Abstract
Appropriate expression of GnRH receptor (GnRHR) is necessary for the correct regulation of the gonadotropins, LH and FSH, by GnRH. GnRHR is primarily expressed in the gonadotrope cells of the anterior pituitary, and a number of regulatory elements important for both basal and hormonal regulation of the gene have been identified. Using the gonadotrope-derived cell line, αT3-1, that endogenously expresses GnRHR, we have identified an ATTA element located at −298 relative to the transcriptional start site that is essential for basal expression of the GnRHR gene. LHX3, a member of the LIM homeodomain family, binds the −298 ATTA site in vitro as well as to the endogenous GnRHR promoter in vivo. Additionally, LHX3 specifically activates through this −298 ATTA site in transient transfection assays. LHX3 is essential for pituitary development and has been implicated in the regulation of a number of pituitary specific genes; however, this is the first report identifying its role in the regulation of GnRHR.
GnRH is critical is critical for reproductive fitness in mammals and expression of its receptor, GnRH receptor (GnRHR), in the gonadotrope cells of the anterior pituitary is essential for its function. GnRH is secreted from neurons in the hypothalamus into the hypophyseal-portal blood circulation. It interacts with its receptor, GnRHR, on the surface of gonadotropes and stimulates synthesis and secretion of LH and FSH. LH and FSH, in turn, control folliculogenesis, ovulation, spermatogenesis, and steroidogenesis (1). Due to the key role this receptor plays in the reproductive axis, understanding the mechanisms by which its expression is regulated is central to our understanding of the normal regulation of reproductive function.
Several critical cis-regulatory elements for both basal expression and hormonal regulation of GnRHR have been identified in the proximal promoter region of the gene. A steroidogenic factor-1 (SF-1) binding site at −181/−173, relative to the major transcriptional start site (2), an activating protein-1 (AP-1) site at −276/−269 (3, 4), and the GnRHR activating sequence (GRAS) at −329/−318 (3) have been shown to be necessary for full basal expression of the gene. The GRAS element is also required for activin and GnRH induction and binds to AP-1, Smad, and FoxL2 proteins (5–7). GnRH activates expression of GnRHR at an element termed sequence underlying responsiveness to GnRH-1 (SURG-1) at −292/−285 and the AP-1 site at −276/−269 (SURG-2) (4). Recently, Oct-1 and NFY were reported to bind SURG-1 and contribute to both basal and GnRH-stimulated expression of GnRHR (8). Additionally, the transcription factor pituitary homeobox-1 (Pitx-1) activates the mouse GnRHR promoter using sequences between −308 and −264 (9).
LHX3 (LIM3/P-LIM) is expressed throughout the developing and adult anterior pituitary as well as in the developing nervous system and is required for normal pituitary development (10–12). The LIM homeodomain family of transcription factors, to which LHX3 belongs, is characterized by a homeodomain that serves as a DNA-binding domain and two amino-terminal LIM domains that serve as protein-protein interaction domains (13). In the Lhx3 null mouse, the pituitary primordium initially forms but fails to grow and differentiate, and four of the five anterior pituitary cell lineages, the gonadotropes, thyrotropes, somatotropes, and lactotropes, are absent (12). Humans with mutations in LHX3 display combined pituitary hormone deficiency, providing further evidence for the functional conservation of LHX3 between species and the critical role this protein plays in pituitary development (14). LHX3 is known to activate expression of several pituitary genes including α-glycoprotein subunit (αGSU), prolactin (Prl), TSHβ, FSHβ, and the POU homeodomain transcription factor, Pit-1 (11, 15). Additionally, an unidentified LIM family member was implicated in binding to the distal enhancer of the rat GnRHR by competition experiments with the pituitary glycoprotein basal element (PGBE) of the αGSU (16), a sequence known to bind both LHX3 and another LIM family member, LHX2, in vitro (11, 17).
In this study, the proximal GnRHR promoter was examined for sequence elements necessary for basal expression. Transient transfection of a mouse GnRHR-luciferase reporter into αT3-1 cells, a gonadotrope-derived cell line that endogenously expresses GnRHR and that has been used extensively to study GnRHR regulation, identified an ATTA element at −298 in the mouse GnRHR promoter that is necessary for basal expression and that binds the transcription factor LHX3 in vitro and in vivo. This study furthers our understanding of the regulation of GnRHR and demonstrates a functional role for the transcription factor LHX3 in this regulation.
Materials and Methods
Reporter and expression plasmids
The GnRHR reporter plasmid contains 1.2 kb of the 5′-flanking region of mouse GnRHR as previously described (18), and it was cloned into the SmaI/XhoI restriction site of PGL3 (Promega, Madison, WI). The site-specific mutations in the 4 ATTA sites were made using the following oligonucleotides: −360 ATTA, 5′-GGATCGGGATTTTTAAcggcCTTTTCTGTATTTCATTTTG-3′; −298 ATTA, 5′-CAACAGTTTTTAGAAAACCTATTCcggcAGGCTAATTGGATG-3′); −278 ATTA, 5′-GGCTAATTGGATGATcggcTGAGTCACTTTCGACATC-3′; and −253 ATTA, 5′-CTTTCGACATCAGAcggcGACTCCCAAGTGTCC-3′ (lowercase letters indicate mutated sequence). Mutagenesis was performed using the QuikChange Mutagenesis Kit (Stratagene, La Jolla, CA) according to the manufacturer’s protocol. The LHX3 expression plasmid, FLAG-Lhx3-pcDNA3, was kindly provided by Dr. Richard Maurer and has been previously described (19). The multimerized −298 ATTA site (−298 multi) was constructed with the following oligonucleotides: 5′-CTAGTATTCATTAAGGCTTATTCATTAAGGCTTATTCATTAAGGCTTATTCATTAGGCT-3′ (top strand) and 5′-CTAGAGCCTTAATGAATAAGCCTTAATGAATAAGCCTTAATGAATAAGCCTTAATGAATA-3′ (bottom strand), and the mutant multimer (−298 mut multi) was created with: 5′-TATTCcggcAGGCTTATTCcggcAGGCTTATTCcggcAGGCTTATTCcggcAT-3′ (top strand) and 5′-GCTAGAGCCTgccgGAATAAGCCTgccgGAATAAGCCTgccgGAATAAGCCTgccgGAATAAGCT-3′ (bottom strand). Oligonucleotides were annealed and ligated into the NheI restriction site of pGL3 upstream of a −81 herpes thymidine-kinase promoter (20).
Cell culture, transient transfections, and luciferase and β-galactosidase assays
αT3-1 cells were grown as previously described (21). The day before transfection, αT3-1 cells were aliquoted to 12-well plates at a density of 200,000 cells per well. FuGENE 6 transfection reagent (Roche Molecular Biochemicals, Indianapolis, IN) was used according to the manufacturer’s protocol. Each well was transfected with 700 ng luciferase reporter plasmid along with 200 ng plasmid containing Rous sarcoma virus promoter controlling β-galactosidase gene expression as a control for transfection efficiency. The cells were harvested 48 h after transfection. For cotransfection experiments, 700 ng expression plasmid or the empty plasmid control was also transfected. Cells were prepared and assayed for luciferase and β-galactosidase activity as previously described (21) with the following exceptions: 60µl lysis buffer was used to lyse the cells, and 20 µl extract was directly transferred to a 96-well plate to be measured for either luciferase or β-galactosidase activity.
Normalization of data and statistics
All experiments were performed in triplicate and were repeated at least three times. To normalize for transfection efficiency, all luciferase values were divided by β-galactosidase, and the triplicate values were averaged. To control for interexperimental variation, the empty plasmid, PGL3, was transfected with RSV-βgal and any relevant overexpression vectors, and the average PGL3/βgal value was calculated. Average GnRHRluc/βgal values were divided by the corresponding PGL3/βgal value. Values obtained from each independent experiment were then averaged, and statistics were performed using the statistical analysis program JMP. Significance was set at P ≤ 0.05. Data are presented relative to PGL3, and error bars represent sem.
EMSA
Nuclear extracts were prepared from αT3-1, LβT2, and NIH3T3 cells as previously described (21). The following probes were end-labeled using T4 Polynucleotide Kinase (New England Biolabs, Beverly, MA) according to the manufacturer’s protocol: wild-type (WT) ATTA, 5′-GAAAACCTATTCATTAAGGCTAATTGGATG-3′; PGBE, 5′-ATATCAGGTACTTAGCTAATTAAATGTGCT-3′; and −298 mut, 5′-GAAAACCTATTCcggcAGGCTAATTGGATG-3′ and were column purified using Micro Bio-Spin Chromatography Columns (Bio-Rad Laboratories, Hercules, CA). Binding reactions were carried out using 2 µg nuclear protein and 4 fmol [32P]-labeled oligonucleotide in a 20-µl reaction containing 5 mm dithiothreitol, 0.025 µg/µl Poly dIdC, 1 mm phenylmethylsulfonylfluoride, 0.25 mg/ml BSA, and binding buffer [50 mm HEPES (pH 7.8), 250 mm KCl, 5 mm EDTA, 25 mm spermidine, 30% glycerol, and 10% Ficol]. For supershift assays, 1.5 µg LHX3 (U.S. Biological, Swampscott, MA) or normal rabbit IgG (Santa Cruz Biotechnology, Santa Cruz, CA) antibody was used. In competitions, 250-fold excess of unlabeled oligonucleotide was used. Reactions were carried out, and the gel was run as described in (21).
Chromatin immunoprecipitation (ChIP) assay
αT3-1 cells were grown to confluency in 15-cm plates, and proteins were cross-linked to DNA by the addition of 1% formaldehyde directly to the cell medium. The nuclear fraction was obtained, and chromatin was sonicated to an average length of 1 kb in sonication buffer [50 mm HEPES, 140 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, and 0.1% sodium dodecyl sulfate (SDS)]. The lysate was diluted with ChIP dilution buffer [0.01% SDS, 1.1% Triton, 1.2 mm EDTA, 16.7 mm Tris (pH 8), and 167 mm NaCl] to a final volume of 3.5 ml and precleared with 100 µl Protein A/G PLUS-Agarose beads (Santa Cruz Biotechnology). Protein-DNA complexes were incubated overnight with either the same LHX3 antibody used in EMSAs or a nonspecific IgG control and precipitated with Protein A/G beads (Santa Cruz Biotechnology). A fraction of the protein DNA was not precipitated but set aside for later examination of the total chromatin content (termed input). Beads were sequentially washed with the following buffers: low-salt wash buffer [0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris (pH 8), and 150 mm NaCl], high-salt wash buffer (low-salt wash buffer but with 500 mm NaCl), LiCl wash buffer [250 mm LiCl, 1% NP-40, 1% Na-deoxycholate, 1 mm EDTA, and 10 mm Tris (pH 8)], and twice with Tris-EDTA. Protein-DNA complexes were eluted with elution buffer (1% SDS and 0.1 m NaHCO3), and cross-links were reversed with the addition of 200 mm NaCl and incubation at 65 C for 4 h. DNA was phenol-chloroform-extracted and ethanol-precipitated. Precipitated DNA was resuspended in 50 µl nuclease-free water, the input fraction was resuspended in 500 µl, and the sequence of interest was amplified by PCR. Primers used in PCR (5′-GCAAAATGCATTTGAAAAGCAATTGTTTTG-3′ and 5′-ATAAAAAGACGGGCCATCTGC-3′) spanned the −441 to −202 region of the mouse GnRHR gene and generated a single 239 band on a 5% PAGE stained with ethidium bromide. PCR conditions were as follows: 1 min at 95 C, 1 min at 63 C, and 1 min at 72 C repeated 26 times followed by an extension of 10 min at 72 C. To ensure that the PCR was still in the linear phase after 26 cycles, a dilution curve of the input was performed. PCR was performed on input concentrations of 0.5×, 1×, 2×, and 3× to test whether the PCR end product reflected the relative amounts of starting material.
Results
An ATTA site located at −298 is critical for basal expression of GnRHR
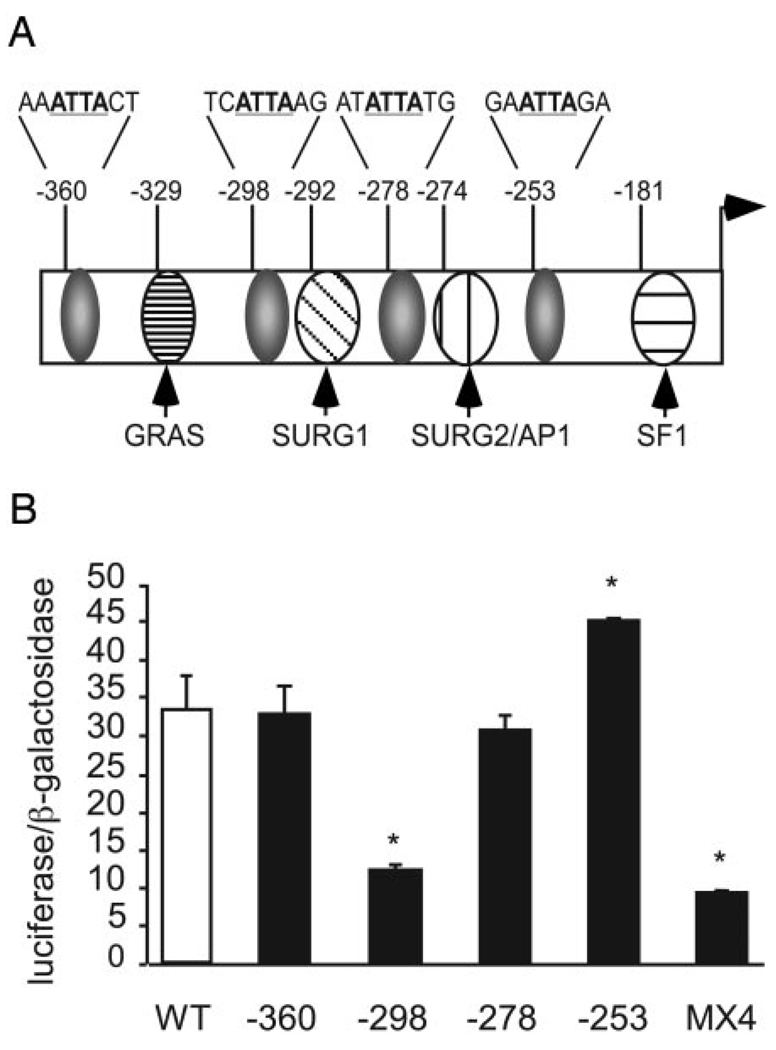
The −365/−232 proximal promoter region of GnRHR is required for full basal expression of GnRHR (9), and within this region, a number of important cis-regulatory elements have been identified (Fig. 1A). This region also contains four uncharacterized ATTA sites located at −360, −298, −278, and −253, relative to the transcriptional start site, and an additional TAAT site at −290 that is part of the SURG-1 element (4, 8). Because the sequence ATTA is known to be the consensus binding site for many homeodomain transcription factors (22), we investigated the functional relevance of these four ATTA sites in the regulation of the mouse GnRHR gene. To determine the importance of these sites for GnRHR gene expression, mutations of the core ATTA sites were created in the context of the 1.2-kb mouse GnRHR-luciferase reporter plasmid. These mutant plasmids were then assayed for luciferase expression in transient transfection experiments in αT3-1 cells. A mutation in the ATTA site at−298 significantly decreased basal expression of GnRHR by approximately 60% relative to the WT GnRHR reporter gene (Fig. 1B). Mutations at the −360 and −278 ATTA sites had no effect, whereas mutation of the −253 ATTA site resulted in an approximate 35% increase in basal expression. When a quadruple ATTA mutant, MX4, containing mutations in all four sites was tested, basal expression of GnRHR was slightly reduced relative to the −298 mutation alone; however, this was not statistically significant. These data indicate that the −298 ATTA site is critical for maintaining the expression of the GnRHR gene.
FIG. 1.
Full basal expression of the GnRHR requires the −298 ATTA site. A, Sites in the GnRHR proximal 5′-flanking region important for transcriptional regulation are shown. Gray ovals represent the four ATTA sites. Numbers above the promoter indicate the location of each regulatory element relative to the transcriptional start site. Nucleotides in bold and underlined indicate bases changed to cggc in subsequent transfection and EMSA experiments. B, Basal expression of luciferase reporter plasmids containing either the WT 1.2-kb region of the mouse GnRHR promoter (white bar, WT) or the indicated mutation (black bars, −360, −298, −278, −253, and the quadruple ATTA mutant, MX4) were assayed by transient transfections in αT3-1 cells. Data are the means ± sem of at least three independent experiments, each performed in triplicate, and were normalized as described in Materials and Methods. A one-way ANOVA followed by the Tukey-Kramer HSD post hoc test was used. *, Values that differ significantly from WT GnRHR (P ≤ 0.05).
Specific nuclear complexes form on the −298 ATTA site
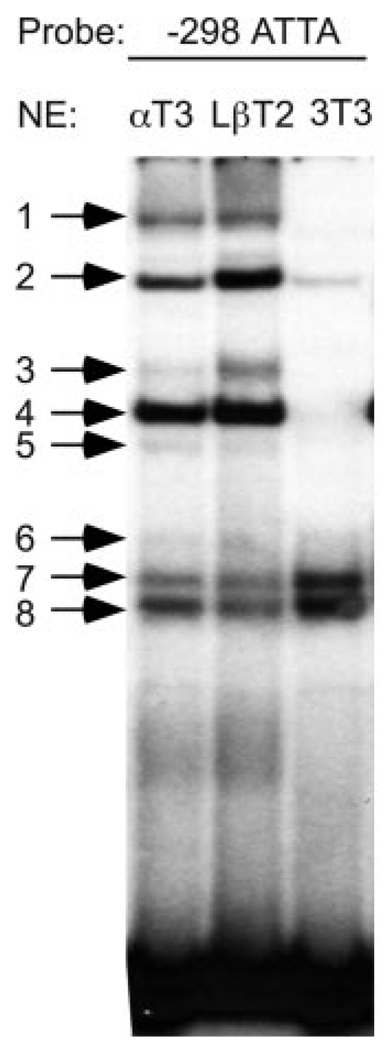
To determine whether the −298 ATTA site is capable of interacting specifically with nuclear protein αT3-1 extracts, EMSAs were performed. An oligonucleotide encompassing the −310/−281 region of the gene was end labeled with 32P and incubated with αT3-1 nuclear extracts. Eight protein complexes were detected (Fig. 2, arrows). To investigate whether these complexes are specific to the αT3-1 cell line, the −298 ATTA probe was also incubated with nuclear proteins from another gonadotrope-derived cell line, LβT2, immortalized at a later developmental time point than theαT3-1 cells (23) or from the fibroblast cell line, NIH3T3 (Fig. 2). As shown in lanes 1 and 2, the same complexes present in the EMSA using αT3-1 extracts were also present using LβT2 nuclear extracts. Although relative levels of protein binding appear to differ between these two cell lines, the mutation of the −298 ATTA site had the same effect in LβT2 cells as in the αT3-1, producing a 60% decrease in basal GnRHR expression (data not shown). Interestingly, complexes bound by the −298 ATTA probe in the NIH3T3 nuclear extracts were significantly different. Complexes 1, 3, 4, and 5 are either not present or are severely diminished in NIH3T3 cells (compare lanes 1 and 3), and complex 2 is significantly reduced. Complex 6 is faintly seen in all three cell types, whereas complexes 7 and 8 are slightly increased in comparison with αT3-1 and LβT2 cells. These results indicate that specific αT3-1 nuclear protein complexes form on the −298 ATTA probe and that these proteins are enriched in the gonadotrope-derived cells.
FIG. 2.
Eight specific nuclear protein complexes bind the GnRHR promoter spanning −298 ATTA. EMSA was performed using the labeled −298 ATTA oligonucleotide probe spanning −310/−281 of the GnRHR promoter and αT3-1 (αT3), LβT2, or NIH3T3 (3T3) nuclear extract as indicated above each lane. Specific protein complexes are indicated by arrows and numbered.
The LIM homeodomain protein LHX3 binds the GnRHR promoter in vitro and in vivo
LHX3 is critical for pituitary development and regulates several pituitary-specific genes including αGSU, Prl, TSHβ, FSHβ, and Pit-1 (11, 15). It is expressed throughout the pituitary and in LβT2 and αT3-1 cells (10, 11), but it is not highly expressed in other cell types. LHX3 is known to bind to the consensus sequence TAATTAATTAT with the second ATTA repeat being the most critical (24). The promoter sequence from −304 to −294 of the mouse GnRHR is ctATTcATTAa. This is a 7- of 11-bp match with the LHX3 consensus-binding sequence, including an exact match to the critical second ATTA repeat (lowercase letters indicate deviation from the consensus sequence). Based on this information and our EMSA data, we chose to test whether LHX3 binds the −298 ATTA site.
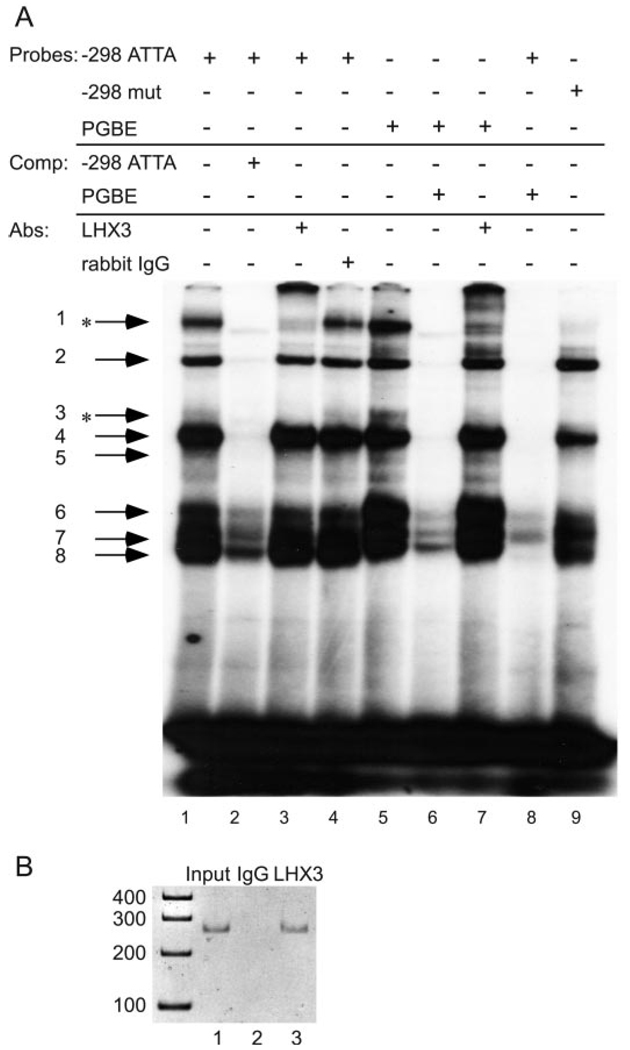
Because LHX3 has previously been shown to bind the PGBE of the mouse αGSU (11), the PGBE was used as a positive control for binding of LHX3. PGBE and −298 ATTA oligonucleotides were radiolabeled and incubated with nuclear extracts from αT3-1 cells, and interacting complexes were compared. Many of the complexes formed on the −298 ATTA probe comigrated with complexes formed on the PGBE probe (compare lanes 1 and 5, Fig. 3A). Addition of excess unlabeled PGBE oligonucleotide (lane 8) efficiently competed for all protein complexes, further suggesting that the same proteins interact with both sites. Inclusion of an antibody specific for LHX3 in the EMSA reaction leads to diminution of complexes 1 and 3 (arrows and asterisk, lane 3). These same complexes were also decreased on the PGBE probe upon inclusion of the LHX3 antibody (lane 7). Inclusion of a nonspecific normal rabbit IgG antibody had no effect (lane 4). Furthermore, when αT3-1 nuclear extracts were incubated with the mutant −298 ATTA probe, complexes 1 and 3 were no longer present (lane 9). These results indicate that the −298 ATTA site is capable of interacting specifically with protein complexes that contain LHX3 and that this element is necessary for LHX3 binding. Because the −298 ATTA probe also contains the −290 TAAT site shown to bind Oct-1 and NF-Y (4, 8), we tested for binding of LHX3 to the −290 TAAT site independent of the −298 ATTA site. Although we could not detect binding of LHX3 to the −290 TAAT under our conditions (data not shown), we cannot eliminate the possibility that the −290 TAAT site could facilitate binding of LHX3 to the −298 probe.
FIG. 3.
LHX3 binds the GnRHR promoter in vitro and in vivo. A, EMSA was performed using labeled −298 ATTA spanning −310/−281 of the GnRHR promoter, PGBE, or mutant −298 ATTA (−298mut) oligonucleotide probe with αT3-1 nuclear extracts. Competitions (comp) were performed using 250-fold excess unlabeled oligonucleotide as indicated in the chart. Antibodies (Abs) directed against LHX3 and normal rabbit IgG were included in the reactions as indicated. Specific protein complexes are indicated by arrows and numbered. Complexes diminished by inclusion of the LHX3 antibody are indicated by asterisks. B, ChIP was performed using cross-linked protein/chromatin from αT3-1 cells and an antibody directed against either LHX3 or, as a negative control, against nonspecific normal rabbit IgG. PCR primers flanking the −298 ATTA site were used to detect precipitation of the genomic DNA. PCR amplification was performed on 0.5% chromatin input (lane 1), and chromatin was precipitated with either IgG (lane 2) or Lhx3 (lane 3). This experiment was repeated at least three times.
To determine whether LHX3 can bind to the regulatory region of the endogenous mouse GnRHR gene in vivo as well as in vitro, we performed ChIP in the αT3-1 cells using an antibody specific for LHX3. PCR primers encompassing the −298 ATTA site were used to detect precipitation of genomic DNA. As expected, these primers amplified a 240-bp region of the mouse GnRHR promoter from input chromatin (Fig. 3B, lane 1). The LHX3 antibody specifically immunoprecipitated the cross-linked protein/DNA region that encompasses the −298 ATTA site (lane 3); however, no immunoprecipitation of this region was detected with a nonspecific rat IgG control antibody (lane 2). To ensure that the PCR conditions used were still semiquantitative, PCR on chromatin input dilutions was performed. At all dilutions, the band intensity of the PCR products correlated with the relative starting amounts, indicating that PCR amplification was in the linear phase (data not shown).
The −298 ATTA site is sufficient for induction by LHX3
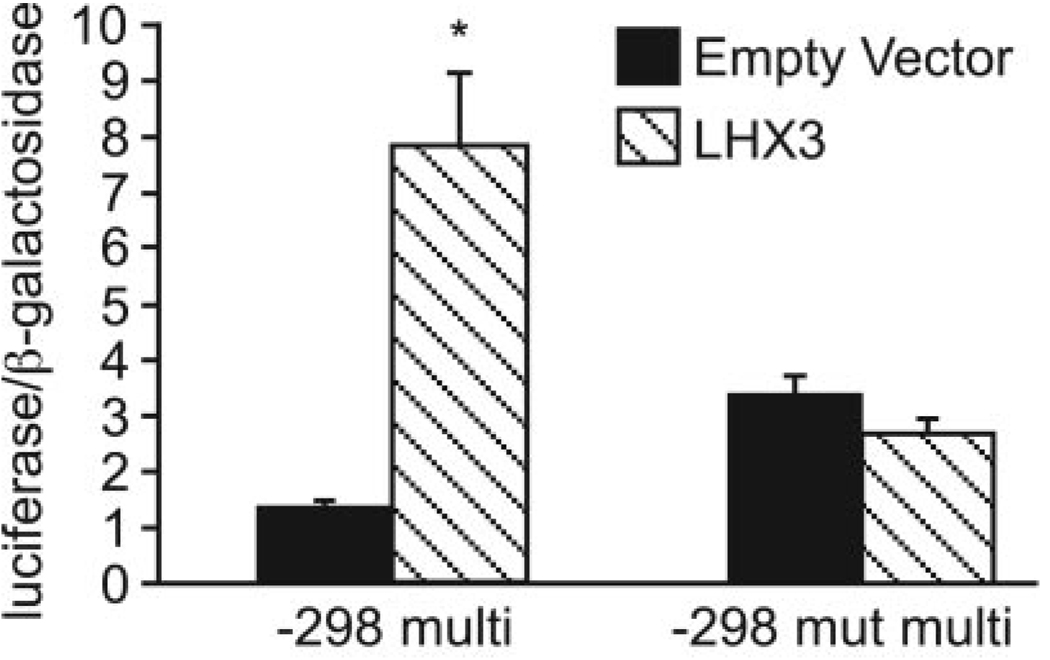
To determine whether the −298 ATTA site is sufficient to confer LHX3 responsiveness to a heterologous promoter, we used a plasmid with four tandem copies of the −298 ATTA site from −303 to −290 controlling luciferase gene expression cloned upstream of a minimal −81-bp thymidine-kinase promoter. Cotransfection of the −298 multimer plasmid and the LHX3 expression plasmid caused a 5- to 6-fold increase in reporter activity in comparison with the multimerized reporter cotransfected with the empty expression plasmid (Fig. 4). In contrast, LHX3 did not activate expression of the multimerized reporter containing the cggc mutation at the −298 site (Fig. 4). Thus, LHX3 can function specifically through the −298 ATTA site to activate reporter gene expression.
FIG. 4.
The −298 ATTA site is sufficient to confer transcriptional activation by LHX3. Transient transfections were performed in αT3-1 cells using a luciferase reporter containing four tandem copies of either the WT (−298 multi) or the mutant −298 ATTA (−298 mut multi) element cloned upstream of a minimal −81 thymidine-kinase promoter. Either an empty overexpression plasmid or one containing the LHX3 cDNA were cotransfected with the multimerized reporter, and reporter gene expression was assayed. Data are the means ± sem of at least three independent experiments, each performed in triplicate, and were normalized as described in Materials and Methods. A one-way ANOVA followed by Student’s t test was performed to analyze statistical significance. *, Significant difference from cells cotransfected with empty vector (P ≤ 0.05).
Discussion
Through GnRHR, GnRH signals to the gonadotropes to synthesize and secrete LH and FSH and thus appropriate expression of this receptor is critical for reproductive function. Four ATTA sites located at −360, −298, −278, and −253 are found within a 5′-flanking region of the mouse gene that has numerous important cis-regulatory elements contributing to the regulation of its expression. Specific protein complexes interact at each of these sites as seen by EMSA. Mutations in two of these sites, those at −298 and −253, have functional consequences on GnRHR gene expression. Mutation of the −298 ATTA site results in a dramatic decrease in GnRHR promoter activity, whereas mutation of the −253 ATTA leads to a slight increase in GnRHR reporter gene expression. Although it is possible that all four ATTA sites play a combinatorial role in regulating expression of GnRHR, either with each other or with other cis-regulatory elements, it is clear that the −298 ATTA is critical for supporting basal expression of the gene.
In EMSA, the −298 ATTA oligonucleotide binds eight distinct complexes in αT3-1 nuclear extracts. Several of these complexes do not form with the NIH3T3 fibroblast extracts, suggesting that expression of these nuclear proteins may be gonadotrope restricted. LHX3 expression is limited to the pituitary and nervous system (10, 11), and it is critical for pituitary development (12). We found that LHX3 binds the GnRHR promoter at the −298 site in vitro as well as to the in vivo promoter. This is in contrast to a recent study by Kam et al. (8). In this study, an EMSA probe that extended from −300 to −277, including both the −298 ATTA and −290 TAAT sites, bound Oct-1 and NF-Y but showed no supershift upon inclusion of an antibody to LHX3 (8). This could be due to the relative position of the −298 ATTA site on the EMSA probe. As mentioned previously, the reported LHX3 consensus site is 11 bp long (24) and would extend from −304 to −294 of the mouse GnRHR promoter and thus would not be entirely contained in the probe used in that study. Because it is likely that bases directly upstream of the −298 ATTA site play a role in stabilizing the binding of LHX3 to the promoter, this finding is not unexpected.
The proximal 5′-regulatory region of the GnRHR is highly active and a number of transcription factors have been identified that are important for its regulation. This multiplicity of factors may be reflected in the complex banding pattern we have observed with the −298 ATTA probe, and it will be interesting to determine what other factors play a role in GnRHR expression at this site. LIM homeodomain proteins are known to interact with a variety of proteins through the LIM domain, and such cofactors could be interacting with LHX3 at the −298 ATTA site (13). Cofactors that have been shown to bind to LHX3 include NL1 (LDB1/CLIM/Chip) (25–27), SLB (19), and RLIM (28). NL1 binds LIM domain-containing proteins, and through it, LHX3 can interact with other LIM family members (29) as well as with non-LIM transcription factors such as the Otx proteins (27). In contrast, SLB selectively binds LHX3 and LHX4 and may specifically modulate their actions (19). RLIM is a transcriptional repressor that functions through recruitment of the Sin3A/histone deacetylase transcriptional repressor complex (28). In addition to these cofactors and their interacting partners, LHX3 has also been reported to interact directly with the LIM-HD protein, Isl1 (29) and Pit-1.
SF-1, AP-1, Smads, FoxL2, NFY, Oct-1, and Pitx-1 are important regulators of basal and hormonal expression of GnRHR (2–9). Not only are all of these transcription factors known regulators of GnRHR, in some cases, there is evidence that they function together to cooperatively regulate GnRHR gene expression. AP-1 and Smads cooperate to mediate GnRH and activin induction of GnRHR at the GRAS element (6). Additionally, Oct-1 and AP-1 binding at the SURG-1 and SURG-2 elements, respectively, appear to interact in supporting basal expression of GnRHR (8). Whether LHX3 functionally interacts with any of these transcription factors remains to be investigated. In other promoter contexts, LHX3 acts synergistically with Pit-1 in regulating transcription of the αGSU, Pit-1, TSHβ, and Prl pituitary genes (11). LHX3 also synergistically interacts with Pitx-1 on the αGSU promoter when expressed with NL1 (27). Additionally, LHX3 and NeuroM have been shown to interact synergistically on a motor neuron enhancer from the Hb9 gene (30).
LHX3 has been found to be involved in the regulation of a number of pituitary genes including Prl, TSHβ, αGSU, FSHβ, Pit-1, and, in this study, GnRHR. Due to the critical nature of LHX3 in pituitary development, it is perhaps not surprising that it plays a role in regulating a number of pituitary-specific genes. Because LHX3 is a highly versatile transcription factor that can potentially interact with a variety of partners, specificity of this regulation is likely due to cell-specific complex formation with interacting transcription factors. Although the αT3-1 cell line represents a gonadotrope cell at an early stage of development, LHX3 expression in the pituitary continues through adulthood (10), and it is likely that LHX3 is responsible for maintaining pituitary gene expression in the adult as well as during development. This study demonstrates an important role for LIM proteins, and potentially for their interacting partners as well, in supporting basal expression of the GnRHR gene, thus expanding our understanding of their critical role in the regulation of the hypothalamic-pituitary-gonadal axis.
Acknowledgments
We thank Richard Maurer for kindly providing the LHX3 expression vector and Djurdjica Coss and Varykina G. Thackray for helpful discussions, advice, and careful reading of the manuscript.
This work was supported by National Institute of Child Health and Human Development/National Institutes of Health (NIH) through a cooperative agreement (U54 HD12303) as part of the Specialized Cooperative Centers Program in Reproduction Research (to P.L.M.). This work was also supported by NIH Grant R37 HD20377 (to P.L.M.). J.S.B was partially supported by National Institutes of Health Grant T32 GM08666, and S.M.M. was partially supported by National Institutes of Health Grant T32 DK07541. R.R. was the recipient of The Endocrine Society Summer Fellowship.
Abbreviations
- AP-1
Activating protein-1
- ChIP
chromatin immunoprecipitation
- GnRHR
GnRH receptor
- GRAS
GnRHR activating sequence
- αGSU
α-glycoprotein subunit
- PGBE
pituitary glycoprotein basal element
- Pit-1
POU homeodomain transcription factor-1
- Pitx-1
pituitary homeobox-1
- Prl
prolactin
- SDS
sodium dodecyl sulfate
- SF-1
steroidogenic factor-1
- SURG
sequence underlying responsiveness to GnRH
- WT
wild type
References
- 1.Burns KH, Matzuk MM. Minireview: genetic models for the study of gonadotropin actions. Endocrinology. 2002;143:2823–2835. doi: 10.1210/endo.143.8.8928. [DOI] [PubMed] [Google Scholar]
- 2.Duval DL, Nelson SE, Clay CM. A binding site for steroidogenic factor-1 is part of a complex enhancer that mediates expression of the murine gonadotropin-releasing hormone receptor gene. Biol Reprod. 1997;56:160–168. doi: 10.1095/biolreprod56.1.160. [DOI] [PubMed] [Google Scholar]
- 3.Duval DL, Nelson SE, Clay CM. The tripartite basal enhancer of the gonadotropin-releasing hormone (GnRH) receptor gene promoter regulates cell-specific expression through a novel GnRH receptor activating sequence. Mol Endocrinol. 1997;11:1814–1821. doi: 10.1210/mend.11.12.0020. [DOI] [PubMed] [Google Scholar]
- 4.Norwitz ER, Cardona GR, Jeong KH, Chin WW. Identification and characterization of the gonadotropin-releasing hormone response elements in the mouse gonadotropin-releasing hormone receptor gene. J Biol Chem. 1999;274:867–880. doi: 10.1074/jbc.274.2.867. [DOI] [PubMed] [Google Scholar]
- 5.Duval DL, Ellsworth BS, Clay CM. Is gonadotrope expression of the gonadotropin releasing hormone receptor gene mediated by autocrine/paracrine stimulation of an activin response element? Endocrinology. 1999;140:1949–1952. doi: 10.1210/endo.140.4.6780. [DOI] [PubMed] [Google Scholar]
- 6.Norwitz ER, Xu S, Xu J, Spiryda LB, Park JS, Jeong KH, McGee EA, Kaiser UB. Direct binding of AP-1 (Fos/Jun) proteins to a SMAD binding element facilitates both GnRH- and activin-mediated transcriptional activation of the mouse GnRH receptor gene. J Biol Chem. 2002;277:37469–37478. doi: 10.1074/jbc.M206571200. [DOI] [PubMed] [Google Scholar]
- 7.Ellsworth BS, Burns AT, Escudero KW, Duval DL, Nelson SE, Clay CM. The gonadotropin releasing hormone (GnRH) receptor activating sequence (GRAS) is a composite regulatory element that interacts with multiple classes of transcription factors including Smads, AP-1 and a forkhead DNA binding protein. Mol Cell Endocrinol. 2003;206:93–111. doi: 10.1016/s0303-7207(03)00235-1. [DOI] [PubMed] [Google Scholar]
- 8.Kam KY, Jeong KH, Norwitz ER, Jorgensen EM, Kaiser UB. Oct-1 and nuclear factor Y bind to the SURG-1 element to direct basal and gonadotropin-releasing hormone (GnRH)-stimulated mouse GnRH receptor gene transcription. Mol Endocrinol. 2005;19:148–162. doi: 10.1210/me.2004-0025. [DOI] [PubMed] [Google Scholar]
- 9.Jeong KH, Chin WW, Kaiser UB. Essential role of the homeodomain for pituitary homeobox 1 activation of mouse gonadotropin-releasing hormone receptor gene expression through interactions with c-Jun and DNA. Mol Cell Biol. 2004;24:6127–6139. doi: 10.1128/MCB.24.14.6127-6139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seidah NG, Barale J-G, Marcinkiewicz M, Mattei M-G, Day R, Chretien M. The mouse homeoprotein mLIM-3 is expressed early in cells derived from the neuroepithelium and persists in adult pituitary. DNA Cell Biol. 1994;13:1163–1180. doi: 10.1089/dna.1994.13.1163. [DOI] [PubMed] [Google Scholar]
- 11.Bach I, Rhodes SJ, Pearse RV, Heinzel T, Gloss B, Scully KM, Sawchenko PE, Rosenfeld MG. P-Lim, a LIM homeodomain factor, is expressed during pituitary organ and cell commitment and synergizes with Pit-1. Proc Natl Acad Sci USA. 1995;92:2720–2724. doi: 10.1073/pnas.92.7.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheng HZ, Zhadanov AB, Mosinger BJ, Fujii T, Bertuzzi S, Grinberg A, Lee EJ, Huang SP, Mahon KA, Westphal H. Specification of pituitary cell lineages by the LIM homeobox gene Lhx3. Science. 1996;272:1004–1007. doi: 10.1126/science.272.5264.1004. [DOI] [PubMed] [Google Scholar]
- 13.Hobert O, Westphal H. Functions of LIM-homeobox genes. Trends Genet. 2000;16:75–83. doi: 10.1016/s0168-9525(99)01883-1. [DOI] [PubMed] [Google Scholar]
- 14.Netchine I, Sobrier ML, Krude H, Schnabel D, Maghnie M, Marcos E, Duriez B, Cacheux V, Moers A, Goossens M, Gruters A, Amselem S. Mutations in LHX3 result in a new syndrome revealed by combined pituitary hormone deficiency. Nat Genet. 2000;25:182–186. doi: 10.1038/76041. [DOI] [PubMed] [Google Scholar]
- 15.West BE, Parker GE, Savage JJ, Kiratipranon P, Toomey KS, Beach LR, Colvin SC, Sloop KW, Rhodes SJ. Regulation of the follicle-stimulating hormone beta gene by the LHX3 LIM-homeodomain transcription factor. Endocrinology. 2004;145:4866–4879. doi: 10.1210/en.2004-0598. [DOI] [PubMed] [Google Scholar]
- 16.Pincas H, Amoyel K, Counis R, Laverriere JN. Proximal cis-acting elements, including steroidogenic factor 1, mediate the efficiency of a distal enhancer in the promoter of the rat gonadotropin-releasing hormone receptor gene. Mol Endocrinol. 2001;15:319–337. doi: 10.1210/mend.15.2.0593. [DOI] [PubMed] [Google Scholar]
- 17.Roberson MS, Schoderbek WE, Tremml G, Maurer RA. Activation of the glycoprotein hormone α-subunit promoter by a LIM-homeodomain transcription factor. Mol Cell Biol. 1994;14:2985–2993. doi: 10.1128/mcb.14.5.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Albarracin CT, Kaiser UB, Chin WW. Isolation and characterization of the 5′-flanking region of the mouse gonadotropin-releasing hormone receptor gene. Endocrinology. 1994;135:2300–2306. doi: 10.1210/endo.135.6.7988412. [DOI] [PubMed] [Google Scholar]
- 19.Howard PW, Maurer RA. Identification of a conserved protein that interacts with specific LIM homeodomain transcription factors. J Biol Chem. 2000;275:13336–13342. doi: 10.1074/jbc.275.18.13336. [DOI] [PubMed] [Google Scholar]
- 20.Coss D, Jacobs SB, Bender CE, Mellon PL. A novel AP-1 site is critical for maximal induction of the follicle-stimulating hormone beta gene by gonadotropin-releasing hormone. J Biol Chem. 2004;279:152–162. doi: 10.1074/jbc.M304697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenberg SB, Mellon PL. An Otx-related homeodomain protein binds an LHβ promoter element important for activation during gonadotrope maturation. Mol Endocrinol. 2002;16:1280–1298. doi: 10.1210/mend.16.6.0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laughon A. DNA binding specificity of homeodomains. Biochemistry. 1991;30:11357–11367. doi: 10.1021/bi00112a001. [DOI] [PubMed] [Google Scholar]
- 23.Alarid ET, Windle JJ, Whyte DB, Mellon PL. Immortalization of pituitary cells at discrete stages of development by directed oncogenesis in transgenic mice. Development. 1996;122:3319–3329. doi: 10.1242/dev.122.10.3319. [DOI] [PubMed] [Google Scholar]
- 24.Bridwell JA, Price JR, Parker GE, McCutchan Schiller A, Sloop KW, Rhodes SJ. Role of the LIM domains in DNA recognition by the Lhx3 neuroendocrine transcription factor. Gene. 2001;277:239–250. doi: 10.1016/s0378-1119(01)00704-1. [DOI] [PubMed] [Google Scholar]
- 25.Agulnick AD, Taira M, Breen JJ, Tanaka T, Dawid IB, Westphal H. Interactions of the LIM-domain-binding factor Ldb1 with LIM homeodomain proteins. Nature. 1996;384:270–272. doi: 10.1038/384270a0. [DOI] [PubMed] [Google Scholar]
- 26.Jurata LW, Kenny DA, Gill GN. Nuclear LIM interactor, a rhombotin and LIM homeodomain interacting protein, is expressed early in neuronal development. Proc Natl Acad Sci USA. 1996;93:11693–11698. doi: 10.1073/pnas.93.21.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bach I, Carriere C, Ostendorff HP, Andersen B, Rosenfeld MG. A family of LIM domain-associated cofactors confer transcriptional synergism between LIM and Otx homeodomain proteins. Genes Dev. 1997;11:1370–1380. doi: 10.1101/gad.11.11.1370. [DOI] [PubMed] [Google Scholar]
- 28.Bach I, Rodriguez-Esteban C, Carriere C, Bhushan A, Krones A, Rose DW, Glass CK, Andersen B, Izpisua Belmonte JC, Rosenfeld MG. RLIM inhibits functional activity of LIM homeodomain transcription factors via recruitment of the histone deacetylase complex. Nat Genet. 1999;22:394–399. doi: 10.1038/11970. [DOI] [PubMed] [Google Scholar]
- 29.Jurata LW, Pfaff SL, Gill GN. The nuclear LIM domain interactor NLI mediates homo- and heterodimerization of LIM domain transcription factors. J Biol Chem. 1998;273:3152–3157. doi: 10.1074/jbc.273.6.3152. [DOI] [PubMed] [Google Scholar]
- 30.Lee SK, Pfaff SL. Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron. 2003;38:731–745. doi: 10.1016/s0896-6273(03)00296-4. [DOI] [PubMed] [Google Scholar]