002/0471142956.cy0114s22. [DOI] [PubMed] [Google Scholar]
- Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, Racz P, Tenner-Racz K, Dalesandro M, Scallon BJ, Ghrayeb J, Forman MA, Montefiori DC, Rieber EP, Letvin NL, Reimann KA. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–60. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- Sopper S, Stahl-Hennig C, Demuth M, Johnston IC, Dorries R, ter Meulen V. Lymphocyte subsets and expression of differentiation markers in blood and lymphoid organs of rhesus monkeys. Cytometry. 1997;29:351–62. [PubMed] [Google Scholar]
- Steinman RM. Some interfaces of dendritic cell biology. APMIS. 2003;111:675–97. doi: 10.1034/j.1600-0463.2003.11107802.x. [DOI] [PubMed] [Google Scholar]
- Tung JW, Heydari K, Tirouvanziam R, Sahaf B, Parks DR, Herzenberg LA. Modern flow cytometry: a practical approach. Clin Lab Med. 2007;27:453–68. v. doi: 10.1016/j.cll.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RL, Johnson RP. Delineation of multiple subpopulations of natural killer cells in rhesus macaques. Immunology. 2005;115:206–14. doi: 10.1111/j.1365-2567.2005.02147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KC, Burdo TH. HIV and SIV infection: the role of cellular restriction and immune responses in viral replication and pathogenesis. APMIS. 2009;117:400–12. doi: 10.1111/j.1600-0463.2009.02450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KC, Corey S, Westmoreland SV, Pauley D, Knight H, deBakker C, Alvarez X, Lackner AA. Perivascular macrophages are the primary cell type productively infected by simian immunodeficiency virus in the brains of macaques: implications for the neuropathogenesis of AIDS. J Exp Med. 2001;193:905–15. doi: 10.1084/jem.193.8.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabel BA, Silverio AM, Butcher EC. Chemokine-like receptor 1 expression and chemerin-directed chemotaxis distinguish plasmacytoid from myeloid dendritic cells in human blood. J Immunol. 2005;174:244–51. doi: 10.4049/jimmunol.174.1.244. [DOI] [PubMed] [Google Scholar]