Abstract
The p53 tumor suppressor protein and the MDM2 oncoprotein form a feedback-control loop that up-regulates cellular MDM2 production, blocks p53 activity, and promotes p53 decay. tsg101 was discovered as a gene whose deficiency results in neoplastic transformation of NIH 3T3 cells and the ability to generate metastatic tumors in nude mice. Its protein product contains a domain, Ubc, characteristic of the catalytic domain of ubiquitin conjugase (E2) enzymes but lacking an active-site cysteine crucial for ubiquitin conjugase activity. Here we report that TSG101 participates with MDM2 in an autoregulatory loop that modulates the cellular levels of both proteins, and also of p53, by affecting protein decay. We show that the Ubc domain of TSG101 interferes with ubiquitination of MDM2, that TSG101 inhibits MDM2 decay and elevates its steady-state level, and that these events are associated with down-regulation of p53 protein. Conversely, pulse–chase and Western blot experiments in wild-type and mutant fibroblasts indicate that elevation of MDM2 by overexpression of wild-type p53, by amplification of the endogenous MDM2 gene, or by transfection of MDM2-expressing constructs promotes TSG101 loss, which we show occurs by 26S proteasome-dependent decay. Our results identify TSG101 as both a regulator of, and target of, MDM2/p53 circuitry.
Keywords: tumorigenesis, ubiquitination, proteolysis
The tsg101 tumor susceptibility gene initially was identified by the reversible neoplasia associated with deficiency of its protein product in mouse fibroblasts (1). Deficiency of TSG101 induced by antisense RNA in NIH 3T3 cells leads to colony formation in 0.5% agar, focus formation in monolayer cell cultures, and the ability to form metastatic tumors in athymic nude mice (1). Turn-off of tsg101-inactivating antisense RNA reverses these features of neoplastic transformation as well as the nuclear, microtubule, and mitotic spindle abnormalities observed in TSG101-deficient cells (1, 2). The steady-state level of TSG101 protein normally is regulated posttranslationally in cells within a narrow range (3), and overexpression of TSG101 from an adventitious promoter can also lead to neoplastic transformation (1). Truncated TSG101 transcripts, which are observed in a variety of human tumors as well as in normal cells (4–7), have been attributed to aberrant or alternative RNA splicing (8) and have been correlated with both cellular stress (4, 5) and mutation of p53 (5). The TSG101 protein contains motifs common to transcription regulators (1) and can modulate transcriptional activation by steroid hormone receptors (9–11).
Sequence analysis has also suggested that TSG101, which is expressed in mammalian cells from the earliest stages of embryonic development and in multiple tissues of adult mice (8), may additionally have a role in the regulation of ubiquitin-mediated proteolysis (12, 13). The N-terminal region of the TSG101 protein contains a domain (Ubc) that resembles the catalytically active region of ubiquitin conjugases (E2 enzymes) but lacks an active-site cysteine residue crucial to the function of these enzymes (12–14), leading to speculation that TSG101 may act as a dominant-negative inhibitor of ubiquitination (12, 13).
p53 is a key tumor suppressor that transcriptionally activates MDM2 as well as other genes implicated in both cell growth and cell death (15–18). MDM2 in turn negatively regulates p53 by promoting its ubiquitin-mediated degradation (19–21). Despite p53/MDM2 feedback control, p53 accumulates in cells soon after DNA damage, hypoxia, and other types of stress, suggesting that the actions of MDM2 and p53 on each other are themselves regulated (16). Several mechanisms for such regulation have been proposed (16, 17) and recent evidence indicates that alteration of MDM2 stability mediated by its interactions with other cellular proteins may have a role in this process (22–24). The studies reported here indicate that TSG101 participates with MDM2 in a separate feedback control loop that affects MDM2 stability and, consequently, p53/MDM2 feedback regulation. We show that TSG101 is, in fact, an a inhibitor of ubiquitination, that MDM2 is a key cellular target of this inhibition, and that degradation of the TSG101 protein is in turn regulated by MDM2.
Materials and Methods
Plasmid and Vector Construction.
Full-length human TSG101 cDNA was inserted into the pLLEXP1 vector (1) between the cytomegalovirus (CMV) promoter and polyadenylation site. HA-tagged (human influenza virus hemagglutinin peptide, YPYDVPDY), Flag-tagged, and c-Myc-tagged TSG101 and TSG101 deletion mutant cDNAs were generated by PCR and were also cloned by using pLLEXP1. Vectors expressing human wild-type p53 (pC53-SN3) (25), human mutant p53 (pC53-Cx21an3, amino acid 175 mutation, Arg to His) (26), human MDM2 (pCHDM1B) (27), and tagged ubiquitin HM-Ub and HM-K48R-Ub (28) have been described. pCMV-GFP (CLONTECH) was used to express green fluorescent protein (GFP).
Cell Culture and Transfection.
Saos-2, U2OS, SJSA-1, and NIH 3T3 cells were obtained from the American Type Culture Collection (ATCC). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) [Saos-2, NIH 3T3, p53−/− mouse embryo fibroblasts (MEFs), and p53−/− and MDM2−/− MEF; ref. 29] or RPMI medium 1640 (U2OS and SJSA-1) supplemented with 10% FBS. Transfections were carried out using either Lipofectamine (Life Technologies) or FuGENE 6 (Roche) as described by the manufacturers.
Immunoprecipitation and Western Blot Analysis.
Immunoprecipitation and Western blot analysis were performed as described (30). Cells were lysed with Nonidet P-40 lysis buffer on ice, and the protein extracts were precleared with prewashed Pierce protein A/G agarose beads (50 μl of 50% slurry per 0.5 ml of protein extract). The precleared protein extracts were incubated with antibody for 8 h to overnight at 4°C on a rotating rocker, and then with the prewashed Pierce protein A/G agarose beads for an additional 2 h (10–15 μl of 50% slurry per 0.5 ml of protein extract). The immunocomplex was washed four times with Nonidet P-40 buffer, dissolved in SDS loading buffer, and fractionated on SDS/10% polyacrylamide gels (Bio-Rad). The proteins were then transferred to NitroPure membrane (Micron Separations, Westboro, MA) and incubated with specific antibody and appropriate horseradish peroxidase (HRP)-coupled secondary antibody (Santa Cruz Biotechnology and Promega). The membranes were washed in phosphate-buffered saline (PBS) and visualized using enhanced luminol reagent (NEN). Autoradiograms of Western blots were scanned with Scanmaster 3 (Howtek, Hudson, NH) and analyzed by using the Quantity One program (Precision Digital Images, Bothell, WA). Antibodies used for immunoprecipitations were rabbit anti-TSG101 (1:200, CLONTECH), anti-p53 (Ab-1, 1:400, Calbiochem), anti-MDM2 (SMP-14, 1:200, Santa Cruz Biotechnology), and anti-HA (1:200, CLONTECH). Antibodies used for Western blots were rabbit anti-TSG101 (1:200, CLONTECH), anti-p53 (DO-1, 1:1000, Santa Cruz Biotechnology), anti-HA (1:500, HRP-labeled, Roche), and anti-Flag (1:500, M2, Kodak), anti-α-tubulin (1:20000, Neomark), anti-rabbit IgG (1:5000, HRP-labeled, Promega) and anti-mouse IgG (1:10,000, HRP-labeled, Santa Cruz Biotechnology). Anti-GFP antibody was obtained from CLONTECH and was used at 1:500 dilution.
In Vivo Ubiquitination of MDM2.
HM-Ub or HM-K48R-Ub was introduced into SJSA-1 cells by cotransfection with vectors expressing TSG101 mutant proteins B or F or with controls lacking an insert or expressing TSG101 cDNA in the antisense direction. Twenty-four hours later, the transfected cell cultures were treated with MG132 (2 μM) for 12 additional hours, lysed with 6 M guanidinium chloride, and sonicated for 20 s. The His-tagged proteins were purified with Ni-nitrilotriacetate (NTA) spin columns (Qiagen), washed four times with 0.8 ml of wash buffer (8 M urea/0.1 M NaH2PO4/0.01 M Tris⋅Cl, pH 6.2), and eluted once with wash buffer at pH 4, and an additional time with pH 4 wash buffer containing 250 mM imidazole. The purified proteins were analyzed by Western blotting as above.
Pulse–Chase Experiments.
NIH 3T3, p53−/− MEF, and p53−/−/ MDM2−/− MEF cells (1 × 106) were seeded onto 100-mm plates for 24 h and were pulse-labeled with [35S]methionine for 2 h, washed twice with prewarmed PBS, and chased by culturing in DMEM supplemented with 10% FBS for 0, 2, 4, 8, and 12 h. Cell lysates from pulse-labeled cells were immunoprecipitated with anti-TSG101 antibody (10 μg/500 μg of total protein), resolved by electrophoresis in SDS/10% polyacrylamide gels, and analyzed with a PhosphorImager (ImageQuant Storm 840, Molecular Dynamics).
Results
Physical and Functional Interaction of TSG101 with p53 and MDM2.
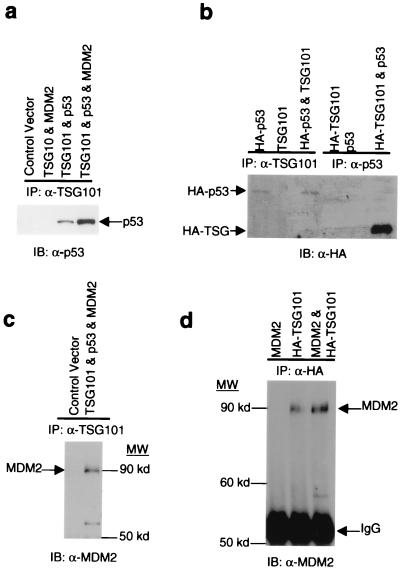
During investigations aimed at identifying physical and functional interactions between TSG101 and proteins previously implicated in tumorigenesis, we found that TSG101 can bind to both p53 and MDM2. This is shown in Fig. 1, which presents results of Western blot analyses of extracts of Saos-2 human osteosarcoma cells cotransfected with constructs expressing combinations of these proteins. Complexes immunoprecipitated from cell extracts by antibody to TSG101 contained p53, either untagged or as fused to an influenza B HA peptide epitope used for detection, and conversely, identified TSG101 in complexes immunoprecipitated with antibody to p53 (Fig. 1 a and b). Similarly, a 90-kDa band detected with anti-MDM2 antibody and representing an MDM2 complex with the small ubiquitin-like protein SUMO-1 (24) was present in Saos-2 cell protein complexes immunoprecipitated with antibodies to native or epitope-tagged TSG101 (Fig. 1 c and d).
Figure 1.
Interaction of TSG101 with p53 and MDM2. Constructs expressing human TSG101, p53, or MDM2 proteins (2 μg of DNA for each plasmid) were introduced by transfection into Saos-2 cells, as indicated in Materials and Methods. Protein extracts from transfected cell populations were immunoprecipitated by the antibodies indicated. IP, immunoprecipitation; IB, immunoblotting; α-, anti-. (a and b) Native or HA-tagged TSG101 or p53 proteins in Western blots were detected by anti-p53 monoclonal antibody (a; AB-1; 1:1000; the secondary antibody was HRP-conjugated goat anti-mouse IgG, diluted 1:1000) or by anti-HA antibody labeled with HRP (b; diluted 1:500). (c and d) Western blot detection of immunoprecipitated proteins analyzed with anti-MDM2 antibody.
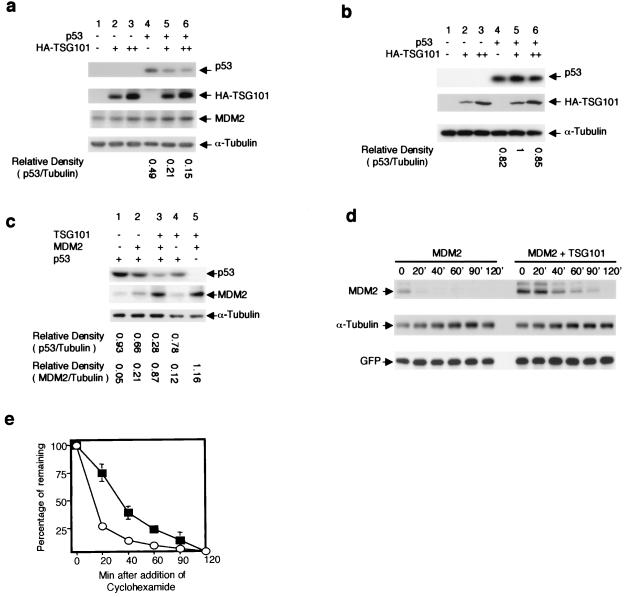
MDM2 is a ubiquitin–protein ligase that mediates its own decay as well as the degradation of p53 (31, 32). That the ability of TSG101 to interact with MDM2 and/or p53 has functional consequences on the steady-state levels of MDM2 and p53 is shown in Fig. 2: TSG101 overexpression has divergent effects on the steady-state cellular levels of both the p53 and MDM2 proteins. As seen in Fig. 2a, overproduction of TSG101 in Saos-2 cells, which synthesize endogenous MDM2 protein but not native p53, reduced the level of p53 expressed from a transfected construct by 70%. However, in identically transfected cells that carry null chromosomal mutations in both p53 and MDM2 and thus lack the ability to synthesize either of these proteins [i.e., p53−/−/MDM2−/− MEFs (29, 33, 34); Fig. 2b], we observed no effect of TSG101 on p53—suggesting that TSG101-mediated reduction of the p53 level requires the presence of MDM2.
Figure 2.
Effect of expression of TSG101 on the cellular level of p53 and the degradation of MDM2. (a) The indicated constructs overexpressing p53 and HA-TSG101 were introduced into Saos-2 cells by transfection, and p53 and HA-TSG101 levels were analyzed by Western blotting 48 h later. + indicates 25 ng of transfected p53 expression vector DNA or 4 μg of HA-TSG101 expression vector DNA; ++ indicates 8 μg of HA-TSG101 expression vector DNA. The density of p53 bands relative to cellular α-tubulin was determined by scanning of exposed films. (b) The identical constructs overexpressing p53 and HA-TSG101 were introduced into p53−/−/MDM2−/− MEF cells by transfection, and p53 and HA-TSG101 levels were analyzed as in a. (c) Constructs overexpressing the proteins indicated were introduced into Saos-2 cells by transfection and p53 and MDM2 levels were analyzed as in a. + indicates 25 ng of transfected p53 expression vector DNA, 1 μg of MDM2 expression vector DNA, or 4 μg of TSG101 expression vector DNA. (d) Saos-2 cells were transfected by an MDM2-expressing construct in the absence or presence of a TSG101-expressing construct. Cellular protein was extracted after addition of cycloheximide at the indicated times (min) and analyzed by Western blotting as in a, using anti-MDM2 antibody. A GFP expression vector was cotransfected to normalize transfection efficiency; the expressed GFP protein was detected with anti-GFP antibody. (e) Plot of degradation of MDM2 for the experiment shown in d, which was representative of five separate experiments. ○, Absence of TSG101; ■, presence of TSG101.
Although less than 50% of the population of Saos-2 cells was transfected by TSG101-expressing constructs under the experimental conditions we used (data not shown), this was sufficient to elevate the MDM2 level in extracts of the entire cell population (Fig. 2a), supporting the notion that TSG101 down-regulates p53 by elevating MDM2. Direct evidence for this conclusion was provided by experiments in which Saos-2 cells were transfected with constructs expressing p53, MDM2, and TSG101, individually or in combination. As others have observed previously (20, 21), overproduction of MDM2 from a cotransfected CMV-based expression vector resulted in a decrease in the level of p53 protein (Fig. 2c, lanes 1 and 2); concurrent overproduction of TSG101 in cotransfected cells was associated with further elevation of the MDM2 protein level (Fig. 2c, lanes 3 and 5) and a prominent further decrease in p53 (lane 3).
Effect of TSG101 on MDM2 Decay.
MDM2 normally has a short half-life of 15–20 min (35). As seen in Fig. 2 d and e, TSG101 inhibits MDM2 degradation and prolongs its half-life. In this experiment, cells transfected with an construct expressing MDM2 from a CMV promoter, or cotransfected with constructs that express both MDM2 and TSG101, were treated with cycloheximide to stop protein synthesis, and MDM2 protein was assayed by Western blot analysis of samples taken at the indicated times. The half-life observed for MDM2 (approximately 15 min), which is consistent with earlier determinations (35), nearly doubled (to 28 min) in cells that concurrently overexpressed TSG101.
The Ubiquitin-Conjugase-Like Ubc Domain of TSG101 Inhibits Ubiquitination of MDM2.
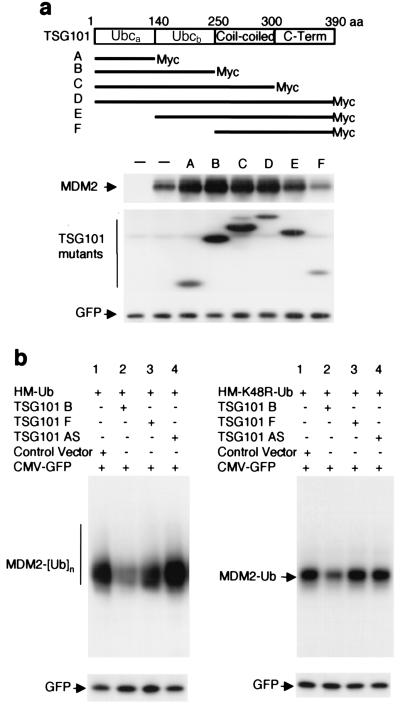
Because the Ubc domain of TSG101 lacks a cysteine residue required for conjugase function (12–14), it previously was speculated that TSG101 may inhibit ubiquitination by forming nonproductive complexes with ubiquitin or its target proteins and consequently interfering with the function of bona fide E2 (12, 13). The results seen in Fig. 3a, which show the effects of mutant TSG101 proteins on the steady-state level of MDM2 expressed from a cotransfected construct, indicate that TSG101's ability to stabilize MDM2 is sharply reduced by deletion of sequences from the Ubc domain. They also show that overexpression of the TSG101 Ubc domain's “a” region, which contains residues bracketing the active-site locus that in functional E2 enzymes contains a cysteine, is sufficient to cause accumulation of MDM2 (construct A). The presence of the “b” region of TSG101's Ubc domain enhanced the effects of the a region and even in the absence of Ubca led to some stabilization of MDM2 (constructs B vs. A and E vs. F).
Figure 3.
Effects of Ubc domains of TSG101 on MDM2 degradation and ubiquitination. (a) The indicated constructs expressing c-Myc-tagged full-length and deletion mutants of TSG101 (A-F, 8 μg), a construct expressing HA-tagged MDM2 (1.5 μg), and a construct expressing GFP (2 ng) were introduced into U2OS cells by transfection. HA-MDM2, c-Myc-tagged TSG101s (A-F), and GFP were detected by Western blotting 48 h after transfection with anti-HA, anti-c-Myc, and anti-GFP antibodies. (b) The indicated constructs express ubiquitin tagged with both His6 and c-Myc (HM-Ub, 5 μg, Left), or a dominant-negative variant ubiquitin tagged with His6 and c-Myc (HM-K48R-Ub, 5 μg, Right). These were cotransfected into SJSA-1 cells with a GFP expression construct (2 ng), constructs expressing TSG101 mutant B or F (4 μg each), a construct expressing antisense TSG101 (4 μg), or a construct containing no DNA insert (4 μg). Protein extracts were applied to Ni-NTA columns, and the ubiquitin-labeled MDM2 was eluted and detected by Western blotting with anti-MDM2 antibody. An aliquot (1/20) of protein extracts was used for the detection of GFP by Western blotting to normalize for transfection efficiency.
Further analysis of truncated TSG101 proteins consisting of largely the Ubc domain (construct TSG101B) or lacking this domain (construct TSG101F) showed that overexpression of the TSG101 Ubc domain interferes with ubiquitination of endogenous MDM2 (Fig. 3b). In the experiment shown in Fig. 3b Left, cellular proteins conjugated to His-tagged ubiquitin were isolated by Ni-NTA column chromatography, and MDM2 was identified in this protein pool by Western blotting using anti-MDM2 antibody. Notwithstanding the ability of TSG101 and its Ubc domain to globally increase the cellular level of both endogenous and adventitious MDM2 (Fig. 2), cells overproducing the TSG101 Ubc domain showed a decrease in ubiquitinated MDM2 vs. controls (Fig. 3b Left, lanes 1 and 2). That the TSG101 Ubc domain can decrease ubiquitination of MDM2 was demonstrated also by an experiment in which ubiquitin chains initiated on cellular proteins were tagged with a ubiquitin variant (K48R) (28) that impedes ubiquitin chain elongation; this preserves tagged proteins and allows the extent of ubiquitin addition to endogenous MDM2 to be evaluated by immunoprecipitation of MDM2 (Fig. 3b Right).
MDM2 Modulates the Decay of TSG101.
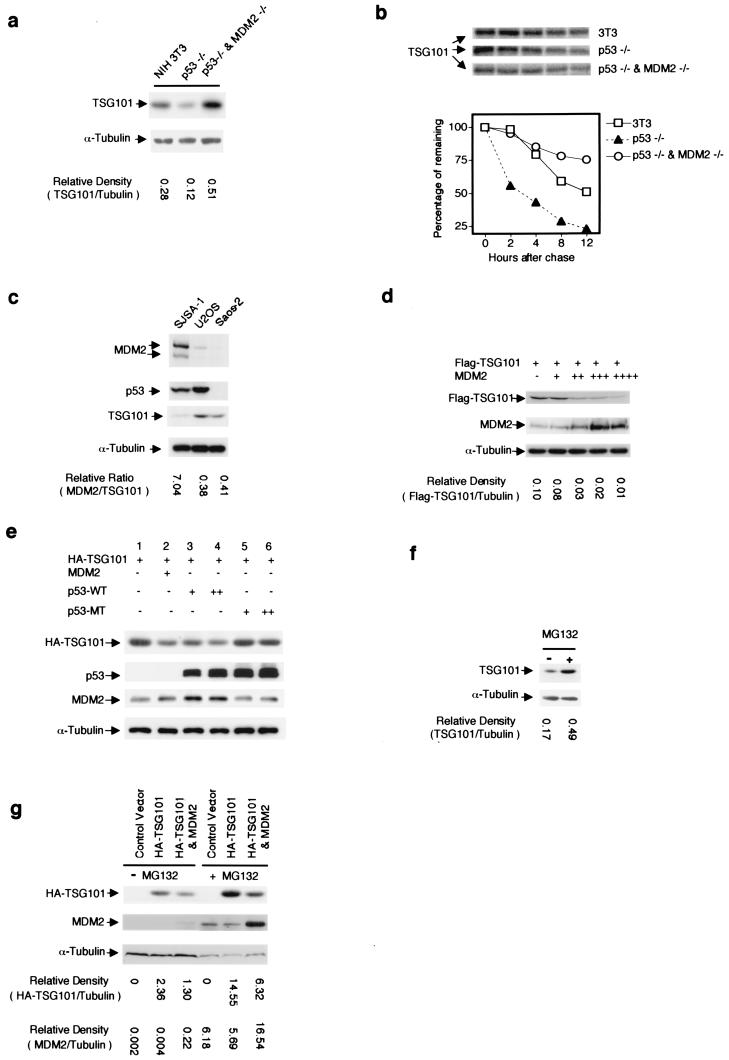
Earlier work has shown that both TSG101 excess and deficiency can lead to abnormal cell growth (1) and that the steady-state level of TSG101 protein normally is regulated within a narrow range by proteolysis (3). Just as TSG101 modulates the MDM2 level by negatively regulating its ubiquitination and decay, we found that MDM2 has a parallel key role in the proteolysis of TSG101. This action of MDM2 was suggested initially by the observation that the intracellular concentration of endogenous TSG101 relative to α-tubulin was markedly higher in p53−/−/MDM2−/− MEFs than in p53−/− MEF cells, which are capable of producing MDM2 (Fig. 4a). The correctness of the notion that MDM2, which as already noted mediates the degradation of both itself and p53, also affects the decay of TSG101 was supported by multiple lines of evidence. First, the pulse–chase experiment seen in Fig. 4b shows that the decay of endogenous TSG101 was accelerated in p53−/− MEFs but was reduced in MEFs doubly mutated in p53 and MDM2. Second, SJSA-1 cells, which contain multiple copies of MDM2 as a result of gene amplification (16), showed a major deficiency of endogenous TSG101, as compared with cells (U2OS and Saos-2) containing a single chromosomal copy of MDM2 (Fig. 4c). Third, a dosage-dependent decrease of Flag-tagged TSG101 protein expressed from the CVM promoter was observed in Saos-2 cells cotransfected with constructs that produce adventitious MDM2 (Fig. 4d); because Saos-2 cells lack p53, this experiment also indicates that MDM2-mediated acceleration of TSG101 decay does not require p53. Finally, while p53 overexpression, which activates endogenous MDM2 production (1, 16), was associated with a decrease in TSG101, overexpression of a mutant p53 protein that lacks the ability to increase endogenous MDM2 had no effect on the TSG101 level (Fig. 4e).
Figure 4.
MDM2-dependent proteolysis of TSG101. (a) Cellular TSG101 protein levels detected by Western blotting with protein extracts (40 μg each) from mouse fibroblast NIH 3T3 cells, p53−/− MEFs, and MEFs mutated in both p53 and MDM2. Relative densities of TSG101 protein bands were calculated after normalization to cellular α-tubulin. (b) Cell cultures of NIH 3T3, p53−/− MEFs and p53−/−/MDM2−/− MEFs were pulse-labeled with [35S]methionine for 1 h, and chased for 0, 2, 4, 8, and 12 h. The 35S-labeled TSG101 was immunoprecipitated by anti-TSG101 antibody, resolved by electrophoresis in an SDS gel, and visualized and quantitated by ImageQuant. (c) Protein extracts from SJSA-1, U2OS, and Saos-2 cells were analyzed by Western blotting using antibodies to MDM2, p53, TSG101, and α-tubulin. The ratio MDM2/TSG101 was determined after normalization with cellular α-tubulin. (d) Vectors expressing Flag-tagged TSG101 and MDM2 were introduced into Saos-2 cells by cotransfection. Transfecting amounts of DNA are designated by + for 0.5 μg of Flag-TSG101 expression vector and + to ++++ for 1 to 4 μg of MDM2 expression vector. (e) Combinations of vectors expressing HA-tagged TSG101, MDM2, and/or wild-type (WT) or mutant (MT) p53 protein were introduced by transfection into Saos-2 cells, and protein extracts of transfectants were analyzed by Western blotting using antibody as indicated. Where indicated, transfectants received 1 μg of plasmid DNA expressing HA-TSG101, 1 μg of plasmid DNA expressing MDM2, and p53-expressing constructs as follows: + and ++, 1 μg and 2 μg, respectively. The p53 mutation replaced the Arg at amino acid 175 with His. (f) Saos-2 cell cultures were treated as indicated with proteasome inhibitor MG132 (2 μM) for 24 h, and cellular protein extracts were analyzed by Western blotting with anti-TSG101 antibody. (g) The indicated constructs were introduced into Saos-2 cells by transfection (concentrations designated as in d). Transfected cells were cultured in the absence or presence of MG132 for 24 h, and protein extracts of cells were analyzed by Western blotting with anti-HA and anti-MDM2 antibody. The intensity of HA-TSG101 and MDM2 protein bands is indicated relative to cellular α-tubulin.
That the effects of MDM2 on TSG101 degradation are, like those of p53, carried out by the 26S proteasome was shown by an experiment in which addition of the proteasome inhibitor MG132 resulted in accumulation of endogenous TSG101 (Fig. 4f). Moreover, in the presence of MG132, overexpression of MDM2—which accumulated to >40× normal levels when the proteasome inhibitor was present—failed to accelerate decay of TSG101, confirming that MDM2-promoted decay of TSG101 requires proteasome action (Fig. 4g).
Discussion
The cellular concentrations of p53 and its negative regulator MDM2 normally are maintained at low basal levels by a well-recognized feedback control loop (1, 15–18, 36). The activation of p53 that follows cellular stress such as DNA damage, hypoxia, oncogene activation, or telomere shortening is mediated at least in part by a decrease in MDM2 from basal levels (for reviews, see refs. 16–18 and 36). The result is slowed p53 decay, an increase in p53 protein level, and consequently, either cell growth arrest or apoptosis. Later, however, accumulation of p53 transcriptionally activates the biosynthesis of MDM2, which shuttles p53 from the nucleus to the cytoplasm and promotes its ubiquitination and degradation by the proteasome (16, 37). As MDM2 turnover normally is extremely rapid (t1/2 ≈ 15–20 min; ref. 35), factors that prolong its half-life by only a few minutes can be expected to importantly alter its steady-state concentration and therefore functioning of the MDM2/p53 loop. Our results show that TSG101, which interferes with ubiquitin-mediated degradation of MDM2, has such effects. Additionally, our results show that TSG101—like p53—is a target of MDM2-mediated decay by the proteasome.
Recent evidence indicates that the ability of MDM2 to function as a ubiquitin ligase (E3) for itself and p53 is modulated by interaction with the small ubiquitin-like protein SUMO-1 at a specific site within the MDM2 RING finger domain essential to the E3 activity of MDM2 (24). Like the interaction of MDM2 with TSG101, sumoylation of MDM2 impedes MDM2 degradation and increases its ability to target p53 for degradation. We have not determined whether sumoylation of MDM2 also affects MDM2's ability to promote decay of TSG101, or conversely whether the ability of TSG101 to stabilize MDM2 is mediated through sumoylation of MDM2.
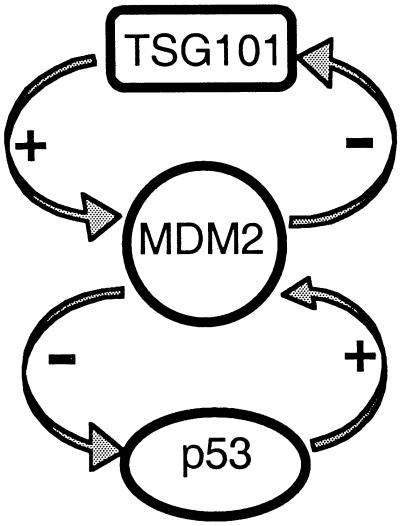
Fig. 5 schematically summarizes both our present findings and certain previously known biological interactions between p53 and MDM2. The data reported here support a model in which the Ubc-like domain of TSG101 interferes with MDM2 ubiquitination, stabilizes MDM2, and leads to MDM2 accumulation; in turn, the accumulated MDM2 protein both negatively regulates p53 and accelerates decay of TSG101. According to this model, the steady-state levels of all three proteins are interrelated through two feedback control loops that include MDM2 as a central participant. We suggest that the balance between TSG101-dependent stabilization of MDM2 and MDM2-mediated proteolysis of both TSG101 and p53 determines how TSG101 will affect p53/MDM2 feedback control. Interestingly, although the E3 activity of MDM2 mediates the decay of both itself and p53, TSG101 interference with proteolysis appears to apply only to MDM2—as MDM2-promoted degradation of p53 is not inhibited by TSG101 overexpression. Further complexity of the functional interactions between TSG101 and the MDM2/p53 loop is evident from the finding that p53 accumulation in TSG101−/− mutant embryos does not lead to the expected activation of MDM2 transcription (38). We speculate that the effects of TSG101 on MDM2 may also influence other transactions of MDM2.
Figure 5.
Model showing functional interactions of the TSG101/MDM2 and p53/MDM2 feedback control loops.
The multifaceted effects of TSG101 on growth regulatory proteins and pathways may explain why overexpression of TSG101 mRNA in either the sense or antisense direction can lead to neoplastic transformation (1)—whereas the total absence of TSG101 in null-mutant mice results in p53 accumulation, cell growth arrest, and early embryonic death (38).
Acknowledgments
We thank Drs. A. Levine, B. Vogelstein, and R. Tjian for p53- and MDM2-expressing plasmids, Dr. R. Kopito for ubiquitin-expressing plasmids, and Drs. G. Lozano and A. Giaccia for p53−/− and MDM2−/− MEFs. We acknowledge the helpful comments and advice of Drs. A. Levine, C. Prives, T. Tlsty, Y. Cao, Y. Zhang, J. Chen, A. Giaccia, and those of A. C. Y. Chang, K. Liu, B. Sohlberg, K. Lee, and other members of the Cohen laboratory. These studies were supported by grants to S.N.C. from the National Foundation for Cancer Research and the California Cancer Research Program.
Abbreviations
- HA
influenza virus hemagglutinin peptide
- GFP
green fluorescent protein
- HRP
horseradish peroxidase
- CMV
cytomegalovirus
References
- 1.Li L, Cohen S N. Cell. 1996;85:319–329. doi: 10.1016/s0092-8674(00)81111-3. [DOI] [PubMed] [Google Scholar]
- 2.Xie W, Li L, Cohen S N. Proc Natl Acad Sci USA. 1998;95:1595–1600. doi: 10.1073/pnas.95.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng G H, Lih C J, Cohen S N. Cancer Res. 2000;60:1736–1741. [PubMed] [Google Scholar]
- 4.Gayther S A, Barski P, Batley S J, Li L, de Foy K A, Cohen S N, Ponder B A, Caldas C. Oncogene. 1997;15:2119–2126. doi: 10.1038/sj.onc.1201591. [DOI] [PubMed] [Google Scholar]
- 5.Turpin E, Dalle B, de Roquancourt A, Plassa L F, Marty M, Janin A, Beuzard Y, de The H. Oncogene. 1999;18:7834–7837. doi: 10.1038/sj.onc.1203196. [DOI] [PubMed] [Google Scholar]
- 6.Lee M P, Feinberg A P. Cancer Res. 1997;57:3131–3134. [PubMed] [Google Scholar]
- 7.Sun Z, Pan J, Bubley G, Balk S P. Oncogene. 1997;15:3121–3125. doi: 10.1038/sj.onc.1201521. [DOI] [PubMed] [Google Scholar]
- 8.Wagner K U, Dierisseau P, Rucker E B, 3rd, Robinson G W, Hennighausen L. Oncogene. 1998;17:2761–2770. doi: 10.1038/sj.onc.1202529. [DOI] [PubMed] [Google Scholar]
- 9.Watanabe M, Yanagi Y, Masuhiro Y, Yano T, Yoshikawa H, Yanagisawa J, Kato S. Biochem Biophys Res Commun. 1998;245:900–905. doi: 10.1006/bbrc.1998.8547. [DOI] [PubMed] [Google Scholar]
- 10.Sun Z, Pan J, Hope W X, Cohen S N, Balk S P. Cancer. 1999;86:689–696. doi: 10.1002/(sici)1097-0142(19990815)86:4<689::aid-cncr19>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 11.Hittelman A B, Burakov D, Iniguez-Lluhi J A, Freedman L P, Garabedian M J. EMBO J. 1999;18:5380–5388. doi: 10.1093/emboj/18.19.5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koonin E V, Abagyan R A. Nat Genet. 1997;16:330–331. doi: 10.1038/ng0897-330. [DOI] [PubMed] [Google Scholar]
- 13.Ponting C P, Cai Y D, Bork P. J Mol Med. 1997;75:467–469. [PubMed] [Google Scholar]
- 14.Hochstrasser M. Science. 2000;289:563–564. doi: 10.1126/science.289.5479.563. [DOI] [PubMed] [Google Scholar]
- 15.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 16.Freedman D A, Levine A J. Cancer Res. 1999;59:1–7. [PubMed] [Google Scholar]
- 17.Prives C. Cell. 1998;95:5–8. doi: 10.1016/s0092-8674(00)81774-2. [DOI] [PubMed] [Google Scholar]
- 18.Oren M. J Biol Chem. 1999;274:36031–4. doi: 10.1074/jbc.274.51.36031. [DOI] [PubMed] [Google Scholar]
- 19.Lane D P, Hall P A. Trends Biochem Sci. 1997;22:372–374. doi: 10.1016/s0968-0004(97)01119-5. [DOI] [PubMed] [Google Scholar]
- 20.Haupt Y, Maya R, Kazaz A, Oren M. Nature (London) 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 21.Kubbutat M H, Jones S N, Vousden K H. Nature (London) 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Xiong Y, Yarbrough W G. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 23.Sherr C J, Weber J D. Curr Opin Genet Dev. 2000;10:94–99. doi: 10.1016/s0959-437x(99)00038-6. [DOI] [PubMed] [Google Scholar]
- 24.Buschmann T, Fuchs S Y, Lee C G, Pan Z Q, Ronai Z. Cell. 2000;101:753–762. doi: 10.1016/s0092-8674(00)80887-9. [DOI] [PubMed] [Google Scholar]
- 25.Baker S J, Markowitz S, Fearon E R, Willson J K, Vogelstein B. Science. 1990;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 26.Levine A J, Wu M C, Chang A, Silver A, Attiyeh E F, Lin J, Epstein C B. Ann NY Acad Sci. 1995;768:111–128. doi: 10.1111/j.1749-6632.1995.tb12115.x. [DOI] [PubMed] [Google Scholar]
- 27.Chen J, Marechal V, Levine A J. Mol Cell Biol. 1993;13:4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ward C L, Omura S, Kopito R R. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 29.McMasters K M, Montes de Oca Luna R, Pena J R, Lozano G. Oncogene. 1996;13:1731–1736. [PubMed] [Google Scholar]
- 30.Fiddler T A, Smith L, Tapscott S J, Thayer M J. Mol Cell Biol. 1996;16:5048–5057. doi: 10.1128/mcb.16.9.5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Honda R, Yasuda H. EMBO J. 1999;18:22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fang S, Jensen J P, Ludwig R L, Vousden K H, Weissman A M. J Biol Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 33.Montes de Oca Luna R, Wagner D S, Lozano G. Nature (London) 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 34.Jones S N, Roe A E, Donehower L A, Bradley A. Nature (London) 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 35.Olson D C, Marechal V, Momand J, Chen J, Romocki C, Levine A J. Oncogene. 1993;8:2353–2360. [PubMed] [Google Scholar]
- 36.Vousden K H. Cell. 2000;103:691–694. doi: 10.1016/s0092-8674(00)00171-9. [DOI] [PubMed] [Google Scholar]
- 37.Roth J, Dobbelstein M, Freedman D A, Shenk T, Levine A J. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruland J, Sirard C, Elia A, MacPherson D, Wakeham A, Li L, de la Pompa J L, Cohen S N, Mak T W. Proc Natl Acad Sci USA. 2001;98:1859–1864. doi: 10.1073/pnas.98.4.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]