Abstract
As another step toward extracting quantitative information from hyperpolarized 3He MRI, airway diameters in humans were measured from projection images and multislice images of the lungs. Values obtained were in good agreement with the Weibel lung morphometry model. The measurement of airway caliber can now be achieved without the use of ionizing radiation. Furthermore, it was demonstrated that 3D airway tree renderings could be constructed from the multislice data. Both the measurement of airway diameters and the rendering of 3D airway information hold promise for the clinical assessment of bronchoconstrictive diseases such as asthma and the associated evaluation of treatment effectiveness. Work is being done to address the uncertainties of the manually intensive methods we have developed.
Keywords: hyperpolarized 3He MRI, asthma, airway tree
Pulmonary diseases of the human airways, such as asthma, chronic obstructive pulmonary disease, and bronchiolitis obliterans, affect many individuals. According to the National Institutes of Health, approximately 10% of the adult population in the United States suffers from asthma, resulting in direct and indirect costs of $11.8 billion for 1998. While the role and relative importance of the distal airways in causing asthma have been controversial (1–3), there is no doubt that their contribution to the manifestation and severity of asthma is significant and thus deserving of further study. Currently, high-resolution computed tomography (HRCT) is the gold standard for quantification of individual airway diameters. However, the radiation exposure inherent to HRCT makes it unviable for repetitive studies or pediatric imaging (4,5). A tool for obtaining airway morphometric information without exposing a patient to harmful radiation would be highly desirable.
The lack of radiation and versatile nature of MRI make it an attractive imaging modality for this purpose, but the heterogeneous composition and the resulting inhomogeneous magnetic environment, plus low water content within the airspaces of the lung have made it difficult to image using proton MRI. Hyperpolarized 3He magnetic resonance imaging (HP 3He MRI) overcomes the difficulty of low proton density within the lung by using hyperpolarized 3He as an imaging signal source (6,7). 3He is hyperpolarized by spin exchange with optically pumped rubidium vapor (8). It is then inhaled by a subject, and pulmonary MR images are acquired. Ventilation images of the lung map out gas distribution within the lung. Some studies have used static ventilation imaging to examine the ventilation defects in asthmatic lungs and their corresponding improvement with treatment (9). However, airway tree structure is poorly assessed using static ventilation imaging because the signal in the distal airways is obscured by the signal from the lung periphery. If, however, the lung is scanned dynamically during inhalation, the airway tree can be imaged before the gas reaches the periphery (10,11). The drawback to this method is that dynamic HP 3He MR images are not conventionally obtained, often requiring specially programmed pulse sequences and/or multiple breaths of 3He. To this end, our group has recently developed a new dynamic imaging method to obtain human airway images with a standard pulse sequence, using only a single inhalation of HP 3He (12). This technique allows for the acquisition of projection and multislice airway images such that airways down to the seventh generation can be distinguished, depending on the chosen flip angle. While with the ability to visualize down to the 5th–7th generation airways, we still cannot conclusively characterize the role of the distal airways; however, characterizing the role of the central airways is nonetheless a step toward the ultimate goal of assigning each airway group its place in the big picture.
Previous works have presented airway diameter values obtained from HP 3He MR images (13,14). However, the methods by which these measurements were obtained were not described, nor was airway diameter measurement a goal of these studies. In this paper, we present a method for measuring the diameters of the airways from generations 0 through 5 using the projection and multislice airway images, and validation of the values obtained by comparing them with a widely accepted morphometric model. While 6th and 7th generation airways are visible, unacceptable errors are expected due to the proximity between pixel dimensions and expected airway diameters. Such a method could potentially be developed into a clinical tool allowing physicians to quantify the amount of bronchoconstriction experienced by a pulmonary patient and the bronchodilation brought about by treatment.
Furthermore, we present the first MRI-generated 3D airway tree rendering of the human lung constructed from multislice HP 3He images. While previous renderings of the 3D airway tree have been performed from the HP 3He MRI data of rodents (15), the multiple-breath imaging protocol involved makes it unfeasible for the same task in humans. Together, these techniques open new avenues into the development of tools for the quantitative assessment of diseased airway morphology in the clinical setting.
METHODS
Details regarding subject recruitment and all polarization and imaging hardware can be found in Tooker et al. (12) Typical polarization values obtained were about 20%. Experimental procedures were carried out with informed consent from all subjects and in compliance with the regulations set forth by the IRB.
HP 3He Imaging Breathing Protocols and Scanning Parameters
Two different types of coronal airway images were acquired during the course of the study. Projection images were acquired with a dynamic projection protocol, and multislice images were acquired with a dynamic multislice protocol. In both cases, coronal proton lung images were first acquired for shimming purposes and to determine appropriate scan start and end locations for the multislice scans. Images acquired from both techniques were used for the measurement of airway diameters, while the dynamic multislice technique was further used for 3D airway tree rendering.
The breathing protocol for the dynamic imaging procedures has also been previously discussed by Tooker et al. (12) The scans were initiated just prior to helium inhalation and lasted throughout and past the entire inhalation. Dynamic projection imaging employed the Fast GRE pulse sequence, with the following parameters: echo time/repetition time (TE/TR) 1.2/4.4 msec, matrix 256 × 128, field of view (FOV) 46 cm, phase FOV 0.75, bandwidth 62.5 kHz, and flip angles 13–19°. The anterior-posterior gradient was turned off, and 50 images were acquired sequentially. These scan parameters resulted in a 423-msec acquisition time per image. Dynamic multislice imaging employed identical scan parameters, with the exception of acquiring 5 temporal repeats per slice, over 13 slices (for a total of 5 × 13 = 65 images), 13-mm slice thickness, and interleaved slice image acquisition order to image all 13 slices during each temporal repeat. All gradients were enabled to allow for slice selective information.
Diameter Quantification
A program was written and run in MATLAB version 6.0 (The Mathworks, Inc., Natick, MA) to quantify the diameters of the airways from dynamic HP 3He MR images. Seven multislice image sets and 10 projection image sets were interrogated to measure airway diameters from airway generations 0 through 5 (0 being the trachea). For multislice image sets, the images that contained the most airway information, generally two or three of the central slices, were employed for airway diameter measurement. For each airway generation, a total of 10 diameters were measured to compute an average generation diameter. Five of the measurements came from the left lobe of the lung, while the other 5 came from the right. Some diameters were measured from different positions along the same airway. The average generation diameter was then compared to the theoretical value as proposed by the Weibel lung model (16).
The Weibel model is a model of the respiratory airway tree. In it, each parent airway, starting with the trachea, splits into two daughter airways. Based on measurements from several cadavers, characteristic branching angles, airway diameters, and lengths for the different airway generations are prescribed. The Weibel model is one of several lung morphometry models generalizing patterns by which airway branching and geometry behave in human anatomy. Morphometry models are instrumental in making pressure–flow analyses of the airway tree tractable, and the Weibel model, with its ease of use, is particularly popular. Airway geometry measurements are often compared to these morphometry models to assess whether the numbers obtained are in the correct neighborhood.
To measure airway diameters, thresholding was first applied to the image under analysis to help distinguish the airways from the lung periphery and background noise. Any pixel with an intensity value above the threshold acquired the maximum intensity while any pixel with an intensity value below the threshold was set to zero intensity. The threshold was set iteratively until the best compromise was obtained between maximizing the number of airways visible, while minimizing the retention of background noise.
Whether working on a projection or multislice image, upon selecting an airway, the user draws a line segment perpendicular to the airway walls. MATLAB then calculates the physical length of the line from the pixel dimensions of the image. In multislice images, ends of airway segments are avoided for diameter measurements as these locations may be where an airway emerges from or fades into adjacent slices, and as a result, presents a misleading airway profile.
3D Airway Tree Rendering
Coronal dynamic multislice data sets were utilized for the construction of three-dimensional human airway trees. For each data set, the images comprising the third (of five) temporal repeat were chosen. These images were then imported into 3D Slicer (Brigham and Women’s Hospital, Surgical Planning Lab), a software program specifically designed for three-dimensional visualization, segmentation, and quantitative analysis of radiologic images. Within 3D Slicer, a threshold value was applied to each slice image and adjusted until the elimination of noise was deemed satisfactory by the user. The airways remaining on each resulting image were then manually traced to segment the airway structures from the lung periphery signal. 3D Slicer then stacked the slices of traced airways, forming the 3D airway tree rendering.
RESULTS
Figure 1 presents airway MR images acquired with the two dynamic imaging protocols. Figure 1a is a coronal projection image, while Figures 1b and c are coronal slices from a multislice data set. A representative coronal projection image alongside the resultant image after applying a threshold value with the best compromise is shown in Figure 2.
FIG. 1.
Representative dynamic coronal MR images. (a) Projection image; (b) Multislice image 7 of 13; (c) Multislice image 9 of 13.
FIG. 2.
Thresholding a dynamic projection image. (a) Before thresholding; (b) After thresholding.
Diameter Quantification
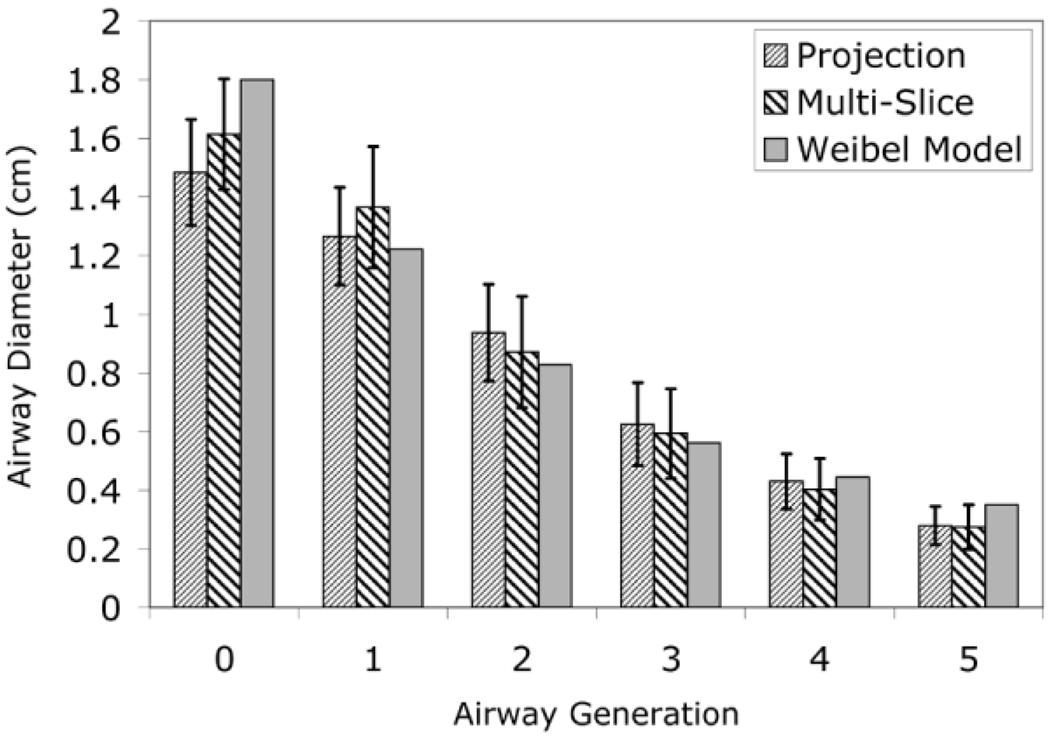
The diameter quantification program has yielded airway diameters comparable to those of the Weibel model for airway generations 0–5. Figure 3 compares the means of the measured diameters against the Weibel model’s values. The error bars represent the SD of the individual measured diameters from the average within each airway generation.
FIG. 3.
Measured and theoretical airway diameters.
Table 1 lists the P values from a t test (calculated using Sigma Plot 2000) to assess the agreement between the diameter values measured from the HP 3He MR projection images and the diameter values of the Weibel model. The large P values (>0.05) indicate that the differences in the calculated and theoretical airway diameters are not statistically significant; therefore the measured diameters are in good agreement with the Weibel model.
Table 1.
P Values Correlating Measured Airway Diameter Values against Weibel Model Predictions
| Gen 0 | Gen 1 | Gen 2 | Gen 3 | Gen 4 | Gen 5 | |
|---|---|---|---|---|---|---|
| Multislice | 0.172 | 0.332 | 0.747 | 0.774 | 0.575 | 0.168 |
| Projection | 0.016 | 0.706 | 0.351 | 0.529 | 0.822 | 0.133 |
Airway Tree Renderings
Three-dimensional airway tree renderings were successfully constructed from the dynamic multislice data sets. A representative 3D airway tree rendering is shown in Figure 4.
FIG. 4.
Airway tree constructed from stacking dynamic multislice data sets in 3D Slicer.
DISCUSSION
We report here a method for quantifying airway diameters from dynamic projection and dynamic multislice images. To our knowledge, this is the first study in which the fidelity of dynamic HP 3He MRI is examined by comparing diameter measurements acquired from MR images to a well-known and generally accepted morphologic airway tree model. Statistical analysis shows that the airway caliber measured are in good agreement with the Weibel morphologic model, thereby supporting the use of dynamic HP 3He MRI to quantify bronchoconstriction and bronchodilation in diseases such as asthma.
When quantifying airway diameters, the choice of the threshold level for filtering of the original airway MR images has a significant impact on the measured airway diameters. If the threshold, chosen to distinguish the airways from the lung periphery and noise, is too high, some legitimate airways may get filtered out. Conversely, some airways cannot be differentiated from the periphery if the threshold is too low. Ideally, with an analog image and an extremely sensitive visual tool, the borders of the airways can be unambiguously identified. The human eye, however, is not sensitive enough to detect the edges of the airways. Furthermore, with the digitization of medical images, the human eye can, at best, identify the pixels containing feature borders, but the exact location of the border within the pixel remains unknown. As a first attempt at measuring airway diameters from HP 3He MR images, no techniques were developed to address these issues. It is well known that mere thresholding doesn’t solve any of these problems. Moreover, since pixels containing airway edges tend to have low pixel intensities, it is likely that these pixels will be filtered out in the thresholding process.
Ideally, the line segment for measuring airway diameter should be drawn perpendicular to the airway for accurate diameter measurement. Fortunately, since the error scales with 1 − cos(θ), where θ is the angle between the drawn segment and true perpendicular, the error only begins to exceed 5% when θ is greater than 18.2°.
The dynamic multislice images acquired in this study were also stacked to yield 3D airway tree renderings. Three-dimensional imaging is an up-and-coming tool in diagnostic care, replacing several endoscopic procedures. Rendering of the 3D airway tree can give pulmonary physicians access to the inside of the airways in a bronchoscope-like paradigm. Besides being noninvasive, these “virtual” endoscopic procedures require minimal patient preparation, making them ideal for screening practices. Such a technique should have advantages over existing morphometric lung models (5,16 –19), which are oversimplifications of real airway geometry, both anatomically inaccurate and lacking a one-to-one correlation with physical lung components. In addition, these models do not provide 3D information.
Data from HRCT have also been used with some success for the rendering of 3D airway trees (19,20). Currently the best 3D model generated from HRCT data contains the first six airway generations.
Presented herein, the current procedure for rendering the 3D airway tree from HP 3He MRI data is somewhat limited by the high aspect ratio of image voxels (1.8 mm pixel dimension by 13 mm slice thickness) and loss of connectivity to legitimate airways. As in the measurement of airway diameters, this being a first attempt, no procedures or techniques were developed to deal with these issues. Similarly, the thresholding process may inadvertently filter out voxels with lower signal, effectively biasing the airway tree volume toward lower values. For a given user measuring airways with a consistent set of criteria, measurements of airway diameter changes resulting from bronchoconstriction or bronchodilation should be rigorous. That is, bias errors may cause airway diameter measurements to be inaccurate, but the bias errors should cancel out when the difference between two measurements is taken. In addition, the nonionizing nature of RF radiation used in MRI allows for further advances in this methodology without subjecting the patient to risks. Visualization of airway trees using 3D renderings generated from HP 3He MRI may one day empower physicians to diagnose the severity of lung diseases and to assess the efficacy of various treatments.
Acknowledgments
Grant sponsor: National Institutes of Health; Grant number: R21-EB1689;
Grant sponsor: National Science Foundation; Grant number: BES-93448;
Grant sponsor: National Aeronautics and Space Administration; Grant number: NAG9–1469.
REFERENCES
- 1.Kraft M, Djukanovic R, Wilson S, Holgate S, Martin R. Alveolar tissue inflammation in asthma. Am J Respir Crit Care Med. 1996;154:1505–1511. doi: 10.1164/ajrccm.154.5.8912772. [DOI] [PubMed] [Google Scholar]
- 2.Carroll N, Cooke C, James A. The distribution of eosinophils and lymphocytes in the large and small airways of asthmatics. Eur Respir J. 1997;10:292–300. doi: 10.1183/09031936.97.10020292. [DOI] [PubMed] [Google Scholar]
- 3.Faul J, Tormey V, Leonard C, Burke C, Farmer J, Horne S, Poulter L. Lung immunopathology in cases of sudden asthma death. Eur Respir J. 1997;10:301–308. doi: 10.1183/09031936.97.10020301. [DOI] [PubMed] [Google Scholar]
- 4.Genier P, Cordeau MP, Beigelman C. High-resolution computed tomography of the airways. J Thorac Imaging. 1993;8:213–229. doi: 10.1097/00005382-199322000-00006. [DOI] [PubMed] [Google Scholar]
- 5.Murata K, Khan A, Rojas KA, Herman PG. Optimization of computed tomography: technique to demonstrate the fine structure of the lung. Invest Radiol. 1988;23:170–175. doi: 10.1097/00004424-198803000-00003. [DOI] [PubMed] [Google Scholar]
- 6.Black RD, et al. In vivo He-3 MR images of guinea pig lungs. Radiology. 1996;199:867–870. doi: 10.1148/radiology.199.3.8638019. [DOI] [PubMed] [Google Scholar]
- 7.Kauczor HU, et al. Normal and abnormal pulmonary ventilation: visualization at hyperpolarized He-3 MR imaging. Radiology. 1996;201:564–568. doi: 10.1148/radiology.201.2.8888259. [DOI] [PubMed] [Google Scholar]
- 8.Happer W, et al. Polarization of the nuclear spins of noble gas atoms by spin exchange with optically pumped alkali metal atoms. Phys Rev A. 1984;29:3092–3110. [Google Scholar]
- 9.Altes TA, Powers PL, Knight-Scott J, Rakes G, Platts-Mills TAE, de Lange EE, Alford BA, Mugler JP, Brookeman JR. Hyperpolarized 3He MR lung ventilation imaging in asthmatics: preliminary findings. J Magn Reson Imaging. 2001;13:378–384. doi: 10.1002/jmri.1054. [DOI] [PubMed] [Google Scholar]
- 10.Chen XJ, et al. MR microscopy of lung airways with hyperpolarized 3He. Magn Reson Med. 1998;39:79–84. doi: 10.1002/mrm.1910390113. [DOI] [PubMed] [Google Scholar]
- 11.Salerno M, Altes TA, Brookeman JR, de Lange EE, Mugler JP., III Dynamic spiral MRI of pulmonary gas flow using hyperpolarized 3He: preliminary studies in healthy and diseased lungs. Magn Reson Med. 2001;46:667–677. doi: 10.1002/mrm.1244. [DOI] [PubMed] [Google Scholar]
- 12.Tooker AC, Hong KS, McKinstry EL, Costello P, Jolesz FA, Albert MS. Distal airways in humans: dynamic hyperpolarized 3He MR imaging—feasibility. Radiology. 2003;227:575–579. doi: 10.1148/radiol.2272012146. [DOI] [PubMed] [Google Scholar]
- 13.Chen BT, Brau ACS, Johnson GA. Measurement of regional lung function in rats using hyperpolarized 3helium dynamic MRI. Magn Reson Med. 2003;49:78–88. doi: 10.1002/mrm.10336. [DOI] [PubMed] [Google Scholar]
- 14.Johnson GA, Cofer GP, Hedlund LW, Maronpot RR, Suddarth SA. Registered 1H and 3He magnetic resonance microscopy of the lung. Magn Reson Med. 2001;45:365–370. doi: 10.1002/1522-2594(200103)45:3<365::aid-mrm1047>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 15.Wang B, Saha PK, Udupa JK, Ferrante MA, Baumgardner J, Roberts DA, Rizi RR. 3D airway segmentation via hyperpolarized 3He gas MRI by using scale-based fuzzy connectedness. Comput Med Imaging Graph. 2004;28:77–86. doi: 10.1016/j.compmedimag.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 16.Weibel ER, Gomez DM. Architecture of the human lung. Science. 1962;137:577–585. doi: 10.1126/science.137.3530.577. [DOI] [PubMed] [Google Scholar]
- 17.Horsfield K, Dart G, Olson DE, Filley GF, Cumming G. Models of the human bronchial tree. J Appl Physiol. 1971;31:202–217. doi: 10.1152/jappl.1971.31.2.207. [DOI] [PubMed] [Google Scholar]
- 18.Kitaoka H, Takaki R, Suki B. A three-dimensional model of the human airway tree. J Appl Physiol. 1999;87:2207–2217. doi: 10.1152/jappl.1999.87.6.2207. [DOI] [PubMed] [Google Scholar]
- 19.Sauret V, Halson PM, Brown IW, Fleming JS, Bailey AG. Study of the three-dimensional geometry of the central conducting airways in man using computed tomographic (CT) images. J Anat. 2002;200:123–134. doi: 10.1046/j.0021-8782.2001.00018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aykac D, Hoffman EA, McLennan G, Reinhardt JM. Segmentation and analysis of the human airway tree from three-dimensional X-ray CT images. IEEE Trans Med Imaging. 2003;22(8):940–950. doi: 10.1109/TMI.2003.815905. [DOI] [PubMed] [Google Scholar]