Abstract
The ribosome filter hypothesis postulates that ribosomes are not simply translation machines but also function as regulatory elements that differentially affect or filter the translation of particular mRNAs. On the basis of new information, we take the opportunity here to review the ribosome filter hypothesis, suggest specific mechanisms of action, and discuss recent examples from the literature that support it.
Keywords: ribosome, rRNA, translation, mRNA, protein
Introduction
In eukaryotes, the translation of mRNA into protein is an important component of gene expression, with significant implications for normal cellular function and development as well as for the ability of cells to respond to stress and disease. The process of translation involves the decoding of mRNA templates into peptide sequences on ribosomes. In eukaryotes, this process begins with recruitment of a 40S ribosomal subunit at either the 5′ m7G cap-structure or an internal ribosome entry site (IRES). In most cases, the ribosomal subunit is then relocated to an initiation codon. Initiation is an important site of regulation that may affect protein synthesis globally or at the level of individual mRNAs. Translation initiation is affected by mRNA-binding factors 1–11 as well as features of the mRNA itself.
A number of observations from our own and other laboratories hinted at yet another level of regulation mediated by the ribosomal subunits themselves and these observations formed the basis for proposing the ribosome filter hypothesis.12 At that time, we made an argument for ribosomal filtering based largely on circumstantial evidence. We suggested that the ribosomal subunits affect the translation of particular mRNAs through mechanisms involving differential mRNA-capture (Figure 1). We now revisit this hypothesis in light of new evidence. Many of the recent examples supporting this notion involve alterations by disease. Additional investigations will be required to determine whether ribosomal filtering is disrupted in these diseases or whether ribosomal filtering occurs predominantly in the disease state.
Figure 1.
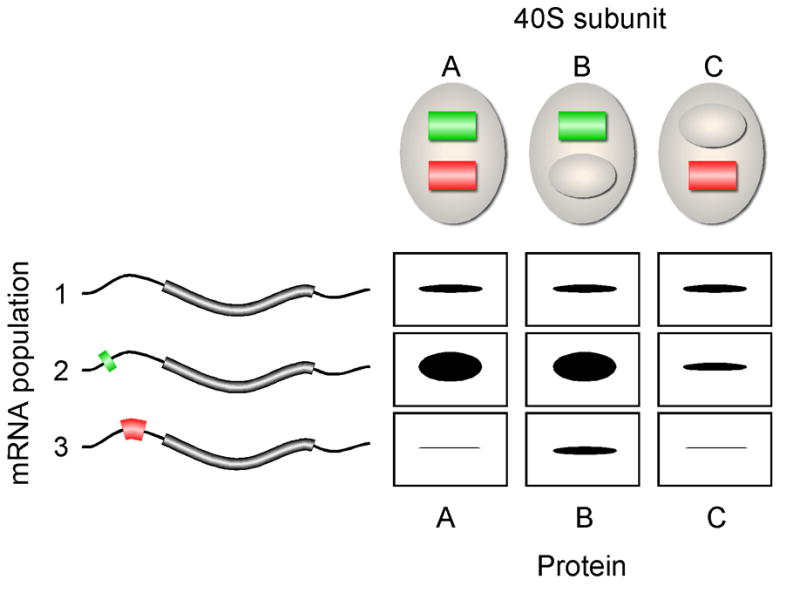
Schematic representation of ribosomal filtering. 40S ribosomal subunits are indicated as large ovals. Subunit A contains two mRNA-binding sites indicated as green and red bars. The red binding site is masked in subunit B and the green binding site is masked in subunit C. The mRNA population shows three types of mRNAs: (#1) lacks mRNA-elements that function as ribosomal binding sites, (#2) contains an mRNA-element (green bar) that enhances translation initiation when it binds to the green binding site in 40S subunits, and (#3) contains an mRNA-element (red bar) that can block translation initiation when it binds to the red binding site the 40S subunits. The amount of protein expressed by the various mRNA-ribosomal subunit combinations are represented by the size of the black bands. Translation of mRNA #1 occurs by a cap-dependent mechanism and is translated with the same relative efficiency by ribosomal subunits A, B, or C. Translation of mRNA #2 is enhanced when translation involves ribosomal subunits A or B, in which the green binding site is accessible. Translation of mRNA #3 is inhibited when translation involves ribosomal subunits A or C, in which the red binding site is accessible.
The ribosome filter hypothesis
A key observation pointing to the hypothesis was the finding that large numbers of mRNAs contain segments that are either similar or complementary to sequences within 18S or 28S rRNAs.12–21 This observation suggested a potential mechanism by which mRNAs might interact directly with ribosomal subunits, e.g. by mRNA-rRNA base pairing between complementary nucleotide sequences. Moreover, several reports suggested that within an organism, eukaryotic ribosomes in various celltypes or during different stages of development (e.g. see reference 22) vary in both ribosomal protein composition and rRNA sequences. These findings raised the possibility that structurally different populations of ribosomes might vary in their ability to translate specific subsets of mRNAs.
Based on these and other observations, we proposed four hypothetical tenets:
The ribosome is a regulatory structure that embodies mechanisms for preferentially translating different subsets of the message population. It was suggested that the ability of ribosomes to filter translation depends on specific interactions in which segments of mRNAs bind to ribosomal proteins or to rRNA. Several of our earlier studies 14–16, 23, 24 focused on demonstrating base pairing between complementary segments of mRNA and rRNA. We suggested that, during initiation of translation, most such interactions would occur with the 40S subunit, but we did not exclude the possibility of interactions with the 60S subunit or with 80S ribosomal complexes.
Ribosomes may display a continuum of regulatory effects. Some interactions may enhance the translation of particular mRNAs while other interactions may sequester certain mRNAs and diminish their rates of translation.
Competition for binding sites in ribosomal subunits may affect the rate of translation of different mRNAs. It was hypothesized that segments of mRNAs that bind to ribosomal subunits may act as competitive modules or domains. This competition for particular binding sites in ribosomal subunits might therefore affect the rate of translation of other mRNAs or groups of mRNAs that are able to bind to these same sites.
The filter may also be modulated as a result of altering or masking particular binding sites on ribosomes. Subsets of mRNA may be translated with different relative efficiency by populations of ribosomes that differ in the accessibility of ribosomal binding sites, for example as a result of ribosome heterogeneity due to differences in ribosomal protein composition, or by interaction with extraribosomal factors, which may include proteins or noncoding RNAs.
The first tenet of the hypothesis suggests two main mechanisms of ribosomal filtering, one involving complementarity between mRNA and rRNA sequences, and the other involving interactions between mRNA sequences and ribosomal proteins. We consider these mechanisms and then discuss recent supporting evidence for them.
Mechanisms of filter action
mRNA-rRNA interactions: evidence for base pairing
A critical feature of the filter hypothesis concerns the ability of different members of a population of ribosomes to interact specifically and selectively with particular mRNAs. In earlier studies we investigated potential mRNA-rRNA base-pairing as an important mechanism underlying this interaction. Our most extensive studies in this regard focused on a 9-nucleotide element located in the mRNA of the Gtx homeodomain protein.16, 23
The 9-nt Gtx element was shown to be an IRES module in studies performed using dicistronic mRNAs.16 We found that linking together individual Gtx elements could lead to an exponential increase in the expression of a reporter protein. Various control experiments indicated that the Gtx elements enhanced expression by increasing the efficiency of translation. To avoid potential limitations associated with the use of dicistronic mRNAs, we also tested Gtx elements in the 5′ leader of a monocistronic mRNA.23 In this context, multiple linked copies of the Gtx element enhanced translation up to 8-fold compared to the activity mediated by the cap-structure alone. RNA analyses showed that this increased activity could not be accounted for by alterations in mRNA levels.
Sequence comparisons revealed a perfect complementary match between the 9-nt Gtx element and nucleotides contained within helix 26 of the 18S rRNA.15 It is notable that these nucleotides in the rRNA are located in the platform of the 40S subunit, in close proximity to the predicted mRNA-binding tract.
These results suggested that the Gtx element affects translation by a mechanism that involves base pairing to complementary nucleotides in the 18S rRNA. To test this notion, we performed a number of functional and biochemical experiments.16, 23, 24 In one set of studies, we altered the length of the complementary match.23 The data showed that translation was maximally enhanced by a specific 7-nt sequence in the 9-nt element and that the degree of enhancement declined progressively as the sequences involved in the complementary interactions became longer or shorter. These observations suggested that the shorter complementary interactions were insufficiently stable for efficient translation initiation and that the longer complementary interactions were too stable. This conclusion was corroborated by biochemical studies which showed that the length of the complementary match was directly correlated with the ability of RNA probes to bind to ribosomes.
To assess the requirement for complementarity in the match, we developed a novel yeast system that enabled us to alter both the mRNA and rRNA sequences.24 The 18S rRNA of the yeast Saccharomyces cerevisiae has a poor complementary match to the Gtx element and we found that the presence of the Gtx elements did not enhance translation of mRNA in these cells.24 To determine whether the Gtx elements were inactive because they could not base-pair to yeast 18S rRNA, we introduced complementary sequences into the yeast 18S rRNA either in the form of mouse-yeast hybrid 18S rRNAs, or directly by introducing appropriate point mutations of the yeast 18S rRNA itself. When constructs containing the Gtx elements were tested in yeast whose rRNAs contained the complementary match, translation efficiencies were dramatically increased over that in cells lacking the complementary match. Any of five different single point mutations within the 7-nt Gtx elements were shown to disrupt translation rates. In each case, activity was restored by a corresponding mutation of the hybrid 18S rRNA designed to restore complementarity.
By showing that the activity of the Gtx elements required an intact complementary match to the 18S rRNA, these studies provided the first evidence in eukaryotes for an mRNA-rRNA base-pairing interaction analogous to the Shine-Dalgarno interaction in bacteria.25–30 This example supports a key mechanism proposed to underlie instances of the ribosomal filter. Furthermore, this example served to demonstrate the principle of ribosomal filtering, albeit synthetically, by showing that an mRNA containing Gtx-elements is translated more efficiently by ribosomes that have a complementary match to the Gtx-element (mouse-yeast hybrid) than by ribosomes lacking such a match (yeast). We expect that in addition to the Gtx-element, there are other mRNA elements that affect translation initiation in eukaryotes by base pairing to 18S rRNA.
Ribosomal protein heterogeneity
Heterogeneity in a ribosomal protein may affect the translation of specific mRNAs that bind to that ribosomal protein. Alternatively, such heterogeneity may modulate the translation of specific mRNAs by altering the accessibility of binding sites contained within the rRNA. Examples of ribosomal heterogeneity were discussed in our earlier publication as a possible mechanism by which different populations of ribosomes differentially regulate the translation of subsets of mRNAs.12 More recent evidence for ribosomal heterogeneity comes from a global survey of protein expression in mice, which was performed using mass spectrometric multi-dimensional protein identification technology (MudPIT).31 We searched the data from this study using a web-browser interface and found that some ribosomal proteins, including S3, S4, and S7, were detected in all tissues examined while other ribosomal proteins were detected at high levels in some tissues but were not detected in others. For example, ribosomal protein S6 was detected at high levels in brain, kidney, and liver, but was detected at only a low level in lung tissue and not at all in the heart. These results complement in situ hybridization results obtained in our laboratory (Prieto, Mauro, and Edelman, unpublished data), which showed that the levels of mRNAs for ribosomal proteins had patterns of expression that were dramatically different in different tissues. In a different study, Angelastro et al. 32 found that the relative abundance of individual ribosomal protein mRNAs varied by over a hundredfold in rat pheochromocytoma (PC12) cells and that the levels of approximately half of these ribosomal protein mRNAs changed (up to 5-fold) when cells were treated with nerve growth factor (NGF).
These findings, together with various other observations (see reference 12), suggest that the ribosomal subunits in different tissues, as well as in different subcellular locations are likely to be heterogeneous with respect to their constituent ribosomal proteins. However, additional studies are required to determine to what degree this heterogeneity affects filtering.
Recent evidence supporting the filter hypothesis
Recent observations over a wide range of biology, including different species and various diseases (see references 33–42) have been specifically suggested by their investigators to be consistent with the function of a ribosome filter. We choose here to discuss a number of examples to illustrate the widespread applicability of filter mechanisms. Some of these examples involve modification of the 18S rRNA or ribosome binding proteins. Other examples involve ribosomal protein heterogeneity.
Pseudouridinylation of rRNA
Strong evidence for a ribosome filter comes from Yoon et al.43, who addressed the possibility of altered ribosomal function in the X-linked dyskeratosis congenita syndrome. This disease results in bone marrow failure and is characterized by premature aging, nail and skin abnormalities, and a predisposition to cancer. The disease is caused by a mutation in the DKC1 gene that disrupts dyskerin, a protein associated with ribonucleoprotein (RNP) complexes, including the H/ACA family of small nucleolar (sno) RNP particles. In these snoRNPs, dyskerin functions as a pseudouridine synthase that modifies specific uridine residues in rRNA. Using a mouse model, the authors showed that global protein synthesis was generally unaffected in cells with a hypomorphic mutation in the DKC1 gene. However, a limited microarray analysis identified three mRNAs with a decreased representation in polysomes of such cells when compared to polysomes of wild type cells: the p27 tumor suppressor, the X-linked inhibitor of apoptosis protein (XIAP), and the mitochondrial ion channel Bcl-xL. All three mRNAs were shown to contain an IRES: Bcl-xL in the Yoon et al., study 43; XIAP and p27 in other studies (see references 44 and 45, respectively).
To assess whether the observed translational effects were directly mediated by the ribosomal subunits, an mRNA containing the cricket paralysis virus (CrPV) IRES was tested. This IRES binds directly to the 40S ribosomal subunit and facilitates translation initiation even in the absence of initiation factors and the initiator Met-tRNA.46 The experiments revealed that translation mediated by the CrPV IRES was impaired in cells with the DKC1 mutation. The data makes accounting for the results by indirect effects unlikely, particularly since pseudouridine does not appear to be found in mRNA.47 It is more likely that ribosomes lacking pseudouridine modifications were impaired in their ability to translate specific IRES-containing mRNAs, even though polysome analyses suggested that these ribosomes were generally unaffected in their ability to translate most other mRNAs. These observations are consistent with the notion that the ribosome is a regulatory structure that embodies mechanisms for preferentially translating particular subsets of the mRNA population.
Methylation of ribosomal protein S24
Studies performed in the slime mold Dictyostelium discoideum showed that the translation of ribosomal protein mRNAs is altered during development by a subset of structurally distinct ribosomes.48, 49 During vegetative growth, it was found that ribosomal protein mRNAs were almost completely associated with polysomes. In contrast, at the start of development, ribosomal protein mRNAs were specifically excluded from polysomes. Mangiarotti (2002)49 reported that these excluded mRNAs were associated with a class of 40S subunits in which ribosomal protein S24 was methylated. This ribosomal protein was not methylated in 40S subunits obtained from later developing cells or growing cells. In vitro experiments using eight ribosomal protein mRNAs showed that methylated ribosomal protein S24 was actually required for the selective binding of these mRNAs to the ribosomal subunits. These results suggest that during slime mold development a ribosomal filtering mechanism regulates the translation of ribosomal protein mRNAs themselves.
Ribosomal proteins S2, S5, and S10
The notion that the ribosome is a regulatory element showing mRNA selectivity and that it is not simply a tape head that converts ribonucleotide sequences into amino acid sequences is illustrated by the translation properties of different eukaryotic ribosomes. For instance, ribosomes from yeast and mammalian cells show differences in both rRNAs and ribosomal proteins. Nevertheless both can translate similar mRNAs with high efficiency, e.g. simple cap-dependent mRNAs with a short 5′ leader. However, not all mRNAs are translated with similar relative efficiencies by yeast and mammalian ribosomes. Indeed high selectivity has been observed in translation mediated by an IRES from hepatitis C virus (HCV).
HCV causes hepatitis C disease, characterized by liver inflammation that can lead to cirrhosis and cancer. Translation of HCV mRNA requires an IRES that binds directly to 40S ribosomal subunits.50 This binding occurs via a small number of ribosomal proteins as revealed by UV cross-linking studies.51 This IRES is inactive in Saccharomyces cerevisiae, apparently because it fails to bind to yeast 40S ribosomal subunits.50, 51 This failure was attributed to large differences between the mammalian and yeast ribosomal subunits in three ribosomal proteins: S2, S5, and S10, which bind the IRES in mammalian cells.51 Together, these observations suggest that the ability of 40S ribosomal subunits to initiate translation via the HCV IRES depends on their ability to bind to these ribosomal proteins.
The co-evolution of mRNA and ribosomal binding sites may explain why the HCV IRES functions in its mammalian hosts, but not in yeast. We expect that other examples of ribosomal filtering, both of cellular and viral mRNAs, will also involve co-evolution of mRNA and ribosomal binding sites.
Other indications of ribosomal filtering
The studies discussed above provide strong support for the notion of a ribosomal filter. Numerous other studies are consistent with this notion but require further analysis to know whether they represent instances of ribosomal filtering. We briefly identify some of the most promising examples below.
Binding of Reaper to 40S ribosomal subunits
The activity of the Reaper protein provides another example consistent with ribosomal filtering.52 In Drosophila, Reaper induces apoptosis through multiple cellular targets, both during normal development and in response to DNA stress.53 One of Reaper’s effects is to inhibit protein synthesis, which is consistent with the results of a recent study reporting that Reaper can bind to the 40S ribosomal subunit.52 This study further showed that the addition of Reaper to a cell-free lysate blocked the translation of two cap-dependent mRNA constructs containing 5′ leader sequences from either HSP-70 or globin mRNAs. However, the translation of a reporter mRNA containing the CrPV IRES in its 5′ leader was less sensitive to the presence of this protein. These results suggest that the differential effects of Reaper on mRNA translation reside with the ribosomes and thus are consistent with a ribosomal filtering mechanism.
Methylation of rRNA
As discussed above, the differential pseudouridinylation of rRNA provides a means for generating ribosomal heterogeneity that affects the initiation of particular mRNAs. We raise the possibility that the differential methylation of specific nucleotides in rRNA may also provide a comparable basis for ribosomal filtering. This notion is strengthened by recent reports indicating that a defect in 18S rRNA-methylation underlies Treacher-Collins syndrome, a disease affecting the development of bones and other tissues in the head and face.54 This disease results from mutations in the nucleolar phosphoprotein treacle, which appears to affect both transcription and 2′-O-methylation of pre-rRNA. To assess whether methylation of ribosomal nucleotides affects ribosomal filtering will require additional studies, for example of the type used to analyze ribosomal function in dyskeratosis congenita.
Mutations in ribosomal proteins S19 and S24
Another potential example of ribosomal filtering comes from Diamond-Blackfan anemia.55 This disease shares some symptoms with dyskeratosis congenita, including bone marrow failure and increased incidence of cancer. Additional symptoms may include short stature as well as other physical defects. Mutations in two ribosomal protein genes, S19 and S24, account for approximately 27% of cases; the remaining 73% of cases are thought to involve mutations in at least two other genes. Ribosomal protein S19 has been found be important in 18S rRNA processing and in the assembly of the 40S subunit. Moreover, when various mutations of the S19 protein associated with the disease (eleven missense mutations and a trinucleotide insertion) were tested in transfected cells, none of the mutated S19 proteins were assembled into mature ribosomes.56 Theories regarding the effects of S19 mutations suggest a non-specific global effect on ribosomal function,57 with the possibility that S19 may have extraribosomal functions.58 However, mutations affecting ribosomal protein S24 give rise to the same disease. Indeed, this ribosomal protein shows high heterogeneity within normal mammalian tissues, suggesting that it is not essential for ribosomal function.59 Interestingly, S24 is the same ribosomal protein which appears to facilitate ribosomal filtering in Dictyostelium discoideum (see above). The fact that mutations affecting two different proteins of the 40S ribosomal subunit give rise to the same disease suggests that Diamond-Blackfan anemia is a ribosomal disorder. However, additional studies are required to determine whether this is a disease of the ribosome filter that differentially affects the translation of mRNAs important for hematopoiesis.
RACK1
RACK1 was recently identified as a ribosomal protein of the 40S subunit (reviewed in reference 60), and various studies have shown that this protein can affect the translation of subsets of mRNAs. For example, in Saccharomyces pombe, cells with a disrupted RACK1 gene showed a small overall reduction in protein synthesis, but a dramatic reduction in the expression of several proteins, including ribosomal protein L25.61 Additional analysis showed that the levels of the L25 mRNA were not altered in cells lacking RACK1, suggesting that the reduced levels of ribosomal protein L25 were caused by a decrease in the translation efficiency of its mRNA. In other experiments performed in Saccharomyces cerevisiae, RACK1 was deleted. Its absence resulted in a general enhancement of protein synthesis with specific increases in the expression of several proteins.62 To determine whether RACK1 affects ribosomal filtering will require showing that the observed effects on translation are mediated by the ribosomal subunits themselves.
New lines of investigation
A recent report on the 2006 Translational Control meeting at Cold Spring Harbor indicated that an emerging line of inquiry involved looking for ribosomal modifications as control points for regulating mRNA translation.63 Two studies were highlighted: the first showed that various manipulations affecting the Saccharomyces cerevisiae 60S ribosomal subunit, including deletion of several ribosomal protein genes or genes of proteins involved in 60S processing or maturation, led to cells with significantly longer life spans. In the second study, ribosomes from poliovirus-infected and uninfected HeLa cells were compared using mass spectrometry. In infected cells, the ribosomal proteins showed increased di-methylation as well as changes in ribosomal protein acetylation. If differences in the composition of the 60S ribosomal subunit or in post-translational modifications of ribosomal proteins can be shown to affect the translation of specific mRNAs these effects may provide examples of ribosomal filtering.
Conclusion
This brief review has identified some studies that provide support for the notion that ribosomes can mediate selective mRNA translation. Other studies are consistent with the filter hypothesis and we look forward to further investigations along these lines. The range of examples reflects the variety of means by which ribosomal filtering may be modulated. We expect that future studies will reveal additional filtering mechanisms. Such knowledge will be valuable for understanding normal cellular function and development. In addition, it should clarify the mechanisms used by some viruses to usurp the host translation, as well as those involved in various ribosomal diseases.
Acknowledgments
We thank Drs. Joseph Gally and Stephen A. Chappell for valuable comments and critical readings of the manuscript. Funding was provided by the National Institutes of Health Grants GM61725 and GM078071 (to V.P.M.), the U.S. Public Health Service Grant NS39837 (to G.M.E.), and the G. Harold and Leila Y. Mathers Charitable Foundation.
Abbreviations, acronyms
- IRES
internal ribosome entry site
- MudPIT
multi-dimensional protein identification technology
- NGF
nerve growth factor
- RNP
ribonucleoprotein
- sno
small nucleolar
- XIAP
X-linked inhibitor of apoptosis protein
- CrPV
cricket paralysis virus
- HCV
hepatitis C virus
Contributor Information
Vincent P. Mauro, Email: vmauro@scripps.edu.
Gerald M. Edelman, Email: edelman@nsi.edu.
References
- 1.Kozak M. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J. 1997;16:2482–92. doi: 10.1093/emboj/16.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geballe AP, Sachs MS. Translational control by upstream open reading frames. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational control of gene expression. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2000. pp. 595–614. [Google Scholar]
- 3.Meijer HA, Dictus WJ, Keuning ED, Thomas AA. Translational control of the Xenopus laevis connexin-41 5′-untranslated region by three upstream open reading frames. J Biol Chem. 2000;275:30787–93. doi: 10.1074/jbc.M005531200. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Asouli Y, Banai Y, Pel-Or Y, Shir A, Kaempfer R. Human interferon-gamma mRNA autoregulates its translation through a pseudoknot that activates the interferon-inducible protein kinase PKR. Cell. 2002;108:221–32. doi: 10.1016/s0092-8674(02)00616-5. [DOI] [PubMed] [Google Scholar]
- 5.Rogers GW, Jr, Edelman GM, Mauro VP. Differential utilization of upstream AUGs in the beta-secretase mRNA suggests that a shunting mechanism regulates translation. Proc Natl Acad Sci USA. 2004;101:2794–9. doi: 10.1073/pnas.0308576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pantopoulos K. Iron metabolism and the IRE/IRP regulatory system: an update. Ann NY Acad Sci. 2004;1012:1–13. doi: 10.1196/annals.1306.001. [DOI] [PubMed] [Google Scholar]
- 7.Bassell GJ, Kelic S. Binding proteins for mRNA localization and local translation, and their dysfunction in genetic neurological disease. Curr Opin Neurobiol. 2004;14:574–81. doi: 10.1016/j.conb.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Vanderklish PW, Edelman GM. Differential translation and fragile X syndrome. Genes Brain Behav. 2005;4:360–84. doi: 10.1111/j.1601-183X.2005.00134.x. [DOI] [PubMed] [Google Scholar]
- 9.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–35. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–6. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 11.Chappell SA, Edelman GM, Mauro VP. Ribosomal tethering and clustering as mechanisms for translation initiation. Proc Natl Acad Sci U S A. 2006;16:16. doi: 10.1073/pnas.0608212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mauro VP, Edelman GM. The ribosome filter hypothesis. Proc Natl Acad Sci USA. 2002;99:12031–6. doi: 10.1073/pnas.192442499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mauro VP, Edelman GM. rRNA-like sequences occur in diverse primary transcripts: implications for the control of gene expression. Proc Natl Acad Sci USA. 1997;94:422–7. doi: 10.1073/pnas.94.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tranque P, Hu MC-Y, Edelman GM, Mauro VP. rRNA complementarity within mRNAs: A possible basis for mRNA-ribosome interactions and translational control. Proc Natl Acad Sci USA. 1998;95:12238–43. doi: 10.1073/pnas.95.21.12238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu MC-Y, Tranque P, Edelman GM, Mauro VP. rRNA-complementarity in the 5′ UTR of mRNA specifying the Gtx homeodomain protein: evidence that base-pairing to 18S rRNA affects translational efficiency. Proc Natl Acad Sci USA. 1999;96:1339–44. doi: 10.1073/pnas.96.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chappell SA, Edelman GM, Mauro VP. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc Natl Acad Sci USA. 2000;97:1536–41. doi: 10.1073/pnas.97.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Owens GC, Chappell SA, Mauro VP, Edelman GM. Identification of two short internal ribosome entry sites selected from libraries of random oligonucleotides. Proc Natl Acad Sci USA. 2001;98:1471–6. doi: 10.1073/pnas.98.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chappell SA, Mauro VP. The internal ribosome entry site (IRES) contained within the RNA-binding motif protein 3 (Rbm3) mRNA is composed of functionally distinct elements. J Biol Chem. 2003;278:33793–800. doi: 10.1074/jbc.M303495200. [DOI] [PubMed] [Google Scholar]
- 19.Matveeva OV, Shabalina SA. Intermolecular mRNA-rRNA hybridization and the distribution of potential interaction regions in murine 18S rRNA. Nucl Acids Res. 1993;21:1007–11. doi: 10.1093/nar/21.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mignone F, Pesole G. rRNA-like sequences in human mRNAs. Appl Bioinformatics. 2002;1:145–54. [PubMed] [Google Scholar]
- 21.Scheper GC, Voorma HO, Thomas AAM. Basepairing with 18S ribosomal RNA in internal initiation ot translation. FEBS Lett. 1994;352:271–5. doi: 10.1016/0014-5793(94)00975-9. [DOI] [PubMed] [Google Scholar]
- 22.Ramagopal S. Are eukaryotic ribosomes heterogeneous? Affirmations on the horizon. Biochem Cell Biol. 1992;70:269–72. doi: 10.1139/o92-042. [DOI] [PubMed] [Google Scholar]
- 23.Chappell SA, Edelman GM, Mauro VP. Biochemical and Functional analysis of a 9-nucleotide RNA sequence that affects translation efficiency in eukaryotic cells. Proc Natl Acad Sci USA. 2004:9590–4. doi: 10.1073/pnas.0308759101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dresios J, Chappell SA, Zhou W, Mauro VP. An mRNA-rRNA base-pairing mechanism for translation initiation in eukaryotes. Nat Struct Mol Biol. 2006;13:30–4. doi: 10.1038/nsmb1031. [DOI] [PubMed] [Google Scholar]
- 25.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–6. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steitz JA, Jakes K. How ribosomes select initiator regions in mRNA: base pair formation between the 3′ terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc Natl Acad Sci USA. 1975;72:4734–8. doi: 10.1073/pnas.72.12.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dunn JJ, Buzash-Pollert E, Studier FW. Mutations of bacteriophage T7 that affect initiation of synthesis of the gene 0.3 protein. Proc Natl Acad Sci USA. 1978;75:2741–5. doi: 10.1073/pnas.75.6.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Politz SM, Glitz DG. Ribosome structure: localization of N6,N6-dimethyladenosine by electron microscopy of a ribosome-antibody complex. Proc Natl Acad Sci USA. 1977;74:1468–72. doi: 10.1073/pnas.74.4.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hui A, De Boer HA. Specialized ribosome system: Preferential translation of a sinle mRNA species by subpopulation of mutated ribosomes in Escherichia coli. Proc Natl Acad Sci USA. 1987;84:4762–6. doi: 10.1073/pnas.84.14.4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacob WF, Santer M, Dahlberg AE. A single base change in the Shine-Dalgarno region of 16S rRNA of Escherichia coli affects translation of many proteins. Proc Natl Acad Sci USA. 1987;84:4757–61. doi: 10.1073/pnas.84.14.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kislinger T, Cox B, Kannan A, Chung C, Hu P, Ignatchenko A, Scott MS, Gramolini AO, Morris Q, Hallett MT, Rossant J, Hughes TR, Frey B, Emili A. Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell. 2006;125:173–86. doi: 10.1016/j.cell.2006.01.044. [DOI] [PubMed] [Google Scholar]
- 32.Angelastro JM, Torocsik B, Greene LA. Nerve growth factor selectively regulates expression of transcripts encoding ribosomal proteins. BMC Neurosci. 2002;3:3. doi: 10.1186/1471-2202-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giavalisco P, Wilson D, Kreitler T, Lehrach H, Klose J, Gobom J, Fucini P. High heterogeneity within the ribosomal proteins of the Arabidopsis thaliana 80S ribosome. Plant Mol Biol. 2005;57:577–91. doi: 10.1007/s11103-005-0699-3. [DOI] [PubMed] [Google Scholar]
- 34.Green-Willms NS, Fox TD, Costanzo MC. Functional interactions between yeast mitochondrial ribosomes and mRNA 5′ untranslated leaders. Mol Cell Biol. 1998;18:1826–34. doi: 10.1128/mcb.18.4.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams EH, Perez-Martinez X, Fox TD. MrpL36p, a highly diverged L31 ribosomal protein homolog with additional functional domains in Saccharomyces cerevisiae mitochondria. Genetics. 2004;167:65–75. doi: 10.1534/genetics.167.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scharf ME, Wu-Scharf D, Zhou X, Pittendrigh BR, Bennett GW. Gene expression profiles among immature and adult reproductive castes of the termite Reticulitermes flavipes. Insect Mol Biol. 2005;14:31–44. doi: 10.1111/j.1365-2583.2004.00527.x. [DOI] [PubMed] [Google Scholar]
- 37.Bachand F, Silver PA. PRMT3 is a ribosomal protein methyltransferase that affects the cellular levels of ribosomal subunits. EMBO J. 2004;23:2641–50. doi: 10.1038/sj.emboj.7600265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim HD, Lee JY, Kim J. Erk phosphorylates threonine 42 residue of ribosomal protein S3. Biochem Biophys Res Commun. 2005;333:110–5. doi: 10.1016/j.bbrc.2005.05.079. [DOI] [PubMed] [Google Scholar]
- 39.Orfali KA, Ohene-Abuakwa Y, Ball SE. Diamond Blackfan anaemia in the UK: clinical and genetic heterogeneity. Br J Haematol. 2004;125:243–52. doi: 10.1111/j.1365-2141.2004.04890.x. [DOI] [PubMed] [Google Scholar]
- 40.Oliver ER, Saunders TL, Tarle SA, Glaser T. Ribosomal protein L24 defect in belly spot and tail (Bst), a mouse Minute. Development. 2004;131:3907–20. doi: 10.1242/dev.01268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mazumder B, Sampath P, Seshadri V, Maitra RK, DiCorleto PE, Fox PL. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell. 2003;115:187–98. doi: 10.1016/s0092-8674(03)00773-6. [DOI] [PubMed] [Google Scholar]
- 42.Shabalina SA, Ogurtsov AY, Rogozin IB, Koonin EV, Lipman DJ. Comparative analysis of orthologous eukaryotic mRNAs: potential hidden functional signals. Nucleic Acids Res. 2004;32:1774–82. doi: 10.1093/nar/gkh313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoon A, Peng G, Brandenburger Y, Zollo O, Xu W, Rego E, Ruggero D. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312:902–6. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- 44.Holcik M, Korneluk RG. Functional Characterization of the X-Linked Inhibitor of Apoptosis (XIAP) Internal Ribosome Entry Site Element: Role of La Autoantigen in XIAP Translation. Mol Cell Biol. 2000;20:4648–57. doi: 10.1128/mcb.20.13.4648-4657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kullmann M, Gopfert U, Siewe B, Hengst L. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR. Genes Dev. 2002;16:3087–99. doi: 10.1101/gad.248902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pisarev AV, Shirokikh NE, Hellen CU. Translation initiation by factor-independent binding of eukaryotic ribosomes to internal ribosomal entry sites. CR Biol. 2005;328:589–605. doi: 10.1016/j.crvi.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 47.Limbach PA, Crain PF, McCloskey JA. Summary: the modified nucleosides of RNA. Nucleic Acids Res. 1994;22:2183–96. doi: 10.1093/nar/22.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steel LF, Jacobson A. Translational control of ribosomal protein synthesis during early Dictyostelium discoideum development. Mol Cell Biol. 1987;7:965–72. doi: 10.1128/mcb.7.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mangiarotti G. Synthesis of ribosomal proteins in developing Dictyostelium discoideum cells is controlled by the methylation of proteins S24 and S31. Biochem Cell Biol. 2002;80:261–70. doi: 10.1139/o02-005. [DOI] [PubMed] [Google Scholar]
- 50.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CUT. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus. RNAs Genes & Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Otto GA, Lukavsky PJ, Lancaster AM, Sarnow P, Puglisi JD. Ribosomal proteins mediate the hepatitis C virus IRES-HeLa 40S interaction. RNA. 2002;8:913–23. doi: 10.1017/s1355838202022057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Colon-Ramos DA, Shenvi CL, Weitzel DH, Gan EC, Matts R, Cate J, Kornbluth S. Direct ribosomal binding by a cellular inhibitor of translation. Nat Struct Mol Biol. 2006;22:22. doi: 10.1038/nsmb1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomenius M, Kornbluth S. Multifunctional reaper: sixty-five amino acids of fury. Cell Death Differ. 2006;13:1305–9. doi: 10.1038/sj.cdd.4401954. [DOI] [PubMed] [Google Scholar]
- 54.Gonzales B, Henning D, So RB, Dixon J, Dixon MJ, Valdez BC. The Treacher Collins syndrome (TCOF1) gene product is involved in pre-rRNA methylation. Hum Mol Genet. 2005;14:2035–43. doi: 10.1093/hmg/ddi208. [DOI] [PubMed] [Google Scholar]
- 55.Flygare J, Karlsson S. Diamond-Blackfan anemia: erythropoiesis lost in translation. Blood. 2007;109:3152–4. doi: 10.1182/blood-2006-09-001222. [DOI] [PubMed] [Google Scholar]
- 56.Angelini M, Cannata S, Mercaldo V, Gibello L, Santoro C, Dianzani I, Loreni F. Missense mutations associated to Diamond-Blackfan Anemia affect the assembly of ribosomal protein S19 into the ribosome. Hum Mol Genet. 2007:20. doi: 10.1093/hmg/ddm120. [DOI] [PubMed] [Google Scholar]
- 57.Ellis SR, Massey AT. Diamond Blackfan anemia: A paradigm for a ribosome-based disease. Med Hypotheses. 2006;66:643–8. doi: 10.1016/j.mehy.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 58.Matsson H, Davey EJ, Draptchinskaia N, Hamaguchi I, Ooka A, Leveen P, Forsberg E, Karlsson S, Dahl N. Targeted disruption of the ribosomal protein S19 gene is lethal prior to implantation. Mol Cell Biol. 2004;24:4032–7. doi: 10.1128/MCB.24.9.4032-4037.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gazda HT, Grabowska A, Merida-Long LB, Latawiec E, Schneider HE, Lipton JM, Vlachos A, Atsidaftos E, Ball SE, Orfali KA, Niewiadomska E, Da Costa L, Tchernia G, Niemeyer C, Meerpohl JJ, Stahl J, Schratt G, Glader B, Backer K, Wong C, Nathan DG, Beggs AH, Sieff CA. Ribosomal protein S24 gene is mutated in Diamond-Blackfan anemia. Am J Hum Genet. 2006;79:1110–8. doi: 10.1086/510020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nilsson J, Sengupta J, Frank J, Nissen P. Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep. 2004;5:1137–41. doi: 10.1038/sj.embor.7400291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shor B, Calaycay J, Rushbrook J, McLeod M. Cpc2/RACK1 is a ribosome-associated protein that promotes efficient translation in Schizosaccharomyces pombe. J Biol Chem. 2003;278:49119–28. doi: 10.1074/jbc.M303968200. [DOI] [PubMed] [Google Scholar]
- 62.Gerbasi VR, Weaver CM, Hill S, Friedman DB, Link AJ. Yeast Asc1p and mammalian RACK1 are functionally orthologous core 40S ribosomal proteins that repress gene expression. Mol Cell Biol. 2004;24:8276–87. doi: 10.1128/MCB.24.18.8276-8287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baker KE, Coller J. The many routes to regulating mRNA translation. Genome Biol. 2006;7:332. doi: 10.1186/gb-2006-7-12-332. [DOI] [PMC free article] [PubMed] [Google Scholar]


