Abstract
Culture independent approaches for accessing small molecules produced by uncultured bacteria are often hampered by the inability to easily clone environmental DNA (eDNA) fragments large enough to capture intact biosynthetic gene clusters that can be used in heterologous expression studies. Here we show that homology screening of eDNA mega-libraries for clones containing natural product biosynthetic genes, coupled with transformation-assisted recombination (TAR) in yeast, can be used to access large, functionally intact, natural product gene clusters from the environment. The eDNA derived gene cluster reported here was functionally reconstructed from two overlapping cosmid clones using TAR. The isolation and structure elucidation of three new fluostatins (F, G and H) produced by this TAR reconstructed gene cluster is described.
Environmental samples are known to contain thousands of bacterial species that are refractory to culturing.1 Culture independent approaches for accessing small molecules produced by these bacteria rely on the cloning and heterologous expression of natural product gene clusters from environmental samples. This approach is often hampered by the inability to easily clone environmental DNA (eDNA) fragments large enough to capture intact biosynthetic gene clusters that can be used in heterologous expression studies. Here we show that homology screening of eDNA mega-libraries for clones containing natural product biosynthetic genes, coupled with transformation-assisted recombination (TAR) in yeast,2 can be used to access large, functionally intact, natural product gene clusters from uncultured soil bacteria (Figure 1). The eDNA derived gene cluster reported here was functionally reconstructed from two overlapping cosmid clones using TAR. Isolation and structure elucidation of metabolites obtained through heterologous expression of this gene cluster identified three new fluostatins that had not been characterized from studies of cultured species.
Figure 1.
a. Outline of the procedure used to recover and reconstruct the eDNA derived fluostatin gene cluster (red: minimal PKS, green: candidate ring cleavage oxygenases). b. Overview of a general approach for functionally accessing large natural product gene clusters present in soil. c. HPLC traces of organic extracts derived from individual clones as well as the TAR reassembled fluostatin gene cluster.
DNA extracted directly from soil collected in the Anzo Borrego (AB) desert of southern California was used to construct a multimillion membered eDNA cosmid library. This eDNA mega-library was screened for clones containing Type II PKS biosynthetic machinery,3 and each minimal PKS containing clone recovered from the library was then shuttled into Streptomyces for heterologous expression studies. Extracts from cultures of Streptomyces albus transformed with clone AB649 were found to contain the tetracyclic polyketides rabelomycin (1) and dehydrorabelomycin (6-hydroxytetrangulol) (2).4 Full sequencing of the eDNA insert found in AB649 suggested that the biosynthetic system responsible for the production of these metabolites might extend beyond the sequence captured on this individual clone. Previous studies have shown that libraries containing over 10,000,000 cosmids, like the Anzo Borrego library used in this study, provide sufficient coverage of soil metagenomes to permit the recovery of large natural product gene clusters on overlapping eDNA clones.5 Clones containing DNA fragments that overlap each end of AB649 were recovered from the AB mega-library and sequenced.
AB1850, a clone overlapping one end of AB649 (Figure 1a), appeared to contain additional biosynthetic machinery that we thought might augment the metabolites produced by AB649 alone. Although together AB649 and AB1850 likely constitute a complete Type II PKS biosynthetic system, the fact that the biosynthetic machinery lies on two separate clones makes functionally accessing the small molecule(s) encoded by this system difficult. We recently showed that transformation-assisted recombination in Saccharomyces cerevisiae could be used to reconstruct large continuous stretches of DNA captured on overlapping cosmid clones.5 To reassemble these two clones into a single DNA fragment that could be used in heterologous expression studies, both clones and an AB649/AB1850 specific yeast capture vector were co-transformed into S. cerevisiae and allowed to recombine. AB649/1850 (GenBank number HM193369), the >70 kb bacterial artificial chromosome (BAC) constructed in this experiment, was completely sequenced and found to be a faithful reconstruction of the overlapping eDNA sequences captured on AB649 and AB1850 (Figure 1). This newly assembled large insert clone was conjugated into S. albus and found to confer the production of at least four additional clone specific metabolites (3 – 6) not seen in cultures transformed with AB649 alone.6
Compounds 3 – 6 have closely related 13C and 1H NMR spectra.7 Common 13C chemical shifts indicate that each metabolite contains a conserved substructure with 2 carbonyls, 12 aromatic carbons, 3 oxygen substituted carbons and one methyl. Common 1H NMR signals indicate that this conserved substructure has four aromatic protons, two deshielded oxygen substituted methine protons and a methyl singlet. In the case of 3, these atoms accounts for all but three hydrogens that are predicted by HRMS to be present in the final structure. Based on this data as well as observed COSY and HMBC correlations, 3 was determined to be the tetracyclic polyketide fluostatin C.8
HRMS data indicates that compound 4 differs from 3 by 14 mass units and that compounds 5 and 6 each differ from 3 by 98 mass units.6 These differences correspond to an additional CH2 in 4 and an additional C6H11O in 5 and 6. It is apparent from both the 13C and 1H NMR spectra that the extra 14 mass units seen in 4 correspond to a methoxy substituent (C13 57.8 ppm, H13 3.49 ppm). An HMBC correlation from the deshielded methyl to the C1 methine carbon allowed us to position the methoxy as shown in Figure 3. 1H-1H COSY spectra for compound 5 and 6 show an additional 5-carbon spin system. HMBC correlations from H14, H15 and H18 to the new carbonyl carbon (C13) confirmed the placement of this carbonyl directly adjacent to the 5-carbon spin system. Additional HMBC correlations to the C13 carbonyl from the H1 methine proton indicate that the branched 6-carbon substructure is linked through an ester bond to C1. Compounds 5 and 6 have identical molecular formulae and show the same set of 2D NMR correlations. Minor differences seen in the chemical shifts for atoms surrounding C14 indicate that compounds 5 and 6 are stereoisomers at C14. Compounds 4, 5 and 6 are novel natural products that have been named fluostatins F, G and H, respectively. Fluostatins F, G an H show moderate antibacterial activity (MIC against Bacillus subtillus 37.5, 37.5, 21.2 µg/ml, respectively). As reported previously, fluostatin C shows no activity at the highest concentrations tested.8
Figure 3.
Fluostatins isolated from cultures of S. albus transformed with the TAR assembled BAC AB649/1850.
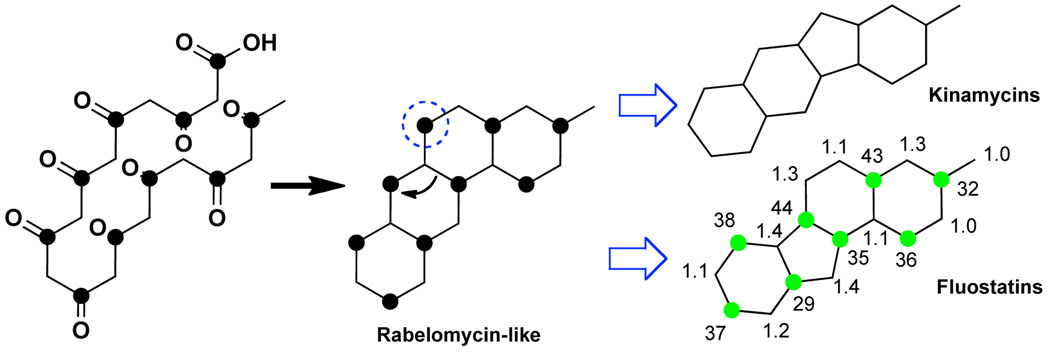
To decipher the origin of the fluostatin skeleton, [1-13C] labeled acetate was fed to cultures transformed with AB649/1850.9 The 13C enrichment pattern seen in fluostatin G isolated from these cultures indicates that like the kinamycins, the fluostatins appear to arise from the excision of C-6 from a rabelomycin-like precursor (Figure 4). In the case of the fluostatins, however, the oxidative excision of C-6 must be accompanied by a rearrangement of the C11a-C6a bond to form the new five-membered ring.
Figure 4.
The fluostatins, like the kinamycins, appear to arise from the excision of C6 from a rabelomycin-like precursor. Ratios of 13C integrations calculated from 13C NMR spectra of fluostatin G isolated from 1-13C acetate fed and unlabeled cultures are shown. Expected positions of 13C enrichment are shown as black circles. Observed sites of 13C enrichment are shown as green circles.
Many families of natural products arise from the oxidation of rabelomycin-like precursors.10 Fluostatin biosynthetic systems appear to couple the C6 oxidation of a rabelomycin-like precursor with a unique rearrangement to generate their distinct carbon skeleton. In addition to the gene cluster described here, we have identified two other eDNA cosmid clones with minimal PKS systems that are closely related to this fluostatin biosynthetic system. The discovery of additional members of this rare family of compounds suggests that culture independent approaches are likely to be a productive method of sampling gene clusters that have not yet been comprehensively explored using culture dependent natural product discovery strategies. This work suggests that TAR will be a generically useful tool for reconstructing large functionally intact biosynthetic gene clusters from the environment, significantly increasing the value of cosmid eDNA mega-libraries to future natural product discovery efforts.
Supplementary Material
Figure 2.
Clone specific metabolites produced by S. albus transformed with cosmid AB649.
ACKNOWLEDGMENT
This work was supported by the NIH GM077516 and The Howard Hughes Medical Institute.
Footnotes
Supporting Information Available: Gene tables, NMR spectra and methods are available via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Rappe MS, Giovannoni SJ. Annu. Rev. Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]; Torsvik V, Goksoyr J, Daae FL. Appl. Environ. Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]; Torsvik V, Ovreas L, Thingstad TF. Science. 2002;296:1064–1066. doi: 10.1126/science.1071698. [DOI] [PubMed] [Google Scholar]
- 2.Larionov V, Kouprina N, Eldarov M, Perkins E, Porter G, Resnick MA. Yeast. 1994;10:93–104. doi: 10.1002/yea.320100109. [DOI] [PubMed] [Google Scholar]; Larionov V, Kouprina N, Graves J, Resnick MA. Proc. Natl. Acad. Sci. U. S. A. 1996;93:13925–13930. doi: 10.1073/pnas.93.24.13925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brady SF. Nat. Protoc. 2007;2:1297–1305. doi: 10.1038/nprot.2007.195. [DOI] [PubMed] [Google Scholar]; King RW, Bauer JD, Brady SF. Angew. Chem., Int. Ed. Engl. 2009;48:6257–6261. doi: 10.1002/anie.200901209. [DOI] [PMC free article] [PubMed] [Google Scholar]; Seow KT, Meurer G, Gerlitz M, Wendt-Pienkowski E, Hutchinson CR, Davies J. J. Bacteriol. 1997;179:7360–7368. doi: 10.1128/jb.179.23.7360-7368.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imamura N, Kakinuma K, Ikekawa N. J. Antibiot. 1982;35:602–608. doi: 10.7164/antibiotics.35.602. [DOI] [PubMed] [Google Scholar]; Liu W-C, Parker WL, Slusarchyk DS, Greenwood GL, Graham SF, Meyers E. J. Antibiot. 1970;23:437–441. doi: 10.7164/antibiotics.23.437. [DOI] [PubMed] [Google Scholar]
- 5.Kim J, Feng Z, Bauer JD, Kallifidas D, Calle PY, Brady SF. Biopolymers. 2010;93:833–844. doi: 10.1002/bip.21450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cultures were grown in ISP-4 containing 5% HP-20 at 30 °C for 2 weeks. A crude extract was eluted (100% MeOH) from resin that had been washed with 50% MeOH. The eluant was partitioned by a silica gel flash chromatography (CHCl3:MeOH step gradient). Individual compounds were purified by reversed phase HPLC (XBridge™ 150 × 10 mm, 5 um) using a linear gradient of 10:90 (H2O:MeOH, 0.1% formic acid) to 100% (MeOH, 0.1% formic acid) over 30 min (7 mL/min). Fluostatins G and H were separated in a second reversed phase HPLC step using a linear gradient of 45:55 (H2O:ACN, 0.1% formic acid) to 50:50 (H2O:ACN, 0.1% formic acid) over 30 min.
- 7.Fluostatin F (4) HRMS-ESI (m/z): [M-H]−: calcd for C19H13O6 337.0712; found 337.0696. 1H NMR (600 MHz, CD3OD) δ 7.31 (H5, s, 1H), 7.14 (H9, dd, 7.1, 8.0 Hz, 1H), 7.06 (H10, d, 7.1 Hz, 1H), 6.89 (H8, d, 8.0 Hz, 1H), 5.82 (H1, d, 2.5 Hz, 1H), 3.89 (H2, d, 2.5 Hz, 1H), 3.49 (H13, s, 3H), 1.55 (H12, s, 3H). 13C (150 MHz, CD3OD) δ 195.8 (C11), 195.7 (C4), 156.4 (C6), 155.3 (C7), 137.8 (C6a), 136.7 (C10a), 133.7 (C11a), 133.5 (C4a), 132.1 (C9), 128.9 (C6b), 127.8 (C11b), 125.8 (C8), 123.3 (C5), 116.1 (C10), 69.8 (C1), 61.3 (C2), 59.3 (C3), 57.7 (C13), 15.2 (C12). Fluostatin G (5) HRMS-ESI (m/z): [M-H]−: calcd for C24H21O7 421.1287; found 421.1257. 1H NMR (600 MHz, CD3CN) δ 7.47 (H5, s, 1H), 7.23 (H9, dd, 7.2, 8.1 Hz, 1H), 7.14 (H10, dd, 7.2, 0.8 Hz, 1H), 7.04 (H8, dd, 8.1, 0.8 Hz, 1H), 6.92 (H1, d, 2.2 Hz, 1H), 3.88 (H2, d, 2.2 Hz, 1H), 2.40 (H14, sext, 7.0 Hz, 1H), 1.54 (H12, s, 3H), 1.32 (H15, m, 2H), 1.26 (H16, m, 2H), 1.07 (H18, d, 7.0 Hz, 3H), 0.78 (H17, t, 7.1 Hz, 3H). 13C NMR (150 MHz, CD3CN) δ 193.7 (C4), 193.3 (C11), 176.5 (C13), 153.9 (C6), 153.1 (C7), 136.8 (C6a), 136.5 (C10a), 133.4 (C14a), 133.3 (C11a), 132.6 (C9), 127.7 (C6b), 126.3 (C11b), 125.2 (C8), 122.1 (C5), 116.9 (C10), 63.9 (C1), 60.7 (C2), 59.4 (C3), 40.0 (C14), 36.5 (C15), 20.8 (C16), 17.2 (C18), 15.0 (C12), 14.1 (C17). Fluostatin H (6) HRMS-ESI (m/z): [M-H]−: calcd for C24H21O7 421.1287; found 421.1248. 1H NMR (600 MHz, CD3CN) δ 7.47 (H5, s, 1H), 7.23 (H9, dd, 7.3, 8.0 Hz, 1H), 7.14 (H10, d, 7.3 Hz, 1H), 7.04 (H8, d, 8.0 Hz, 1H), 6.91 (H1, d, 2.2 Hz, 1H), 3.89 (H2, d, 2.2 Hz, 1H), 2.39 (H14, sext, 7.0 Hz, 1H), 1.54 (H12, s, 3H), 1.36 (H15, m, 2H), 1.28 (H16, m, 2H), 1.06 (H18, d, 7.0 Hz, 3H), 0.82 (H17, t, 7.1 Hz, 3H). 13C NMR (150 MHz, CD3CN) δ 193.7 (C4), 193.4 (C11), 176.7 (C13), 154.0 (C6), 153.2 (C7), 136.9 (C6a), 136.6(C10a), 133.5 (C4a), 133.4 (C11a), 132.6 (C9), 127.7 (C6b), 126.3 (C11b), 125.2 (C8), 122.1 (C5), 117.0 (C10), 64.0 (C1), 60.8 (C2), 59.4 (C3), 39.9 (C14), 36.7 (C15), 20.9 (C16), 17.4 (C18), 15.0 (C12), 14.2 (C17).
- 8.Yamashita N, Harada T, Shin-Ya K, Seto H. J. Antibiot. 1998;51:79–81. doi: 10.7164/antibiotics.51.79. [DOI] [PubMed] [Google Scholar]; Schneider K, Nicholson G, Strobele M, Baur S, Niehaus J, Fiedler HP, Sussmuth RD. J. Antibiot. 2006;59:105–109. doi: 10.1038/ja.2006.15. [DOI] [PubMed] [Google Scholar]; Yu M, Danishefsky SJ. J. Am. Chem. Soc. 2008;130:2783. doi: 10.1021/ja7113757. [DOI] [PubMed] [Google Scholar]
- 9.10 equal portions of [1-13C] acetate were added daily from day 3 through day 12 to attain a final acetate concentration of 10mM.
- 10.Hertweck C. Angew. Chem., Int. Ed. Engl. 2009;48:4688–4716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.