Abstract
GnRH gene expression is restricted to a tiny population of neurons scattered throughout the mediobasal hypothalamus. The combination of a 300-bp enhancer and the 173-bp promoter from the rat GnRH gene can confer this narrow specificity in transgenic mice and in transfections of hypothalamic GT1–7 cells. In the present study, we identify repeated CAATT elements in the 3′ region of the rat GnRH enhancer that bind a tissue-restricted protein complex and play a significant role in cell-restricted expression of the GnRH gene. Deletions of multiple repeats demonstrate their importance in transcriptional activity. In fact, even mutation of a single repeat reduces expression. This reduction can be compensated by the conserved GnRH promoter, which also contains such elements and binds this protein complex. In Southwestern analysis, three proteins from GT1–7 nuclear extract bind to the CAATT element, and these proteins are not found in NIH3T3 cells. This cell-specific protein complex has properties of the Q50 homeodomain family of transcription factors and binds to as many as seven binding sites in the enhancer and promoter to play a key role in GnRH gene expression in the hypothalamus.
Dispersed through several areas of the hypothalamus, the GnRH neurons are responsible for reproductive function. The defining attribute of these neurons is that they produce GnRH and release it in a pulsatile fashion. Changes in the pulsatility of GnRH release are related to key events in reproductive life including entry into puberty, normal cycling of menstruation and estrous, and entry into menopause.
Because of the dispersed nature and small numbers [~800 in the mouse (1)] of the GnRH neuron population, it is difficult to study the molecular mechanisms of transcription, translation, and secretion of GnRH in vivo. To address the need for a large, pure population of GnRH neurons, targeted oncogenesis was used to create immortal, differentiated cell lines expressing GnRH (2). Three kilobases of the 5′ regulatory region of the rat GnRH (rGnRH) gene were fused to the coding region of large Simian virus 40 T antigen and used for the creation of transgenic mice. A hypothalamic tumor isolated from a transgenic mouse was cultured and then subcloned to create three cell lines: GT1–1, GT1–3, and GT1–7.
Initial study of the rGnRH 5′ regulatory region in GT1–7 cells led to the identification of an approximately 300-bp enhancer from −1863 to −1571 relative to the start site and a conserved 173-bp promoter just upstream of the transcriptional start site (3). The 173-bp promoter region is highly conserved between rat, mouse, and human (4). A number of proteins bind and activate in the promoter region, including the POU homeodomain octamer-binding protein Oct-1 and the neuron-restricted homeodomain protein Otx2 (ortho-denticle-related homeobox protein) (5, 6). The rGnRH enhancer displays the defining properties of an enhancer, such as activity in either orientation and at varying distances from the promoter (3). It confers a 50- to 100-fold transcriptional activation over the promoter alone, exclusively in GT1–7 cells. The enhancer coupled with the promoter is sufficient for proper targeting and developmental expression of the GnRH gene in transgenic mice (7). However, substitution of the GnRH promoter with a heterologous one disrupts the targeting in vivo, demonstrating the essential nature of the enhancer/promoter interaction.
Eight regions of the GnRH enhancer bind proteins from GT1–7 nuclear extract as determined by deoxyribonuclease I protection assay. Removing more than 40 bases from either end of the 300-bp enhancer or removing any one of a number of single footprinted regions greatly decreases activity (3). Within the 300-bp enhancer, specific binding sites for a number of transcription factors have been identified. Two central AT-rich regions (AT-a and AT-b) bind Oct-1, and mutation of one, AT-a, has a dramatic effect on transcription in GT1–7 cells (8). The homeodomain proteins Pbx (pre-B lymphoblastic leukemia homeobox protein) and Prep1 (pbx-related protein) also bind to both the rGnRH enhancer and promoter and interact with Oct-1 (Rave-Harel, N., and P. L. Mellon, manuscript in preparation). The zinc finger protein GATA-4 binds to one site within the GnRH enhancer (GATA-b) (9). GATA-4 (10) and Pbx1 (Rave-Harel, N., and P. L. Mellon, manuscript in preparation) are coexpressed embryonically with GnRH in vivo; therefore, they may play an important developmental role in the differentiation of GnRH neurons. Although these factors are known to activate the GnRH enhancer, their patterns of expression are relatively broad and cannot account for the tight hypothalamic specificity of this enhancer in transient transfection assays and in transgenic mice. In this study, we identify a striking repeated element in the essential 3′ region of this enhancer that is comprised of six tandem copies of the sequence CAATT. We demonstrate the essential contribution of these repeated CAATT sequences to neuron-specific transcription of the GnRH gene. Furthermore, we show that the cell-specific protein complex binding to this CAATT sequence has the characteristics of a Q50 homeodomain protein that is restricted to the hypothalamic GT1–7 cell line.
RESULTS
The 3′ Repeat Region of the GnRH Enhancer Contributes to Transcriptional Activity
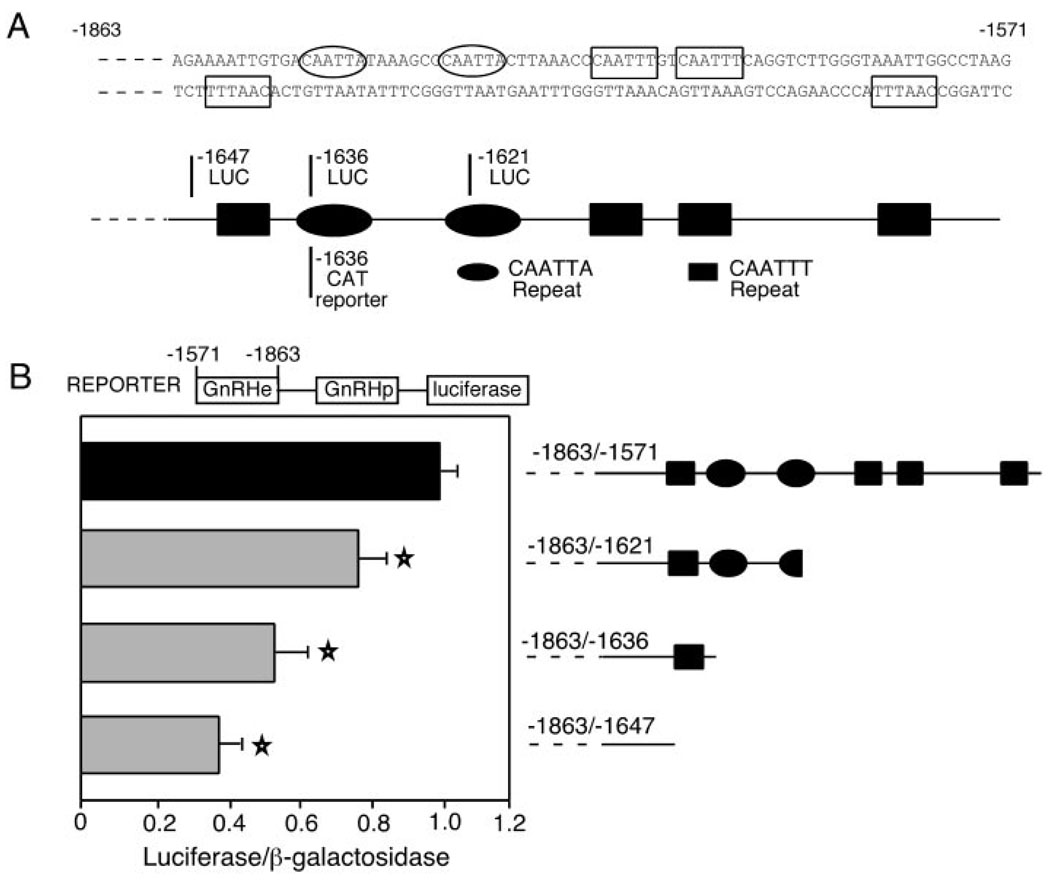
Previous studies of the rGnRH enhancer (−1863/−1571) have shown the importance of the 3′ footprinted region but have left it relatively uncharacterized. Earlier truncation analysis demonstrated that removing 65 bp from the 3′ end (leaving −1863/−1636) reduces transcriptional activation to approximately 25% (3). Clearly, this large footprinted region is essential for transcriptional activation. To better understand the individual elements within this 3′ region of the enhancer, we created additional 3′ truncations (Fig. 1A). The progressive 3′ deletion of these sequences causes a progressive reduction in transcriptional activity. Truncation to −1621 lowers activity to 78% of wild-type activity, whereas truncation to −1636 lowers activity to 54% and truncation to −1647 lowers activity to 39% (Fig. 1B). Here, all of these truncations are linked to the luciferase (luc) reporter gene whereas the earlier analysis was performed with the chloramphenicol acetyl transferase reporter gene (3). It is possible that the difference in reporter genes explains the slightly larger reduction observed previously with the −1636 truncation (3). The pattern of decreasing activity with more extensive truncations indicates either that separate activating elements are located within each of these deleted regions or that repeats of the same element exist in the 3′ region of the rGnRH enhancer.
Fig. 1. The 3′ Region of the GnRH Enhancer Is Essential for Transcriptional Activity.
A, The 3′ portion of the rGnRH enhancer sequence (−1648/−1571) is shown. Repeats in the 3′ region are indicated by boxes (for CAATTT repeats) and ovals (for the two CAATTA repeats). Below the sequence, a schematic of the repeats is shown. The base at which the truncations terminate are shown by a line, with one previously published (3) shown below the schematic and those in the current report shown above the schematic. B, The results of transient transfections of the 3′ truncations of the rGnRH enhancer are shown. To the right of each bar, a schematic indicates the CAATT repeats present in each of the enhancer truncations and the length of the enhancer present in the luc expression vector used. Transient transfections were performed with the rGnRH enhancer/rGnRH promoter (rGnRHe/rGnRHp-luc), either wild-type or truncated as indicated, directing luc expression, normalized with cotransfected RSV-β-gal internal control plasmid. Values are normalized to the full-length rGnRHe/rGnRHp (−1863/−1571), which is set to 1. Error bars indicate ± sem. Each bar represents the mean for at least three separate experiments performed in triplicate (at least n = 9). Truncations that cause a statistically significant difference from wild type, as determined by a one-way ANOVA, are indicated by a star (P < 0.02).
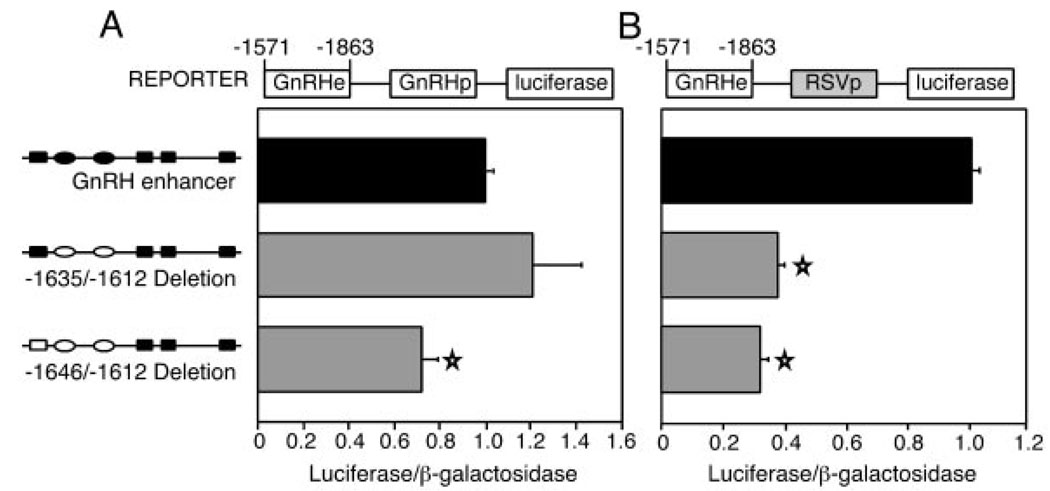
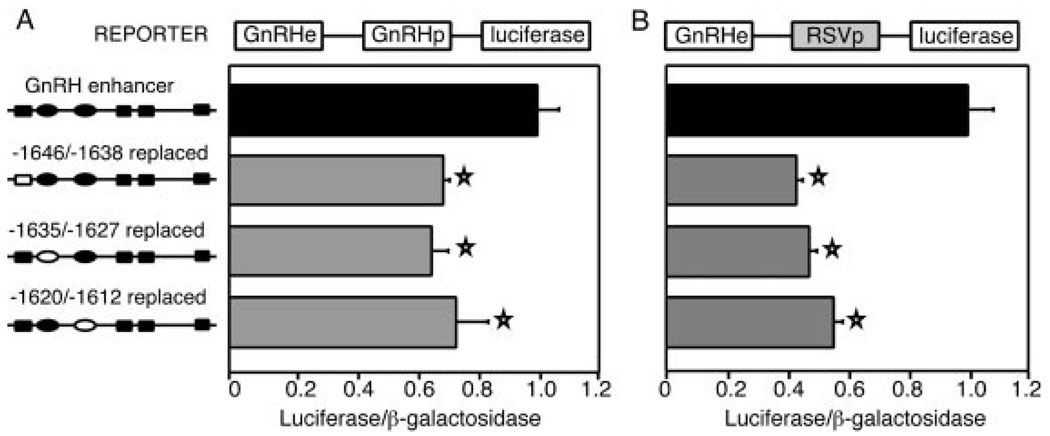
To examine this region in detail, we created small internal deletions. Three separate linker replacement mutations were employed to create two internal deletions (diagrammed to the left of the bar graphs in Fig. 2). When inserted adjacent to the GnRH promoter, the enhancer with an internal deletion from −1646/−1612 results in a decrease to 72% of wild-type levels, whereas the −1635/−1612 deleted enhancer increases slightly to 120%, although this change is not statistically significant (Fig. 2A). In contrast, when these internal deletions of the enhancer are placed adjacent to the heterologous Rous sarcoma virus (RSV) long terminal repeat promoter, both deleted enhancers result in similar decreases to 32% and 38%, respectively (Fig. 2B).
Fig. 2. The GnRH Promoter Compensates for Deletion of Enhancer CAATT Repeats.
Transfection results are displayed for internal deletions of the rGnRH enhancer fused to the native rGnRH promoter (A) or to a heterologous RSV promoter (B). Schematics of the CAATT repeats are shown to the left of the bar graphs. The solid boxes and ovals indicate the wild-type CAATTT and CAATTA repeats, respectively. The hollow boxes and ovals represent those repeats that were deleted or replaced by linker sequence. Transfections were performed as described in Fig. 1B. Stars indicate results statistically significant from wild type, as determined by one-way ANOVA (P < 0.003).
The dramatic decrease in transcription observed when the internally deleted enhancers are placed upstream of the RSV promoter implies that the rGnRH enhancer and promoter cooperate in transcriptional activation. One possibility is that there is sequence similarity between portions of the enhancer and promoter and that these regions bind the same proteins in a redundant fashion. When these repeat sequences are deleted from the enhancer, similar elements in the promoter might compensate.
Each of the CAATT Repeats Binds a Similar Protein Complex
Inspection of the sequence in the 3′ enhancer region and the promoter yields the identification of a CAATTA repeat. This repeat exists at −1636/−1631 and −1623/−1618 in the enhancer (Figs. 1A and 3A). Additionally, a closely related repeat of CAATTT occupies the bottom strand at −1645/−1640 and −1578/−1583, and the top strand at −1609/−1604 and −1601/−1596 (Fig. 3A).
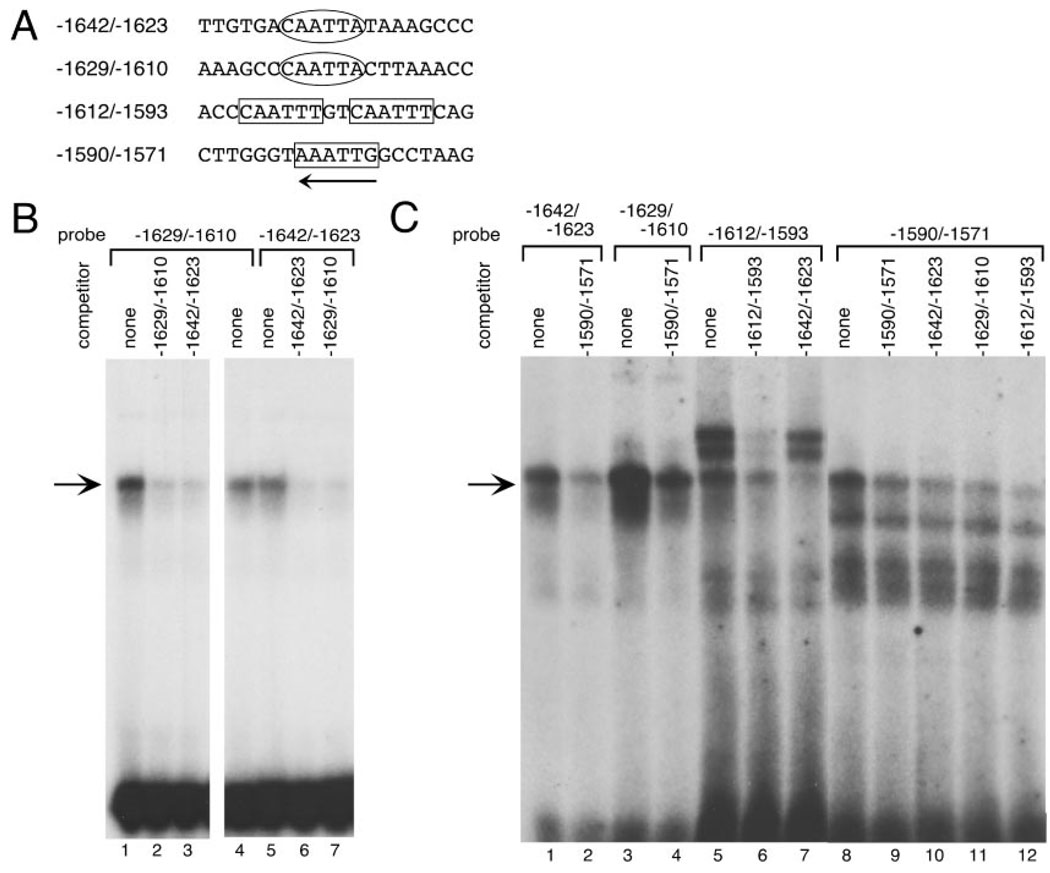
Fig. 3. Multiple CAATT Repeats in the rGnRH Enhancer/Promoter Bind the Same Complex.
A, A summary of the oligonucleotides used as probes and competitors in EMSA. B, GT1–7 nuclear extract was incubated with the end-labeled oligonucleotide indicated (−1629/−1610 in lanes 1–4 and −1642/−1623 in lanes 5–7) and eletrophoresed on a nondenaturing polyacrylamide gel. Competitions were as follows: lanes 1, 4, and 5, no competition; lanes 2 and 7, 100-fold molar excess of −1629/−1610; lanes 3 and 6, 100-fold molar excess of −1642/−1623. The arrow marks the position of the major complex. C, Oligonucleotides including regions of the rGnRH enhancer with CAATTA or CAATTT repeats were tested in EMSA, as indicated above the gel (CAATTA repeats: −1642/−1623 in lanes 1–2 and −1629/−1610 in lanes 3 and 4; CAATTT repeats: −1612/−1593 in lanes 5–7 and −1590/−1571 in lanes 8–12). Inclusion of unlabeled competitor oligonucleotides is indicated above each lane. The arrow marks the position of the major complex.
To test the similarity of the complexes that bind to the CAATT repeats in the rGnRH enhancer and promoter, oligonucleotides including the repeats were designed for use in EMSA. Radiolabeled oligonucleotides were incubated with nuclear extracts from GT1–7 cells. The major complex formed on each of two enhancer CAATTA repeats comigrates (Fig. 3B, lanes 4 and 5) and cross-competes (Fig. 3B, lanes 3 and 7). Self-competition abolishes the complex demonstrating that this complex is specific (Fig. 3B, lanes 2 and 6). The same complex also forms with the two downstream CAATTT repeats (probes −1612/−1593 and −1590/−1571, Fig. 3C, lanes 5 and 8, respectively) and is specific because it is self-competed (Fig. 3C, lanes 6 and 9). They also cross-compete with each other (Fig. 3C, lane 12). The upstream CAATTT element, however, failed to form detectable complexes in EMSA (data not shown). The −1612/−1593 oligonucleotide probe also contains a site for the binding of the homeodomain protein partners Pbx1 and Prep1. This sequence is TTGTCAATTT, overlapping the upstream CAATTT and encompassing the downstream CAATTT within this oligonucleotide. The upper doublet of bands binding to this probe (Fig. 3C, lanes 5, 6, and 7) contains these, more ubiquitous factors and their splicing variants (data not shown).
The downstream CAATTT oligonucleotides also were effective as competitors with the CAATTA repeats as shown for −1590/−1571 in Fig. 3C, lanes 2 and 4. Inversely, CAATTA repeat competition with CAATTT oligonucleotides as probes, also decreases formation of the cell-specific complex (arrow; Fig. 3C, lanes 7, 10, and 11), confirming that the nucleoprotein complex bound by these oligonucleotides is the same.
Evolutionary Conservation of CAATT Repeats
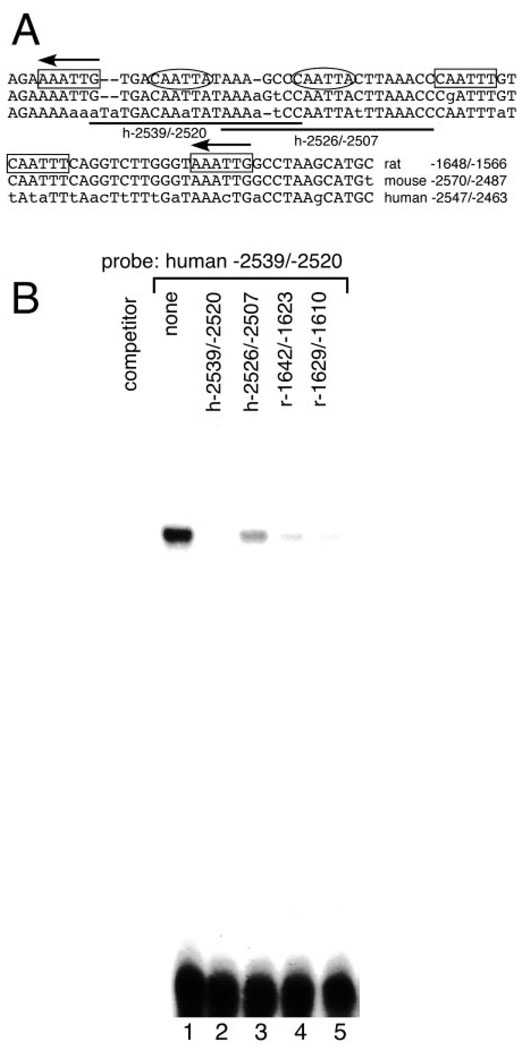
Despite the extreme divergence of the upstream regions of the rat, mouse, and human GnRH genes, we have identified regions of the mouse and human genes that show remarkable conservation with the rat enhancer. These are located more than 2 kb upstream of the transcriptional initiation sites (Fig. 4A). We examined these analogous sequences within the mouse and human upstream regulatory regions for CAATT repeats. The CAATTA elements are found not only within the rat enhancer but also within the mouse and human upstream regulatory regions (Fig. 4A). When tested in EMSA, the repeats in the upstream regulatory region of the rGnRH gene compete (Fig. 4B) and comigrate (data not shown) with those from the human sequence, despite a 1-bp change to CAAaTA in the human gene (Fig. 4A). Therefore, the repeats within the human and rat upstream sequence are likely binding to the same protein complexes. These similarities in sequence and protein-binding properties across divergent species underscore the importance of these sequences for gene regulation.
Fig. 4. The CAATT Repeated Elements Are Evolutionarily Conserved.
A, The 3′ AT-rich nucleotide sequences of the rat, mouse, and human conserved GnRH enhancer regions are shown. Mismatches are indicated in lowercase letters. Spacing changes required to produce the alignment are indicated by dashes within the sequence. Ovals indicate the CAATTA repeats, and boxes indicate the CAATTT repeats. Arrows indicate repeats on the opposite strand. The two probes from the human sequence used in Fig. 4B are underlined and named below the lines. B, An oligonucleotide probe including the sequence of the human GnRH enhancer equivalent to the upstream of the two rat CAATTA repeats (−2539/−2520) was competed by each of the rGnRH enhancer CAATTA repeats (−1642/−1623 and −1629/−1610; lanes 4 and 5) and the two equivalent sequences from the human gene (−2539/−2520 and −2526/−2507; lanes 2 and 3) in EMSA, performed as in Fig. 3.
Individual CAATT Repeats Are Necessary for Activity
To determine the impact of individual repeats on transcriptional activity, transient transfections were performed using the GnRH enhancer carrying linker replacement mutations in three of the individual 3′ CAATT repeats. The repeat altered by each mutation is diagramed as a hollow shape in Fig. 5. Each of the individual linker replacement mutations in this region (used to create the internal deletions shown in Fig. 2) reduces transcription, with the −1646/−1638 replacement reducing transcription to 69%, the −1635/−1627 replacement to 65%, and the −1620/−1612 replacement to 74% when fused to the GnRH promoter (Fig. 5A). These data indicate that each of these three repeats plays an important role in transcriptional activation.
Fig. 5. Individual CAATT Repeats Are Essential for GnRH Enhancer Activity.
A, rGnRH enhancer elements containing linker replacement mutations in the CAATT repeats fused to the rGnRH promoter were transfected into GT1–7 cells. A schematic of the 3′ portion of the enhancer (the CAATT repeat region) is shown to the left of the bar graphs. The solid shapes indicate the repeats present in each plasmid, and the hollow shapes indicate those replaced by linker sequence. Transfections were performed as described in Fig. 1B. A star on a bar signifies a statistically significant decrease from wild-type levels, as tested by a one-way ANOVA (P < 0.05). B, The same linker replacement mutations were placed instead on the heterologous RSV promoter and used in transient transfections. A star on a bar signifies a statistically significant decrease from wild-type levels, as tested by a one-way ANOVA (P < 2 × 10−7).
When the enhancers carrying these linker replacement mutations are analyzed utilizing the heterologous RSV promoter, the −1646/−1638 and −1635/−1627 linker replacement mutations each cause significantly greater decreases in enhancer activity, whereas the degree of reduction caused by the −1620/−1612 linker replacement mutation is similar on both promoters (Fig. 5B). The higher activity of these two mutations in the context of the GnRH promoter indicates that the promoter repeat partially compensates when certain enhancer repeats are disrupted.
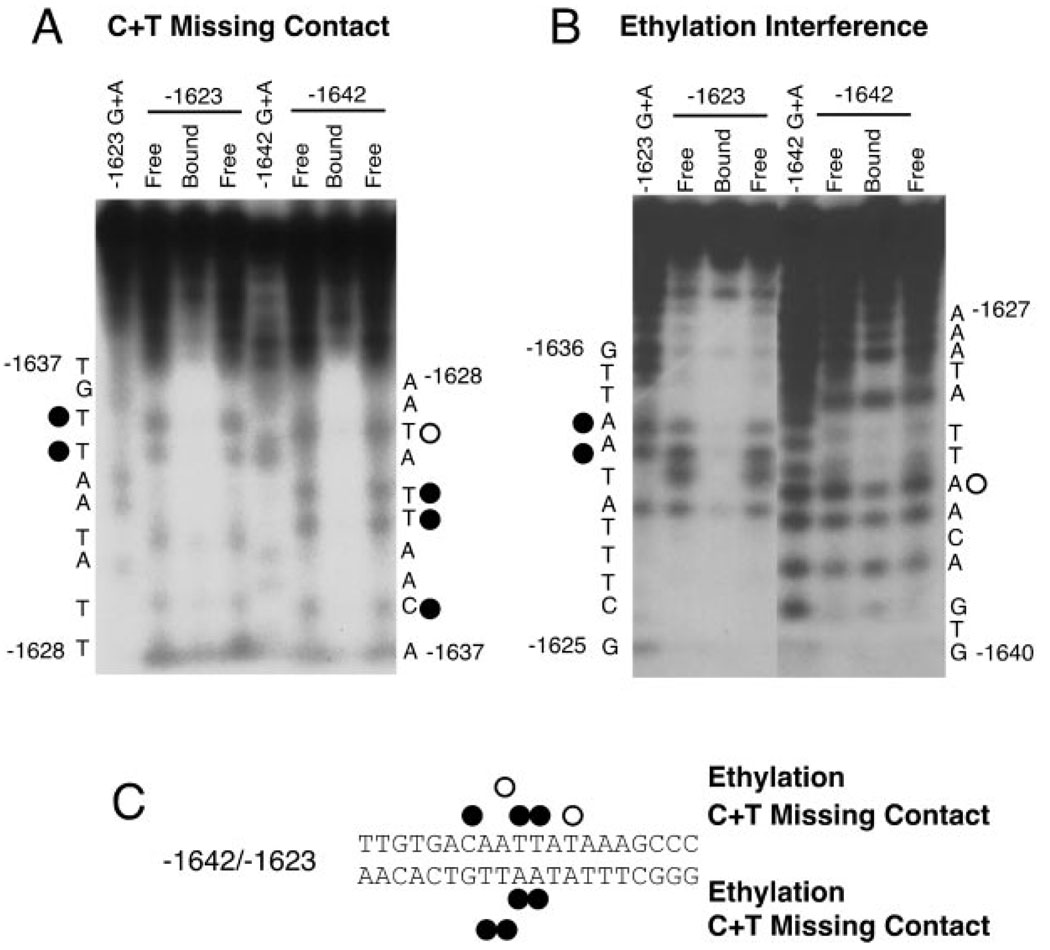
The Protein Complex Contacts the CAATT Nucleotides
To define the bases within the CAATT repeat sequence that participate in binding, ethylation interference and missing contact probing were performed. Representative autoradiograms of the C + T missing contact probing and ethylation interference assay for the rGnRH gene −1642/−1623 repeat are shown in Fig. 6. The combined results for ethylation interference and missing contact probing confirm that the protein strongly contacts the CAATT portion of −1642/−1623 (see summary, Fig. 6C). Note that the base pair that differs between the CAATTA and the CAATTT repeats does not show protein contacts.
Fig. 6. Specific Base Pairs in the CAATTA Sequence Are Required for Protein Binding.
A, A representative C + T missing contact probing autoradiogram is shown. The bottom strand is shown on the left with two identical free probe lanes flanking the bound probe lane. The absence of a band in the bound probe lane is considered to be evidence for participation of that base in protein binding. B, A representative ethylation interference autoradiogram is shown. The labeling is as in A. C, Summary of the bases contacted by protein from GT1–7 nuclear extract bound to −1642/−1623 from the rGnRH enhancer. Closed circles indicate complete contact (complete protection of the band), whereas the open circles (partial elimination of the band) indicate partial contact.
Mutational Analysis of CAATT Repeats
To validate the above findings regarding the binding site for the protein, mutations in the −1642/−1623 region from the rGnRH enhancer were tested in EMSA. In the −1636/−1631 CAATTA repeat, a mutation as small as a single base pair within the repeat region (T to C at −1633) eliminates binding of the specific complex (Fig. 7, lane 2). The double point mutations also completely abolish binding (Fig. 7, lane 3). These results support the above finding that the individual bases within the CAATT repeat region are essential for binding the protein complex.
Fig. 7. Point Mutations in the CAATTA Repeats Prevent in Vitro Binding.
A, A summary of the mutation sequences for both CAATTA repeats and the effects in EMSA. B, EMSA with the wild-type (lane 1), single (lane 2), and double-point (lane 3) mutations in the −1642/−1623 oligonucleotide and GT1–7 nuclear extract. The arrow marks the position of the cell-restricted complex.
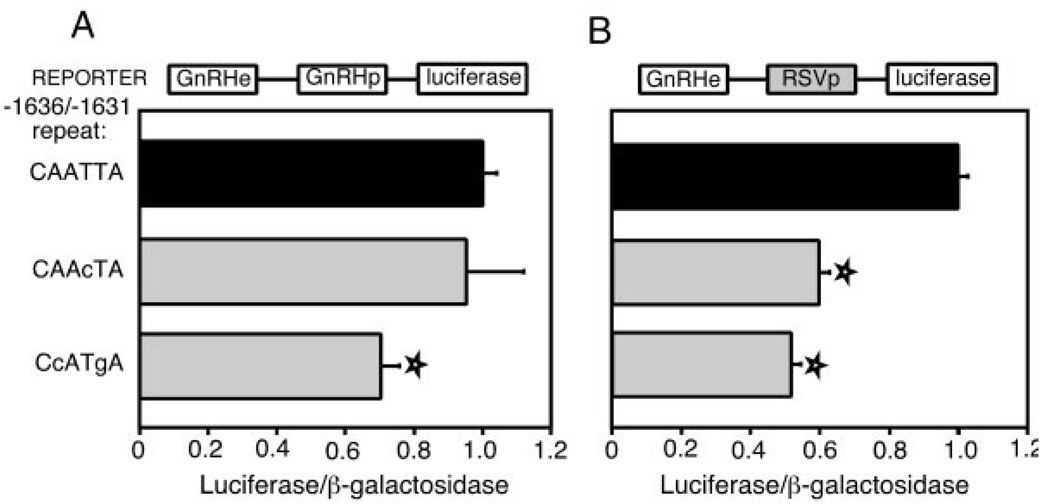
Single or Double Base-Pair Mutations Reduce Transcriptional Activation by the Cell-Restricted Complex
The mutational analysis with EMSA established that single or double base pair mutations affect the binding of the cell-restricted complex to the CAATTA repeat. These specific mutations in the −1636/−1631 CAATTA repeat were used in transient transfections in GT1–7 cells to examine their effects on transcription. A single point mutation at −1633 (changing the repeat to CAAcTA), does not affect transcriptional activation when fused to the rGnRH promoter (Fig. 8A, bar 2). The 2-bp mutation at bases −1635 and −1632 (changing the repeat to CcATgA), decreases transcriptional activity significantly to 70% (bar 3). When either of these mutations is placed on the RSV promoter, each decreases activity to approximately half (Fig. 8B). This effect is similar to that seen with the linker replacement mutations in the two promoter contexts (Fig. 5), where more dramatic effects can be seen in the presence of a heterologous promoter.
Fig. 8. Point Mutations in the −1636/−1631 CAATTA Repeat Decrease Transcriptional Activity.
A, The results of transient transfections utilizing enhancers with a single point (−1633c) or a double point (−1635c/−1632 g) mutation in the −1636/−1631 CAATTA repeat are presented. A diagram of the plasmid is shown above the bar graphs. B, The activities of enhancers containing these same mutations on the RSV promoter are shown. Transfections are as in Fig. 1B. Stars indicate a statistically significant decrease from wild-type values, as determined by a one-way ANOVA (P < 0.05).
The Protein Complex Binding to the CAATT Repeats Has Restricted Expression
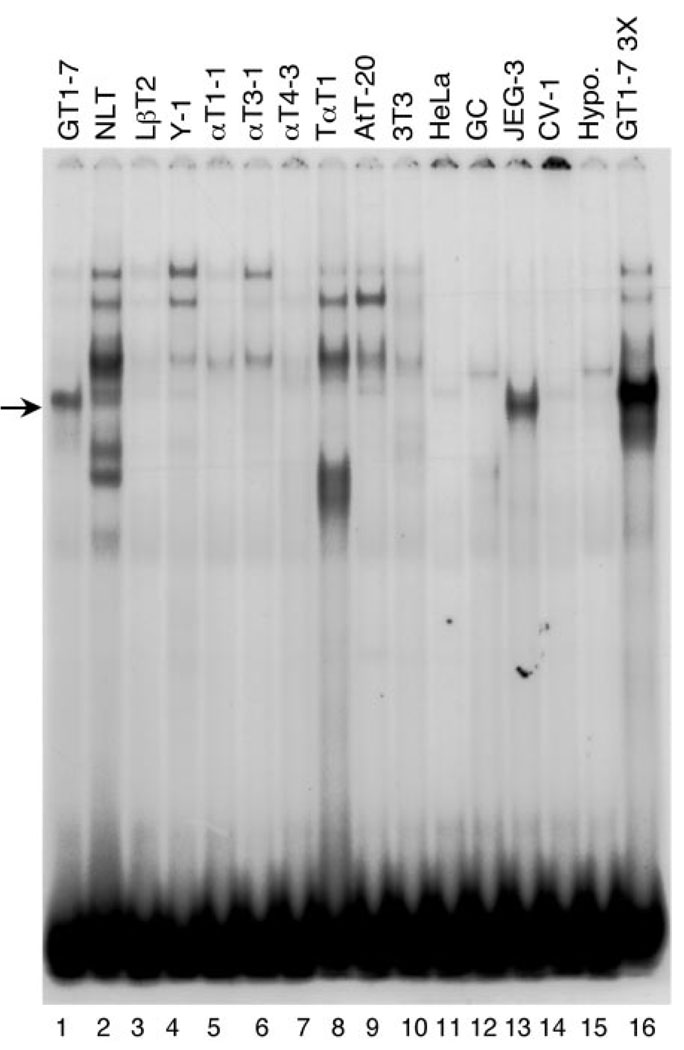
To investigate the expression pattern of the protein(s) in this complex, an extensive panel of nuclear extracts from various cell types was used in EMSA with the −1642/−1623 probe (Fig. 9). The complex formed with GT1–7 nuclear extract on the −1642/−1623 oligonucleotide (Fig. 9, lane 1) is unique. No other cell type shows a band that migrates similarly. A complex of slightly lower mobility is formed with nuclear extract from NLT cells (lane 2), a mouse cell line immortalized with the human GnRH 5′ regulatory region (11), whereas a complex of slightly higher mobility forms with nuclear extract from JEG-3 cells (a human placental cell line). A band with lower mobility exists in the lanes with nuclear extract from LβT2 (mouse pituitary gonadotrope, produced with targeted oncogenesis, lane 3), Y1 (mouse adrenal tumor, lane 4), AtT-20 (mouse pituitary corticotrope, lane 9), HeLa (human epithelial cervical carcinoma, lane 11), and CV-1 (monkey fibroblast, lane 14) cell lines. Because only 800 GnRH neurons are found in the hypothalamus, it is not unexpected that it fails to show a complex. Therefore, it appears that the protein complex formed with the −1642/−1623 probe is restricted in its expression.
Fig. 9. The CAATTA-Binding Complex Is GT1–7 Cell Restricted.
Nuclear extracts from a variety of cell lines, as noted above the gel, were incubated with radiolabeled −1642/−1623 oligonucleotide probe in EMSA. An arrow points to the GT1–7 cell-restricted complex (lanes 1 and 16). NLT cells (lane 2) are a mouse cell line immortalized with the human GnRH 5′ regulatory region (11). LβT2, αT1–1, αT2–1, αT4–1, and TαT1 are all mouse pituitary cells, produced with targeted oncogenesis in transgenic mice (lanes 3–8). Y1 is a cell line from a mouse adrenal tumor (lane 4). AtT-20 cells are mouse pituitary corticotropes (lane 9). NIH3T3 cells are mouse fibroblasts (lane 10). HeLa cells are a human epithelial cervical carcinoma (lane 11). GC cells are rat pituitary cells (lane 12). JEG-3 cells (lane 13) are a human placental cell line. CV-1 cells are monkey fibroblasts (lane 14). Lane 15 has nuclear extracts from mouse hypothalamic tissue.
CAATT Repeats Participate in Restricting GnRH Expression
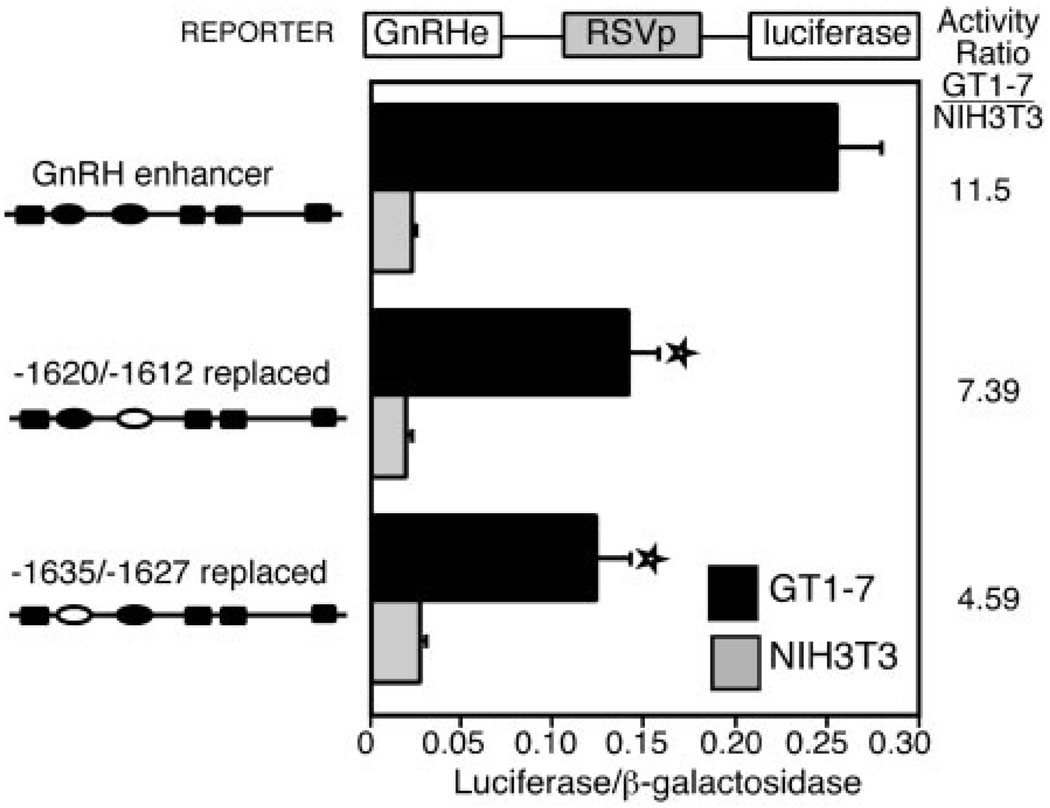
Because the protein complex binding to the repeats has limited expression, we investigated whether the repeat elements play a role in specification of GnRH gene expression. Cell-restricted gene expression can be effectively examined using chimeric plasmids in parallel transient transfections of two distinct cell types, one with highly specific transcription of GnRH, such as GT1–7 cells, and one that has no endogenous expression of GnRH, such as NIH3T3 cells. A chimeric plasmid, in this case with the GnRH enhancer on the RSV promoter, can isolate the effects of the tissue-specific enhancer on restricted gene expression. To control for differences in metabolic factors between distinct cell types, a heterologous RSV enhancer and promoter are linked to the luc gene (RSVe/RSVp-luc), and the same heterologous enhancer and promoter are linked to the β-galactosidase (β-gal) gene (RSVe/RSVp-gal) to serve as the internal control. The RSVe/RSVp-luc plasmid is transfected into cells in parallel with the expression vectors of interest (and all are cotransfected with the internal control plasmid, RSVe/RSVp-gal). The ratio of the RSVe/RSVp-luc value to the RSVe/RSVp-gal value is set to one within each cell type to allow comparison between the cell types.
These transfections reveal that the enhancer confers a high degree of specificity for GT1–7 cells. Furthermore, enhancers carrying either the −1635/−1627 or the −1620/−1612 linker replacement mutations provide relatively lower expression in GT1–7 cells as compared with that in NIH3T3 cells, normalized to wild-type enhancer activity levels (Fig. 10). There is a statistically significant decrease in activity levels from wild type for each of the two linker replacement mutations in GT1–7 cells (shown with a star) but no significant change in activity in the NIH3T3 cells. The activity ratio decreases from 11.5 times higher in the GT1–7 cells compared with NIH3T3 cells for the wild-type rGnRH enhancer to 7.4 times for the −1620/−1612 linker replacement mutation and 4.6 times for the −1635/−1627 linker replacement mutation. These data indicate that these two repeats play a role in the tissue-restricted expression of the GnRH gene.
Fig. 10. Both Enhancer CAATTA Repeats Contribute to Tissue-Specific Expression.
A, GT1–7 cells were transfected with plasmids containing the rGnRH enhancer on the RSV promoter in parallel with the RSVe/RSVp-luc plasmid (and all are cotransfected with the internal control plasmid, RSVe/RSVp-gal). Values are expressed relative to RSVe/RSVp-luc (normalized to the internal control RSVe/RSVp-gal) set to one to allow direct comparisons between the two cell types. Linker replacement mutations replace either of the two CAATTA repeats as shown in the diagrams at the left. Transfections are as in Fig. 1B. Stars indicate a statistically significant decrease from wild-type rGnRHe/RSVp values in GT1–7 cells, as determined by a one-way ANOVA (P < 0.002).
Characterization of the Cell-Restricted Protein Complex
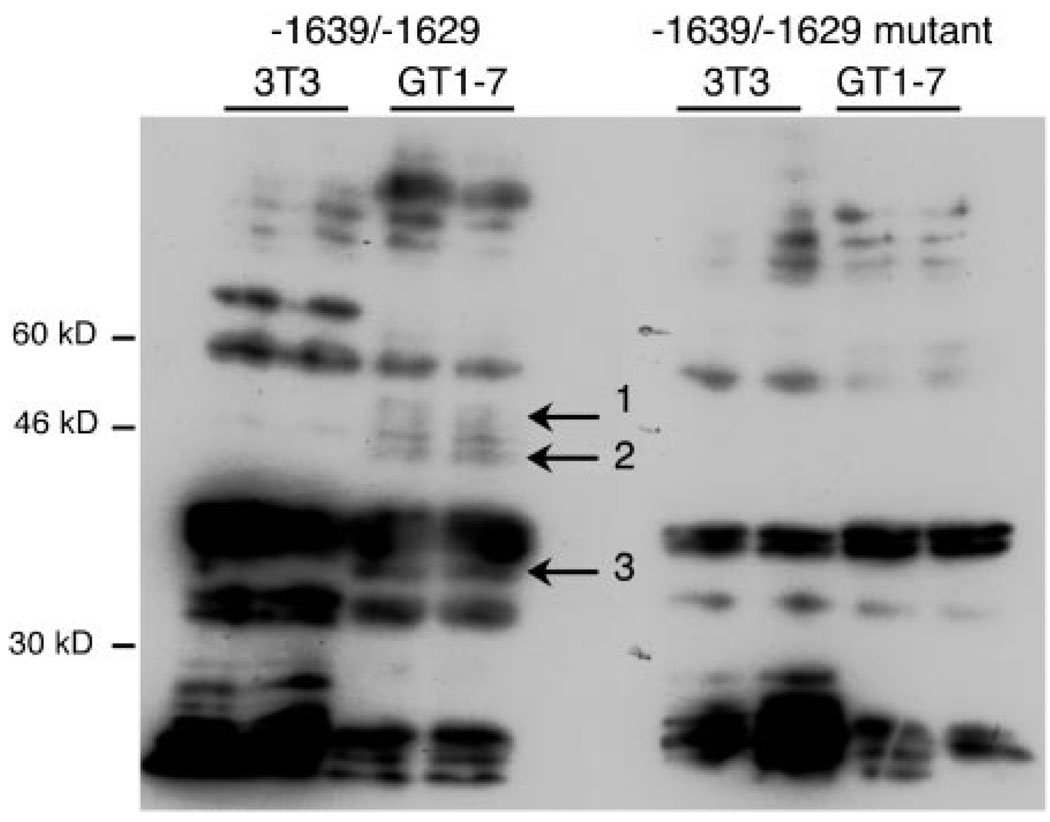
To determine additional characteristics of the cell-restricted protein(s), we used Southwestern analysis to compare binding in the GT1–7 nuclear extract to the NIH3T3 nuclear extract. Because Fig. 9 shows no complex forming in NIH3T3 cells, nuclear extracts from these cells serve as control. A shorter sequence (−1639/−1629) has been shown in EMSA to bind the same tissue-restricted complex as the −1642/−1623 probe and results in a much lower background in Southwestern analysis (data not shown). There are bands present in the GT1–7 nuclear extract lanes that are labeled specifically by a dimerized probe of −1639/−1629 (Fig. 11, indicated by arrows) but not with a dimerized mutant probe (with the CAATTA changed to CAgTTg). One set of bands is approximately 46 kDa, on either side of a band in common with NIH3T3 nuclear extract (proteins 1 and 2). The third band specific to GT1–7 extract appears at about 36 kDa beneath a common doublet (protein 3). Thus, there may be multiple proteins in the cell-restricted protein complex.
Fig. 11. GT1–7 Proteins Bind Specifically to the −1636/−1631 CAATTA Repeat.
Southwestern analysis with NIH3T3 and GT1–7 nuclear extracts, as indicated above the lanes. Duplicate lanes are loaded next to one another. The left half of the figure shows the membrane hybridized with the wild-type −1639/−1629 probe, whereas the right half shows the hybridization with the mutant −1639/−1629 probe (mutations are CAgTTg). The probes are composed of a 2-fold tandem repeat of the sequence named. Size markers are shown on the left of the figure. The arrows in the middle indicate three proteins that are present in the GT1–7 lanes probed with the wild-type probe but not with NIH3T3 nuclear extract, nor with the mutant probe.
The CAATT Repeats Bind a Q50 Homeodomain Protein-Related Complex
The core CAATTA sequence from the two enhancer repeats tested above perfectly matches the DNA-binding consensus sequence for homeodomain proteins with glutamine at amino acid 50 (Q50 homeodomain proteins). The most detailed binding analysis has been performed on the Drosophila proteins fushi-tarazu and engrailed. Engrailed and fushi-tarazu share the core ATTA consensus binding site with other homeodomain proteins but also have preferred binding for CAATTA sites (12, 13). In fact, the consensus site for engrailed binding is TCAATTAAAT, and the −1637/−1628 sequence is ACAATTATAA (7/10 match).
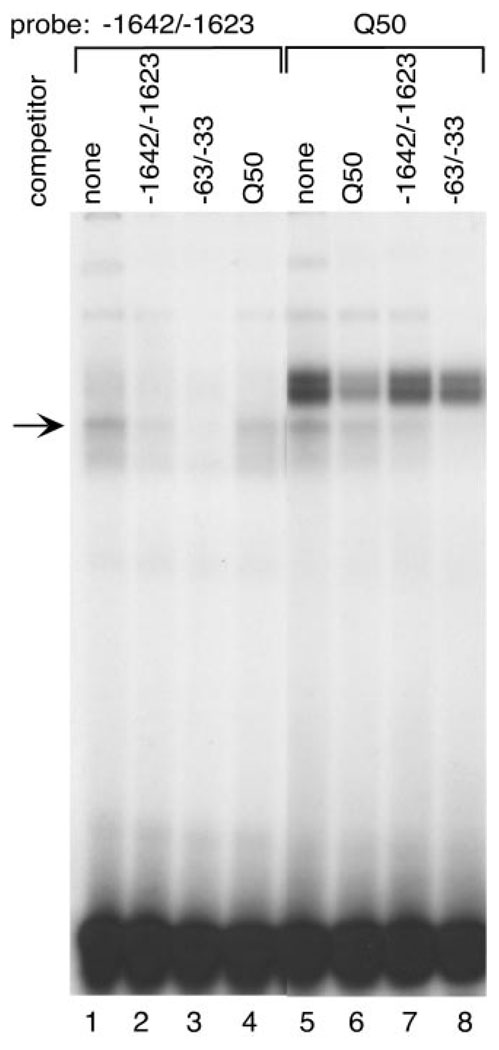
A radiolabeled oligonucleotide with the Q50 consensus site (Q50) incubated with GT1–7 nuclear extract forms a complex that comigrates with the cell-restricted band of the 3′ transcriptional element (−1642/−1623) in EMSAs (Fig. 12, lanes 1 and 5). Cross-competition between the CAATTA repeat probe (−1642/−1623) and the Q50 consensus probe confirms the similarity between these complexes (Fig. 12, lanes 4 and 7). The specificity of the complex is shown by self-competition of each probe (Fig. 12, lanes 2 and 6). The upper doublet forming on the Q50 consensus probe contains the homeodomain proteins Pbx1 and Prep1, mentioned earlier as binding to one of the oligonucleotide probes containing a CAATTT element (Fig. 3C).
Fig. 12. The Same Cell-Restricted Complex Forms on an Q50 Homeodomain Consensus Site and an rGnRH CAATTA Repeat.
EMSA with radiolabeled Q50 homeodomain consensus site (Q50, lanes 5–8) and −1642/−1623 probes (lanes 1–4) with GT1–7 nuclear extract. Molar excess (100-fold) of unlabeled oligonucleotide was included for competition as follows: −1642/−1623, lanes 2 and 7; GnRH promoter CAATTA repeat region −63/−33, lanes 3 and 8; Q50 homeodomain consensus binding site, lanes 4 and 6. An arrow indicates the cell-restricted complex.
Close inspection of the GnRH promoter region reveals a similar CAATTA element located at −55/−50 (6), which is conserved in evolution as well. We have previously shown that this element also binds a homeodomain protein complex related to the Q50 family. To determine whether this element cross-competes with the enhancer repeat elements we performed EMSA with the promoter element as a competitor. The GnRH promoter CAATTA repeat competes both the enhancer CAATTA and the Q50 consensus probes equally well (−63/−33, Fig. 12, lanes 3 and 8). Furthermore, it comigrates with this complex (data not shown). In addition, this Q50 consensus fully competed the same complex from the human CAATTA repeat (−2539/−2520; data not shown).
DISCUSSION
The specification of GnRH gene expression to the select population of 800 hypothalamic neurons in the mouse involves a combination of transcription factors, some known and some remaining to be elucidated. The transcription factors already known to contribute to transcriptional activity, none of which are restricted to GnRH neurons alone, may together create a unique combination that specifies GnRH gene expression to the appropriate neurons. This scheme of combinatorial control by a number of transcription factors binding to a regulatory region facilitates integration of diverse signals, yielding appropriate gene expression (14, 15). On the other hand, one or more transcription factors with restricted expression patterns may limit GnRH gene expression to the appropriate neurons.
The regulation of GnRH gene transcription involves several transcription factors with broad expression patterns, such as Oct-1 (8, 16), CCAAT/enhancer binding protein-β (17), and Pbx/Prep1 (Rave-Harel, N., and P. L. Mellon, manuscript in preparation), and a few factors with relatively restricted expression patterns, such as Otx2 and GATA-4 (5, 9). The brain-specific homeodomain protein, Otx2, acts through footprint 6 of the proximal GnRH promoter to provide GT1–7-cell specific expression (5). Knockout mice homozygous for a null allele of Otx2 are embryonic lethal and have severe phenotypes, including complete absence of forebrain, midbrain, and anterior hindbrain (18–20). The gene dosage of Otx2 is exquisitely sensitive because even heterozygous Otx2 knockout mice can lack olfactory epithelium, olfactory bulb, and the vomeronasal organ (20). The GnRH neurons are born in the olfactory placode and migrate from the presumptive vomeronasal organ through the olfactory bulb to the hypothalamus (21). The Otx2 homeoprotein was shown to colocalize with GnRH in embryonic mouse brain (22) and in the GnRH neurons of the adult mouse hypothalamus (5). Otx2 expression in adult is very rare, indicating that Otx2 may be important in the development of the GnRH neuron and/or in the maintenance of GnRH expression in the adult mouse hypothalamus. The presence of GATA-4 in GT1–7 cells is novel for a neuronal cell type. GATA-4 is present as early as e6 and is necessary for yolk sack development in the mouse (23). Later in development, GATA-4 is in tissues of the developing heart, gut, gonad (24), and pituitary gonadotrope (25). Limited expression of GATA-4 has been reported in the nasopharyngeal arch at embryonic d 9.5 (26), which is along the developmental migratory pathway of the GnRH neurons. GnRH neurons in vivo immunostain for both GATA-4 and GnRH as early as embryonic d 12 (10), but by adulthood the expression is gone. Though these factors are restricted in their spatial and temporal expression, it is unlikely that they are sufficient to target GnRH gene expression to only the 800 hypothalamic neurons found to express GnRH in the mouse brain.
Our studies of the repeated CAATT elements in the 3′ end of the enhancer described herein, provide evidence for another tissue-restricted nuclear protein necessary for GT1 cell-specific expression of the GnRH gene. Though it was known that the entire GnRH 300-bp enhancer is necessary for full activity from the minimal promoter (3), the 80 bp composing the 3′ region of the enhancer had yet to be characterized. Internal deletions involving three of the CAATT repeats were used to examine the collective contribution of these repeats to transcriptional activity. These internal deletions in the context of the GnRH enhancer and RSV promoter show that without these repeated elements, transcriptional activity is reduced by approximately half. Remarkably, deletion of individual repeats reproduces this reduction in activity. Furthermore, single- or double-point mutations in a single repeat can reproduce this reduction. Thus, the integrity of the region containing the repeated CAATT elements is essential for transcriptional activity of the GnRH enhancer. In further support of the relevance of this repeated structure, the series of CAATT elements is conserved in sequence and relative spacing between rat, mouse, and human.
A single major nuclear protein complex binds to the repeat element. Each of the elements, including two from the conserved repeat region from the human gene, bind to the same complex, which is found only in GT1–7 cells from among 14 cell lines. Mutations that reduce expression also prevent this protein complex from binding. Southwestern analysis detected three potential proteins that are found in GT1–7 cells but not NIH3T3 cells that bind the intact but not the mutated CAATTA element. The protein(s) in the complex contact primarily the nucleotides comprising the CAATTA sequence itself. This CAATTA sequence matches the consensus for Q50 homeodomain proteins perfectly and a Q50 consensus element competes for the binding of the GT1–7 cell-specific complex. Elements with single base changes in the second position failed to bind, strongly indicating that this complex contains a Q50 homeodomain protein.
Within the promoter, we have previously described a tissue-restricted factor acting through a CAATTA sequence in footprint 2 (−26/−76) (6). This promoter element binds a complex that comigrates with the complex formed on the CAATT repeats from the enhancer and competes with the Q50-homeodomain protein binding site to a similar extent as the CAATT repeats, indicating that they bind the same (or at least a highly similar) protein. Transfections with a variety of mutations in the enhancer repeats support the hypothesis that the enhancer and promoter CAATT repeats interact and compensate for each other. For example, each of the linker replacement mutations showed the same decrease in transcriptional activity when placed adjacent to the RSV promoter, but different effects when tested on the GnRH promoter, indicating that interactions of the CAATT repeats from the enhancer with the GnRH promoter element may play a role in transcriptional activity.
The rGnRH regulatory regions have other elements repeated in both the enhancer and promoter, including multiple Oct-1 (16) and Pbx/Prep1 binding sites (Rave-Harel, N., and P. L. Mellon, manuscript in preparation). In fact, a Pbx/Prep1 binding site is found near the more 3′ CAATT repeats in the enhancer and near the CAATTA repeat in the GnRH promoter. Although there are several binding sites for each of these transcription factors, not all of the DNA regions are equally important for transcriptional activation in GT1–7 cells. For example, there are two Oct-1 binding sites in the enhancer but mutation of only one of them has an effect on transcription (8). The repeated nature of these important factor binding sites may help ensure that activation from a particular protein will occur and could indicate cooperativity between the enhancer and promoter (because sites exist in both regions). Both the enhancer and the promoter are necessary for proper targeting of a reporter gene to GnRH neurons in transgenic mice (7), supporting the hypothesis that elements from the enhancer and promoter can work together to achieve proper gene expression. In fact, replacing the GnRH promoter with the RSV promoter in reporter transgenes, prevents the GnRH enhancer from targeting to the hypothalamus in vivo in transgenic mice (7).
The identity of the transcription factor(s) that bind the CAATT repeats remains unknown. The clear similarity between the Q50 homeodomain protein consensus site and the GnRH gene repeats led us to consider whether proteins closely related to engrailed or fushi-tarazu might be the protein of interest. There is no known mammalian homolog of fushi-tarazu, but engrailed has been shown to play a role in mammalian transcription from both a Purkinje cell-specific promoter and an intronic area of fibroblast growth factor-8 (27, 28). Engrailed 2 is 41 kDa and could possibly be one of the two bands found in the Southwestern analysis near 46 kDa (29). We investigated the involvement of engrailed with all of the available reagents. Engrailed 2 is expressed in GT1–7, as detected by both Northern and Western analyses (data not shown), but in vitro transcribed/translated engrailed 2 protein fails to bind to the −1642/−1623 oligonucleotide, though it did bind to the consensus probe. From the current studies, it seems unlikely that engrailed is the protein of interest. A number of other Q50 homeodomain candidates for the tissue-restricted transcription factor were investigated. For example, Nkx2.2, was found by Western analysis to be expressed in a positive control of embryonic d 9.5 embryo protein but not in forebrain or in GT1–7 cells, eliminating it as a candidate. Therefore, the identity of the protein complex binding these CAATT repeats within the GnRH gene regulatory region is as yet unknown but likely belongs to the Q50 homeodomain class of proteins.
In summary, we have demonstrated that a repeated structure in the 3′ end of the rGnRH enhancer is crucial for neuron-specific expression of the GnRH gene. This structure contains six copies of a Q50 homeodomain consensus-binding site that is found within the proximal promoter as well and is conserved in mammalian evolution. Disruption of even one of the binding sites is sufficient to reduce expression by half. These elements all bind to a single protein complex found only in GT1–7 cells among 14 cell lines tested. Thus, we find that this complex contains a Q50 homeodomain protein with a pattern of expression that is highly restricted and binding of which is crucial for GnRH neuron-specific gene expression.
MATERIALS AND METHODS
Plasmid Cloning and Mutagenesis
The reporter plasmid rGnRHe/rGnRHp-luc contains the rGnRH enhancer (3) (positions −1571 to −1863 relative to the transcription start site) fused in reverse orientation to the GnRH minimal promoter (positions −173 to −112 relative to the transcription start site) upstream of the luc reporter gene. The wild-type enhancer and mutant enhancers are always in reverse orientation to avoid any potential independent transcriptional start sites within the enhancer. The −173 (SmaI) to −112 (BglII) rGnRH promoter (3) was cloned into pGL3 basic (Promega Corp., Madison, WI). The enhancer fragment (−1571 to −1863) from ENH-173 (30) was obtained by cutting with SmaI and SalI and filling it in with Klenow. It was then inserted in the reverse orientation upstream of the rGnRHp-luc plasmid, cut with the same enzyme.
The RSVe/RSVp-luc plasmid includes the RSV enhancer fused to the RSV promoter in the pGL3 basic plasmid (Promega Corp.). The coding region for luc was replaced with the coding region for β-gal to create the RSVe/RSVp-gal plasmid.
The procedure generating the plasmids with block replacement of sequence was described previously (3). These plasmids were digested with BglII and SalI and inserted into rGnRHe/rGnRHp-luc, which had been digested with the same enzymes, replacing the wild-type enhancer sequence with linker scanner mutation sequence. To create internal deletions, we digested two plasmids, each with a different linker scanner mutation, with BamHI and ligated the two halves together. Thus, we achieved deletion of the intervening sequence (diagrammed in Fig. 2A).
Single and double point mutagenesis were carried out with the Transformer Site-Directed Mutagenesis Kit according to the manufacturer’s directions (CLONTECH Laboratories, Inc., Palo Alto, CA). The oligonucleotides used for mutagenesis included 13–15 bases of wild-type sequence on the 5′ end of the mutation and 12–18 bases of wild-type sequence on the 3′ end of the mutation. The rGnRH enhancer sequence of each of these mutated plasmids was sequenced for the surrounding 200 bp to ensure that no other mutations were introduced.
Cell Culture and Transfections
GT1–7 cells (2) and NIH3T3 cells were used in transient transfections. Cells were maintained in DMEM with 10% fetal bovine serum, penicillin (100 U/ml), streptomycin (0.1 mg/ml), and 4.5% of glucose in an atmosphere with 5% CO2. Transient transfections were performed with FuGENE 6 reagent (Roche Molecular Biochemicals, Indianapolis, IN) using 2 µg reporter plasmid and 1 µg internal plasmid (either thymidine kinase β-gal or RSV β-gal) in 6-cm Petri dishes. Cells were incubated for 24 h with the DNA-Fugene mix before harvesting with a rubber policeman into 0.15 m NaCl, 1 mm EDTA, and 40 mm Tris (pH 7.4). Cell pellets were obtained by centrifugation. Resuspension in lysis buffer (100 mm potassium phosphate, pH 7.8; 0.2% Triton X-100) yielded cellular proteins. β-gal assays were performed as directed by the manufacturer (Tropix, Bedford, MA). Luciferase assays were performed as previously described (8). Luciferase activity was divided by the internal control β-gal values to control for transfection efficiencies between plates. Values were normalized to the activity of the wild-type GnRH regulatory region.
Nuclear Extracts and EMSA
Nuclear extracts for EMSA were prepared according to Clark and Mellon (8). Annealed oligonucleotides (1 pmol, Operon Technologies, Alameda, CA) were phosphorylated with [γ-32P]ATP (6000 Ci/mmol; NEN Life Science Products, Boston, MA) and T4 polynucleotide kinase. Then, probes were purified over a MicroSpin G-50 column (Amersham Pharmacia Biotech, Piscataway, NJ). The 10-µl reaction mixes contained 1 µg of protein and 1 fmol probe in 100 mm KCl, 37 mm HEPES (pH 7.9), 2.5 mm EDTA, 1.25 mm dithiothreitol, 12.5% glycerol, and 4 mm phenylmethylsulfonylfluoride. In the appropriate reactions, 100 fmol (100× labeled probe) of competitor was added 10 min before the addition of probe. The probe was added and incubated for 5 min before loading onto a 5% polyacrylamide nondenaturing gel that had been pre-run for 30 min. After 2 h at 200 V, gels were dried under vacuum and then exposed to film at room temperature for 1–3 d.
The Q50 site is 5′-AAATGTCAATTAAATATCAA-3′ (12). The sequences for all GnRH oligonucleotides are shown in the figures.
Ethylation Interference, Missing Contact Probing, and Southwestern Analyses
Ethylation interference and missing contact probing were performed according to the methods previously described (30, 31).
The method used for Southwestern analysis was described by Horn et al. (32) with the following modifications: Crude GT1–7 nuclear extract was separated on a denaturing SDS-PAGE and transferred to a nitrocellulose filter on a semidry apparatus. The nitrocellulose filter was subjected to denaturation/renaturation before the incubation with the probe according to the following protocol. The filter was washed for 5 min with 6 m guanidinium-hydrochloride in binding buffer [20 mm HEPES (pH 7.9), 50 mm KCl, 1 mm dithiothreitol, 10% glycerol, 0.1% Nonidet P-40]. Half of the solution was removed and replaced with plain binding buffer. This dilution of the guanidinium-hydrochloride was repeated six times. The membrane was soaked in binding buffer alone for 5 min and then incubated with binding buffer/5% (wt/vol) dry nonfat milk for 10 min to block. The filter was washed three times in binding buffer, 5 min each. The incubation overnight took place at 4 C in a seal-a-meal bag with a 5–8 ml binding buffer, 1 µg/ml deoxyinosine:deoxycytidine, and radiolabeled oligonucleotide (prepared as above in EMSA) (106 cpm/ml), all filtered through a syringe and acrodisc. The rest of the procedure continued as previously published (32).
Acknowledgments
The authors thank Melody Clark and Mark Lawson for discussions.
This work was supported by NIH Grant DK-44838 (to P.L.M.). C.G.K. was a predoctoral fellow of the Howard Hughes Medical Institute, N.R.-H. was supported by a Lalor Foundation Postdoctoral Fellowship, M.L.G. was partially supported by the NIH Training Grant T-32-DA-07315, and S.B.N. was partially supported by NIH Training Grant T-32-AG-00216.
Abbreviations
- β-gal
β-Galactosidase
- luc
luciferase
- rGnRH
rat GnRH
- rGnRHe
rGnRH enhancer
- rGnRHp
rGnRH promoter
- RSV
Rous sarcoma virus
- RSVe
RSV enhancer
- RSVp
RSV promoter
REFERENCES
- 1.Wray S, Grant P, Gainer H. Evidence that cells expressing luteinizing hormone-releasing hormone mRNA in the mouse are derived from progenitor cells in the olfactory placode. Proc Natl Acad Sci USA. 1989;86:8132–8136. doi: 10.1073/pnas.86.20.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mellon PL, Windle JJ, Goldsmith P, Pedula C, Roberts J, Weiner RI. Immortalization of hypothalamic GnRH neurons by genetically targeted tumorigenesis. Neuron. 1990;5:1–10. doi: 10.1016/0896-6273(90)90028-e. [DOI] [PubMed] [Google Scholar]
- 3.Whyte DB, Lawson MA, Belsham DD, Eraly SA, Bond CT, Adelman JP, Mellon PL. A neuron-specific enhancer targets expression of the gonadotropin-releasing hormone gene to hypothalamic neurosecretory neurons. Mol Endocrinol. 1995;9:467–477. doi: 10.1210/mend.9.4.7659090. [DOI] [PubMed] [Google Scholar]
- 4.Eraly SA, Mellon PL. Regulation of GnRH transcription by protein kinase C is mediated by evolutionarily conserved, promoter-proximal elements. Mol Endocrinol. 1995;9:848–859. doi: 10.1210/mend.9.7.7476968. [DOI] [PubMed] [Google Scholar]
- 5.Kelley CG, Lavorgna G, Clark ME, Boncinelli E, Mellon PL. The Otx2 homeoprotein regulates expression from the gonadotropin-releasing hormone proximal promoter. Mol Endocrinol. 2000;14:1246–1256. doi: 10.1210/mend.14.8.0509. [DOI] [PubMed] [Google Scholar]
- 6.Nelson SB, Lawson MA, Kelley CG, Mellon PL. Neuron-specific expression of the rat gonadotropin-releasing hormone gene is conferred by interactions of a defined promoter element with the enhancer in GT1–7 cells. Mol Endocrinol. 2000;14:1509–1522. doi: 10.1210/mend.14.9.0521. [DOI] [PubMed] [Google Scholar]
- 7.Lawson MA, MacConell LA, Kim J, Powl BT, Nelson SB, Mellon PL. Neuron-specific expression in vivo by defined transcription regulatory elements of the gonadotropin-releasing hormone gene. Endocrinology. 2002;143:1404–1412. doi: 10.1210/endo.143.4.8751. [DOI] [PubMed] [Google Scholar]
- 8.Clark ME, Mellon PL. The POU homeodomain transcription factor Oct-1 is essential for activity of the gonadotropin-releasing hormone neuron-specific enhancer. Mol Cell Biol. 1995;15:6169–6177. doi: 10.1128/mcb.15.11.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawson MA, Whyte DB, Mellon PL. GATA factors are essential for activity of the neuron-specific enhancer of the gonadotropin-releasing hormone gene. Mol Cell Biol. 1996;16:3596–3605. doi: 10.1128/mcb.16.7.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawson MA, Mellon PL. Expression of GATA-4 in migrating GnRH neurons of the developing mouse. Mol Cell Endocrinol. 1998;140:157–161. doi: 10.1016/s0303-7207(98)00044-6. [DOI] [PubMed] [Google Scholar]
- 11.Radovick S, Wray S, Lee E, Nicols DK, Nakayama Y, Weintraub BD, Westphal H, Cutler GB, Jr, Wondisford FE. Migratory arrest of gonadotropin-releasing hormone neurons in transgenic mice. Proc Natl Acad Sci USA. 1991;88:3402–3406. doi: 10.1073/pnas.88.8.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desplan C, Theis J, O’Farrell PH. The sequence specificity of homeodomain-DNA interaction. Cell. 1988;54:1081–1090. doi: 10.1016/0092-8674(88)90123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laughon A. DNA binding specificity of homeodomains. Biochemistry. 1991;30:11357–11367. doi: 10.1021/bi00112a001. [DOI] [PubMed] [Google Scholar]
- 14.Wolberger C. Combinatorial transcription factors. Curr Opin Genet Dev. 1998;8:552–559. doi: 10.1016/s0959-437x(98)80010-5. [DOI] [PubMed] [Google Scholar]
- 15.Wolberger C. Multiprotein-DNA complexes in transcriptional regulation. Annu Rev Biophys Biomol Struct. 1999;28:29–56. doi: 10.1146/annurev.biophys.28.1.29. [DOI] [PubMed] [Google Scholar]
- 16.Eraly SA, Nelson SB, Huang KM, Mellon PL. Oct-1 binds promoter elements required for transcription of the gonadotropin-releasing hormone gene. Mol Endocrinol. 1998;12:469–481. doi: 10.1210/mend.12.4.0092. [DOI] [PubMed] [Google Scholar]
- 17.Belsham DD, Mellon PL. Transcription factors Oct-1 and C/EBP β (CCAAT/enhancer binding protein-β) are involved in the glutamate/nitric oxide/cyclic guanosine 5′-monophosphate-mediated repression of gonadotropin-releasing hormone gene expression. Mol Endocrinol. 2000;14:212–228. doi: 10.1210/mend.14.2.0418. [DOI] [PubMed] [Google Scholar]
- 18.Acampora D, Mazan S, Avvantaggiato V, Barone P, Tuorto F, Lallemand Y, Brulet P, Simeone A. Epilepsy and brain abnormalities in mice lacking the Otx1 gene. Nat Genet. 1996;14:218–222. doi: 10.1038/ng1096-218. [DOI] [PubMed] [Google Scholar]
- 19.Ang S-L, Jin O, Rhinn M, Daigle N, Stevenson L, Rossant J. A targeted mouse OTX2 mutation leads to severe defects in gastrulation and formation of axial mesoderm and to deletion of rostral brain. Development. 1996;122:243–252. doi: 10.1242/dev.122.1.243. [DOI] [PubMed] [Google Scholar]
- 20.Matsuo I, Kuratani S, Kimura C, Takeda N, Aizawa S. Mouse OTX2 functions in the formation and patterning of rostral head. Genes Dev. 1995;9:2646–2658. doi: 10.1101/gad.9.21.2646. [DOI] [PubMed] [Google Scholar]
- 21.Schwanzel-Fukuda M, Jorgenson KL, Bergen HT, Weesner GD, Pfaff DW. Biology of normal luteinizing hormone-releasing hormone neurons during and after their migration from olfactory placode. Endocr Rev. 1992;13:623–634. doi: 10.1210/edrv-13-4-623. [DOI] [PubMed] [Google Scholar]
- 22.Mallamaci A, DiBlas E, Briata P, Boncinelli E, Corte G. OTX2 homeoprotein in the developing central nervous system and migratory cells of the olfactory area. Mech Dev. 1996;58:165–178. doi: 10.1016/s0925-4773(96)00571-0. [DOI] [PubMed] [Google Scholar]
- 23.Soudais C, Bielinska M, Heikinheimo M, MacArthur CA, Narita N, Saffitz JE, Simon MC, Leiden JM, Wilson DB. Targeted mutagenesis of the transcription factor GATA-4 gene in mouse embryonic stem cells disrupts visceral endoderm differentiation in vitro. Development. 1995;121:3877–3888. doi: 10.1242/dev.121.11.3877. [DOI] [PubMed] [Google Scholar]
- 24.Arceci RJ, King AAJ, Simon MC, Orkin SH, Wilson DB. Mouse GATA-4: a retinoic acid-inducible GATA-binding transcription factor expressed in endodermally derived tissues and heart. Mol Cell Biol. 1993;13:2235–2246. doi: 10.1128/mcb.13.4.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steger DJ, Hecht JH, Mellon PL. GATA-binding proteins regulate the human gonadotropin α-subunit gene in placenta and pituitary. Mol Cell Biol. 1994;14:5592–5602. doi: 10.1128/mcb.14.8.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heikenheimo M, Scandrett JM, Wilson DB. Localization of transcription factor GATA-4 to regions of the mouse embryo involved in cardiac development. Dev Biol. 1994;164:361–373. doi: 10.1006/dbio.1994.1206. [DOI] [PubMed] [Google Scholar]
- 27.Sanlioglu S, Zhang X, Baader SL, Oberdick J. Regulation of a purkinje cell-specific promoter by homeodomain proteins: repression by Engrailed-2 vs. synergistic activation by Hoxa5 and Hoxb7. J Neurobiol. 1998;36:559–571. doi: 10.1002/(sici)1097-4695(19980915)36:4<559::aid-neu9>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 28.Gemel J, Jacobsen C, MacArthur CA. Fibroblast growth factor-8 expression is regulated by intronic engrailed and Pbx1-binding sites. J Biol Chem. 1999;274:6020–6026. doi: 10.1074/jbc.274.9.6020. [DOI] [PubMed] [Google Scholar]
- 29.Hanks M, Wurst W, Anson-Cartwright L, Auerbach AB, Joyner AL. Rescue of the En-1 mutant phenotype by replacement of En-1 with En-2. Science. 1995;269:679–682. doi: 10.1126/science.7624797. [DOI] [PubMed] [Google Scholar]
- 30.Lawson MA, Buhain AR, Jovenal JC, Mellon PL. Multiple factors interacting at the GATA sites of the gonadotropin-releasing hormone neuron-specific enhancer regulate gene expression. Mol Endocrinol. 1998;12:364–377. doi: 10.1210/mend.12.3.0082. [DOI] [PubMed] [Google Scholar]
- 31.Sturm R, Baumruker T, Franza BR, Herr W. A 100-kD HeLa cell octamer binding protein (OBF100) interacts differently with two separate octamer-related sequences within the SV40 enhancer. Genes Dev. 1987;1:1147–1160. doi: 10.1101/gad.1.10.1147. [DOI] [PubMed] [Google Scholar]
- 32.Horn F, Windle JJ, Barnhart KM, Mellon PL. Tissue-specific gene expression in the pituitary: the glycoprotein hormone α-subunit gene is regulated by a gonadotrope-specific protein. Mol Cell Biol. 1992;12:2143–2153. doi: 10.1128/mcb.12.5.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]