Abstract
Aerobic organisms are faced with a dilemma. Environmental iron is found primarily in the relatively inert Fe(III) form, whereas the more metabolically active ferrous form is a strong pro-oxidant. This conundrum is solved by the redox cycling of iron between Fe(III) and Fe(II) at every step in the iron metabolic pathway. As a transition metal ion, iron can be “metabolized” only by this redox cycling, which is catalyzed in aerobes by the coupled activities of ferric iron reductases (ferrireductases) and ferrous iron oxidases (ferroxidases).
Keywords: Iron Metabolism, Metalloenzymes, Oxidation-Reduction, Protein-Metal Ion Interaction, Transport Metals
Introduction
Following the first crystallization of a protein, jack bean urease, reported by Sumner in 1926 (1), the evolution of protein chemistry remained a slow process, hampered by limitations in techniques for protein resolution and analysis. Not surprisingly, many proteins obtained in relatively pure form were those with prosthetic metal ion groups that imparted a visible absorbance to guide the purification. Some were copper proteins whose Cu(II) spectral properties gave their protein host a greenish blue to bright blue hue. Two of these were from mammalian whole blood: hemocuprein (cupric species from erythrocytes, superoxide dismutase) and ceruloplasmin (cerulean blue species from plasma). Superoxide dismutase was first isolated in 1938 by Mann and Keilin (5); Cp2 was first isolated in 1948 by Holmberg and Laurell (6). Both proteins play essential roles in aerobic homeostasis. In the case of ceruloplasmin and the other copper proteins that share its substrate specificity, this role is to manage the redox state of iron in the metabolism of this essential yet potentially cytotoxic transition metal ion. These proteins, all of which are members of the MCO family (7–9), and the mechanism by which they provide this function are a focus of this minireview.
Background
Ataxia (Fig. 1) has long been seen as enzootic (endemic) in animal husbandry, e.g. in sheep; “swayback” was used to characterize the side-to-side (swaying) motion of the hindquarters in the gait of animals presenting with this disorder. The first report of this phenotype was in 1917 when Gaiger (10) described what in Peru was known as “renguera” or limp. Thought possibly linked to heavy metal poisoning (lead), systematic chemical analysis and nutritional studies by Bennetts and Chapman (11) in 1937 demonstrated that this pathology was due to a copper deficiency in the soils of Western Australia; the condition was easily correctable by copper supplementation of the salt licks. Numerous studies from around the world on other domesticated animals confirmed these findings (12). In a series of reports beginning in 1952, Maxwell Wintrobe3 and co-workers demonstrated that copper deficiency in swine caused a systemic iron deficiency and a deficit in a variety of hematologic parameters. For example, cytochrome c oxidase activity was seen to be lower in these copper-deficient animals, as was the oxidase activity in the plasma toward p-phenylenediamine, an activity that was associated with the level of copper-replete active ceruloplasmin (13–15). Similar results were obtained in studies on rats and humans, although human swayback has only recently been recognized clinically as a copper deficiency myelopathy (16, 17).
FIGURE 1.
Enzootic swayback ataxia in sheep. Copper deficiency in animal husbandry, particularly in sheep, was identified in 1937 as the physiologic basis for the “swaying-back” gait phenotypic of this ataxia. The molecular link between the hypocupremia and motor dysfunction was confirmed 20 years later to be a systemic iron deficiency due in part to a decline in activity of the multicopper ferroxidase Cp. This image is from LandLearn NSW.
A principal finding was that iron absorption across the intestinal epithelia was strongly reduced in copper-deficient animals; this observation allowed the Wintrobe group to propose that copper was involved in the mobilization of iron from tissues and not in the biosynthesis of, for example, heme or the formation of red blood cells (18). In other words, copper was required for normal iron metabolism. How copper, ceruloplasmin, and iron trafficking in mammals were connected was not clarified by these experiments. In 1966, however, Osaki, Johnson, and Frieden connected the dots (19); they concluded their Journal of Biological Chemistry article, “The Possible Significance of the Ferrous Oxidase Activity of Ceruloplasmin in Normal Human Serum,” with the following.
While the name ceruloplasmin, coined by its discoverers, Holmberg and Laurell, will always retain its historical importance, we anticipate that the name ferroxidase may be more useful than designating this enzyme as a sky-blue substance from plasma.
Illustrating the truth of this prediction is an objective of this minireview as well.
Kinetic Argument for Ceruloplasmin as a Physiologic Ferroxidase
In 1961, Curzon (20) had noted an apparent “stimulation” of hCp reactivity toward p-phenylenediamine by Fe(II); however, he explicitly discounted any reactivity of hCp with Fe(II). Earl Frieden and colleagues (19) subsequently demonstrated that hCp in fact catalyzed what they then called the ferroxidase reaction (Equation 1), in which hCp coupled the 4-electron reduction of O2 to 2H2O to the oxidation of 4Fe(II).
Quantifying low μm Km values for O2 and Fe(II) (21) in the hCp reaction, Osaki et al. (19) showed that only by hCp catalysis could Tf maintain the iron saturation observed in plasma. They demonstrated that this level of holo-Tf could not be achieved if it depended on the simple autoxidation of Fe(II) by dissolved O2. They calculated that this latter non-enzymatic “delivery” of Fe(III) to apo-Tf would amount to ∼100 μmol/24 h, well below the estimated daily turnover of Tf iron of ∼600 μmol. This shortfall is due entirely to the kinetic difference between uncatalyzed autoxidation of Fe(II) and hCp-catalyzed ferroxidation. With Fe(II)free and [O2]dissolved in the 10–30 μm range in plasma, as a second-order reaction, autoxidation would be far too slow to support the flux of ferric iron formation needed to maintain the level of holo-Tf found in plasma. In contrast, with low μm Km values for both substrates, hCp turnover would be operating at a significant fraction of Vmax, which is 550 min−1; this is equivalent to ∼1000 μmol of Fe(III) delivered to apo-Tf, well above that required for normal iron handling in a day. Furthermore, in vitro ferroxidase catalysis of iron release from the liver was demonstrated, as was the formation of holo-Tf from a mixture of Fe(II) and apo-Tf (22–24). Recent experiments show that this ferroxidase-dependent release of iron from cells is associated with Fpn iron transport catalyzed by Cp or Hp (25). Hp is the paralog of Cp found in mammals that, as a type I membrane protein, localizes its ferroxidase-active catalytic MCO domain on the extracellular surface of an Hp-expressing cell (26–29).
These kinetic primarily in vitro based arguments were not, however, proof that the ferroxidase activity of Cp (or Hp) actually functioned to maintain normal Tf saturation; a negative control was needed to show this directly. These biologic controls would be the knock-outs that became available routinely in lower eukaryotes such as Saccharomyces cerevisiae and subsequently in mice. However, there were numerous naturally occurring genetic traits that less or more directly provided this minus Cp or Hp “control” for how ferroxidases supported iron trafficking in mammals. These included Wilson and Menkes diseases; aceruloplasminemia; and, although this was not clear at the time, the sla mouse, which carries a mutation in the Hp gene.
Genetic Arguments for Ferroxidases as a Key to Iron Homeostasis
Menkes and Wilson diseases are defects in copper metabolism in that each disorder maps to a gene that encodes a specific Cu-ATPase, a copper transporter fueled by the hydrolysis of ATP. The Menkes protein is encoded by ATP7A, whereas the Wilson protein is encoded by ATP7B. ATP7A is on the X chromosome (Menkes disease is therefore X-linked) (30–32), whereas the Wilson gene is on chromosome 13 (33–35). These paralogs are eight-transmembrane domain proteins localized to the vesicular trafficking pathway. Each protein has a cytoplasmic loop containing the ATP-binding domain and a cytoplasmic N-terminal domain decorated by a series of Cys-rich motifs demonstrated to be essential to the proteins' copper transport activity (36–40). They differ in their tissue-specific expression patterns, and these differences relate directly to the differing etiologies of the two disorders.
Irrespective of the tissue of expression, however, both proteins function in the transport of cytoplasmic copper into a vesicular compartment. They share also the substrate for this trafficking; it is not “free” copper, but Cu(I) bound to the copper chaperone HAH1 (human ATX1 homolog 1) (41). The gene encoding this activity was first cloned in S. cerevisiae and designated ATX1 (42). The connection between mutations in either of these two gene classes and a pattern of deficits in iron handling was real but obscure. As summarized below, the broad outlines of the connection between copper and iron trafficking in eukaryotes were revealed eventually by studies in S. cerevisiae (8, 42–47). In the case of Atx1/Hah1 and the ATP7 Cu-ATPases, this connection was the role these latter proteins played in supplying the copper ion prosthetic group to the eukaryotic ferroxidases required for normal iron handling. These are Cp and Hp in mammals, the Fet3 homologs in fungi, and the FOX1 proteins in algae.
ATP7A, the Menkes ATPase (MNK), is expressed primarily in epithelial cells: in the intestine, the placenta, the eye, and blood capillaries in the brain that, together with astrocytes, form the blood-brain barrier. In contrast, ATP7B, the Wilson protein (WND), is expressed primarily in the liver and other endothelial cell-rich organs. The function each protein serves is strongly dictated by the function of the expressing cell type. The liver is the primary source of circulating Cp and is the headwaters of the bile; therefore, the WND protein is responsible for ensuring a supply of copper to hCp that is in the plasma and for excretion of copper from the body via the bile. The intestinal epithelia are primarily responsible for producing membrane-bound Hp, and the MNK protein supplies copper to this ferroxidase. As noted, Hp is required for iron release from the intestinal epithelia into the circulation via the ferrous iron permease Fpn (25). Wilson disease patients typically exhibit a deficit in circulating hCp activity and increased hepatic iron accumulation. In contrast, Menkes disease patients exhibit a systemic copper deficiency that can now be linked to a block in release of copper from the intestinal epithelia at their basolateral surface into the circulation. With respect to brain metabolism of iron, MNK patients exhibit a decline in CNS iron, whereas WND patients exhibit pathophysiology due to a failure to properly manage iron that is already in the CNS.
Aceruloplasminemia (48) and the genetic defect in the sla mouse (49) result in deficiencies in the amount or localization/function of hCp and mouse Hp, respectively, and thus more directly link these enzymes to iron homeostasis. Over 30 different mutant hCp alleles have been identified (50); some lead to a reduction of hCp protein that is otherwise wild-type, and some lead to a wild-type level of protein that is inactive or nearly so (50–52). In those probands that exhibit strongly reduced hCp activity, the common pathology is retinal degeneration, extensive iron deposition in the CNS, decreased glial cell counts, and, in older patients, a marked ataxia and dystonia (51).
There are no known genetic disorders in humans linked to Heph, the gene encoding Hp. The sla mouse, however, represents a sex-linked anemia that maps to a Heph allele carrying an in-frame mutation leading to expression of a protein missing 194 internal residues (26). This protein does exhibit ferroxidase activity but does not traffic to the basolateral membrane of enterocytes in support of iron efflux from the gut into the circulation (53, 54). sla mice are anemic as a result even though they hyperaccumulate iron in their intestinal epithelia. To summarize, there are gene-linked defects in the trafficking of copper to multicopper ferroxidases; in the expression, activation, or stability of a ferroxidase; or in its localization, defects that consistently link to a mismanagement of iron handling in the genetic carrier. These data strongly support the conclusion that MCO ferroxidases are required for iron homeostasis in mammals.
Structure and Function in MCO Ferroxidases
Ferroxidases are essential to iron homeostasis in lower eukaryotes also, and studies in these “model” organisms have primarily provided the structural insights needed to answer the question, “What makes a ferroxidase?” The most significant of these studies have been those in S. cerevisiae and on the ferroxidases expressed by this and other fungi.
In companion papers published in 1994, the Kaplan and Klausner groups reported the cloning of two genes, unique at that time, encoding proteins that physiologically linked copper and iron metabolism. These were FET3 and CTR1 in S. cerevisiae, encoding an MCO that was phenotypically linked to iron uptake (43) and a PM copper transporter whose activity was required for the copper dependent iron uptake supported by Fet3 (44), respectively. Fet3 was shown subsequently to be a ferroxidase (45) whose role in iron uptake was to provide Fe(III) to the iron permease Ftr1 (47). This Fet3-Ftr1 high affinity iron uptake system is encoded in all archived fungal genomes. Physiologically epistatic to this ferroxidase-permease complex is a ferrireductase (Fre1) whose function is to supply Fe(II) substrate to Fet3 (Fig. 2C) (55, 56). Thus, iron uptake at the fungal membrane is a paradigm for the cycle of reduction and oxidation characteristic of aerobic iron metabolism (57).
FIGURE 2.
Iron redox cycling in aerobic iron metabolism. A, iron uptake and efflux from the intestinal enterocyte. Fe(II) is the substrate for transport, whereas the substrate for input and output from this metabolic pathway is Fe(III). B, Fe(II) is both the input and output substrate from Ft redox cycling; Fe(III) is the metabolic intermediate in this pathway. C, iron uptake at the fungal PM inverts the Ft cycle in that Fe(III) is the input and output substrate with Fe(II) as the intermediate. D, iron cycling in the yeast vacuole (Vac) is a combination of Ft and fungal PM iron cycling. The Fe(II) is the input substrate, and Fe(III) is the output substrate with a Ft-like intravacuolar cycling between Fe(II) and Fe(III). All reductions are 1e−. Reductions associated with transmembrane iron trafficking involve NADPH-dependent reductases mediated by cytochrome heme centers; the reducing equivalents associated with Ft iron mobilization have not been characterized. All oxidations are catalyzed by a ferroxidase center and consume dioxygen. The question marks indicate that ionic iron speciation in the cytoplasm is largely uncharacterized.
Like Hp, Fet3 is a type I membrane protein with its catalytic ferroxidase active site projected into the exocytoplasmic space (58–60). Cp lacks the C-terminal transmembrane domains that tether Fet3 proteins and Hp to their respective PMs but, like both proteins, contains the canonical three types of copper sites unique to MCO proteins: the type 1 “blue” copper, the type 2 “normal” copper, and the type 3 binuclear copper cluster. The latter three copper atoms are assembled into a triangular array called the trinuclear cluster. In all of these proteins, residues at the T1 Cu(II) sites recruit Fe(II) as an e− donor; fully reduced by 4Fe(II), the MCO “delivers” these 4e− to the O2, which is bound at the trinuclear cluster, generating 2H2O (9). Important to cell viability in the presence of dissolved O2, this 4e− reduction bypasses the 1e− dioxygen reduction intermediates that are collectively designated as ROS; as a group, ROS are demonstrably cytotoxic.
Whereas the ferroxidase reaction (Equation 1) supported by Hp (and Cp) serves to mobilize Fe(II) being transported out of a cell via Fpn to bind to Tf as Fe(III) (Fig. 2A), the Fe(III) produced by Fet3 is a ligand for transport into the cell via Ftr1 (Fig. 2C). A variety of data indicate that Fet3 and Ftr1 form a complex in the PM of the fungal cell (47, 60). Kinetic data support a model of Fe(III) channeling from Fet3 to Ftr1 in fungal iron uptake, i.e. the ferric iron product of Fet3 ferroxidation traffics directly to Ftr1 without entering bulk solvent (63). A similar associative iron-trafficking mechanism with regard to the Hp-catalyzed release of Fpn-bound iron from the intestinal epithelium has not been demonstrated (Fig. 2A) (25). Although Hp and Fpn co-localize to the basolateral surface of the enterocyte (25, 61, 62), no data have demonstrated as yet the protein interaction that would serve as the physical basis for this mechanism.
In some tissues, particularly in the glia, alternative splicing of the Cp pre-mRNA produces a protein product that is substrate for C-terminal modification by GPI (64). GPI provides a membrane anchor for the Cp so that, when “secreted” from the glial cell, the Cp remains tethered to the exocytoplasmic surface in an orientation equivalent to that of Hp in the enterocyte or Fet3 in the yeast PM. The precise role of GPI-Cp in the CNS is not fully understood, although it is linked to its role in iron efflux from astrocytes (65); what is clear, however, is that its absence correlates to a variety of neuropathologies, including neuronal death (66, 67).
Ferrireductases and Ferroxidases Are the Iron Metabolic Pathway
As Frieden and Osaki presciently noted (57), whereas ferroxidases handle the oxidation, metalloreductases are responsible for the reduction. Fig. 2 illustrates these patterns of iron cycling in the intestinal enterocyte (panel A), in fungal iron uptake (panel C), and in the yeast vacuole (panel D). Fe(III) is the dominant form of iron in the food we eat and is mobilized for transport by reduction via Dcytb, which, like all metalloreductases, is a b-type cytochrome and likely NADPH- dependent (68–71). The Fe(II) product is substrate for the transporter DMT1 (divalent metal transporter 1) (72). The cytoplasmic Fe(II) can efflux from the enterocyte at the basolateral surface via the Fe(II) permease Fpn. As noted above, this efflux is dependent on the ferroxidation due to Hp (27, 29, 54, 61, 73, 74); in plasma, the product of this coupled permeation-oxidation is likely ferric Tf. The redox cycling in Ft (Fig. 2B) involves a similar cycle of ferroxidation and 1e− reduction. Ft has endogenous ferroxidase sites that line the entry channels into the core where iron is stored as Fe(III) (75, 76). Mobilization of iron from Ft is stimulated by e− donors, although their identity in vivo has not been determined; there is no evidence that an NAD(P)H-dependent metalloreductase is involved, however. Not shown here is the essential role that the mammalian Steap3 reductase plays in providing Fe(II) for export out of the endosome in which Fe(III) was released from Tf (77, 78). Positionally cloned as a spontaneous mouse allele that correlated with a severe hypochromic microcytic anemia, Steap3 is now known as one of four Steap reductases (79). Steap2–4 have both cupri- and ferrireductase activities (80) comparable with the corresponding Fre family of reductases in S. cerevisiae (81).
The iron cycling into and out of the yeast vacuole is a paradigm for aerobic iron metabolism (Fig. 2D). Cytoplasmic free iron, whose speciation is uncharacterized, is likely to be Fe(II), given the approximately −250 mV potential of the eukaryotic cytoplasm (E0 (pH 7.0) for Fe(III)/Fe(II) = +400 mV) (82). This assumption is consistent with the fact that the vacuolar iron importer Ccc1 is specific for divalent metals (83). However, as in Ft, iron is likely stored in the vacuole as a hydrated Fe(III) phosphate species (84, 85), and as in Ft, its mobilization for efflux when environmental iron is growth-limiting requires 1e− reduction. Fre6 is the metalloreductase catalyzing this reaction (86).4 What follows recapitulates the Fe(II) ferroxidation/Fe(III) permeation pathway that iron followed in its uptake (Fig. 2, compare D with C); not surprisingly, the proteins handling this vacuolar iron trafficking and efflux are close paralogs of those found in the fungal PM that catalyze iron uptake (87). The reader can recognize also the close parallel between transmembrane iron trafficking in fungi (Fig. 2, C and D) and the iron efflux from the basolateral face of the enterocyte (Fig. 2A).
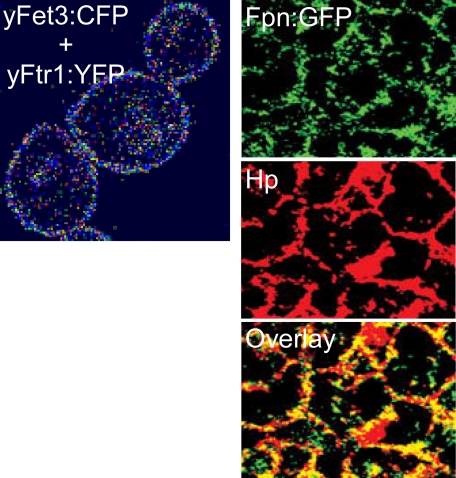
What these schematics do not reflect is the mechanism of this iron trafficking, particularly between the ferroxidase-permease pairs. The contextual basis for the mechanism is illustrated by the fluorescence images in Fig. 3. Most compelling is the FRET image shown in the left panel derived from the Fet3-cyan fluorescent protein + Ftr1-yellow fluorescent protein complex in the yeast PM; quantification of the FRET signal indicates that it comes from a protein complex (60). The yeast ferroxidase-permease pair Fet5-Fth1 (Fig. 2D) can be co-immunoprecipitated, indicating their association in the vacuolar membrane (87). The images in the right panels in Fig. 3 demonstrate the co-localization of the mammalian version of the fungal Fet3-Ftr1 complex, the Fpn-Hp complex at the basolateral surface of polarized Caco-2 cells (29). Evidence for the physical contiguity or interaction of Fpn and Hp has not been reported, however.
FIGURE 3.
Ferroxidase-iron permease proteins in the eukaryotic PM. Left panel, FRET between yeast Fet3-cyan fluorescent protein (yFet3:CFP) and yeast Ftr1-yellow fluorescent protein (yFtr1:YFP) in the yeast PM. The computer-generated image scores the efficiency of donor quenching by the acceptor from white (∼15%) to magenta (∼50%) (N. Kaur and D. J. Kosman, unpublished data). Right panels, Fpn-green fluorescent protein (Fpn-GFP; upper panel), indirect immunofluorescence of Hp (middle panel), and overlay of images from polarized Caco-2 cells at their basolateral surface (lower panel). This figure was modified from Ref. 29 with permission.
Kinetic analysis of 55Fe uptake through the Fet3-Ftr1 complex has supported a model of iron trafficking in which the Fet3-generated Fe(III) is channeled to the permease, Ftr1, without dissociation into bulk solvent (60, 63). This same kinetic feature appears true in the FOX1-FTR1 iron uptake system in the green alga Chlamydomonas reinhardtii.5 Although C. reinhardtii FTR1 is a homolog of yeast Ftr1, C. reinhardtii FOX1 is a homolog of Cp (88–90); in other words, the iron uptake system in C. reinhardtii is assembled from a mammalian ferroxidase and fungal permease while exhibiting a iron-trafficking mechanism characteristic of the lower eukaryote.
The Why of Redox Cycling
As R. J. Williams (91, 92) and Robert Crichton (93, 94) have emphasized, the common era “metallome” is, in essence, the art of the possible, and for redox-active metal ions like iron and copper, it is the art of what is possible in a tidal pool containing 260 μm dissolved O2. Starting ∼2.4 gigayears before the common era (95, 96), the redox potential of the geosphere began its slow oxidation from negative (−400 mV as a lower limit) to positive (+400 mV as an upper limit), and the dominant redox-active first-row transition metal dissolved in the oceans went from Fe(II) to Cu(II). Thus, organisms that had evolved to use Fe(II) as nutrient and cofactor (FeS clusters, mono- and diiron active sites, heme) in support of intermediary metabolic conversions and in terminal electron transfer (to nitrogen and sulfur oxides coming from thermal vents) were faced with a declining ready supply of the trace element upon which they had come to rely. The adaptive response was the reduction-oxidation cycles that now define aerobic iron metabolism. These cycles are supported by heme-dependent metalloreductases and the metallo-oxidases, both linked to ferri- and ferro-handling proteins, transporters, and carriers. At the center of this new iron metabolism were the oxidases that relied on the now readily available Cu(II) as a prosthetic group, the MCO family of proteins. Ferroxidases are now as essential to aerobic iron metabolism as cytochrome c oxidase (another adaptation to the “appearance” of soluble copper and O2) is to the production of ATP driven by a respiration-dependent proton gradient.
Like the heme/copper terminal oxidases, MCOs possess four 1e− redox centers that provide these two enzyme families the unique ability to reduce dioxygen by 4 electrons to 2H2O, thus bypassing ROS. They share also the kinetic property of a low μm or smaller Km for O2; thus, even at, for example, 1 μm dissolved O2, they function at a reasonable fraction of their Vmax values. Put into the geo-evolutionary context, these enzymes would have been efficient at a partial pressure of atmospheric O2 that was as little as 0.5% of what it is at present. Clearly, they were an early addition to the proteome of organisms adapting to thrive in the presence of this toxin and, perhaps, even in those organisms responsible for generating it. These latter were the photosynthetic cyanobacteria, whose presence can be dated approximately coincident with the initial atmospheric oxygenesis (95, 96). The importance of ferroxidases to micro-aerobic homeostasis is illustrated also by the up-regulation of copper delivery to them in ischemia (97) and the increased reliance on them in cellular iron export under the same conditions (98).
The interplay between ferrireductase and ferroxidase in managing the recalcitrant aqueous and redox chemistry of iron is exemplified clearly in S. cerevisiae. Growing aerobically, fungi are presented with Fe(III) in some tightly bound form (“rust,” siderophore, Tf, heme). One strategy is to internalize the iron complex by a receptor-mediated process or by simple fluid-phase endocytosis and then to mobilize the iron by chelate degradation and/or metalloreduction (94). The other is to mobilize the iron from the chelate by exocytoplasmic ferrireduction and then to use the Fe(II) as a substrate for iron permeation. This is the strategy employed at the apical surface of our enterocytes involving Dcytb and DMT1.
Fungi couple this ferrireduction to ferroxidation, however, and then the ferroxidation to ferripermeation. How does this curious redox cycling solve the “iron conundrum?” First, what are the components of that conundrum? They are as follows. 1) Fe(III) is what is available, but it is exchange-inert; for example, its solubility at pH 7.0 is 10−18 mol/liter. 2) Fe(II) is soluble and exchanges its ligands readily, but it is a strong pro-oxidant, generating hydroxyl radicals via what is known as the Fenton reaction (93). Yeast, as do we, solve the first problem by ferrireduction, which mobilizes Fe(II) for subsequent metabolism. However, this reaction brings the second problem into play, one solved by the ferroxidation step in the yeast iron uptake pathway. That this ferroxidation protects the yeast cell from oxidative stress is demonstrated by sensitivity toward metal- and oxygen-dependent toxicity exhibited by yeast cells that produce the ferrireductase but lack the ferroxidase (99).
What this result implies is that Fe(II) generation must be tightly coupled to subsequent Fe(II) metabolism, whether that be permeation into the −250 mV redox potential of the eukaryotic cytoplasm (via DMT1) or “immediate” ferroxidation, as at the yeast PM. Yes, but how does the fungal pathway solve the first problem all over again? Why doesn't the ferroxidase-generated Fe(III) simply hydrolyze and precipitate out of solution? The data indicate that the ferroxidase-permease pair avoids that futile cycle by providing a “channel” for the Fe(III), a transfer mechanism that kinetically competes with dissociation of the Fe(III) into bulk solvent, where it would react with H2O and precipitate as iron (hydr)oxide (63). The fungal system certainly represents a “how to” handle iron in an aerobic environment; time will tell the extent to which it represents iron handling in higher eukaryotes as well.
Supplementary Material
Acknowledgments
This minireview is dedicated to my students and my collaborators, who did all of the work.
This work was supported by National Institutes of Health Grants 53820 and 77826 from NIDDK. This is the first article in the Thematic Minireview Series on Metals in Biology 2010. This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
Dr. Wintrobe, who was Chair of Medicine at the University of Utah, received the first extramural grant awarded by the National Institutes of Health in 1945; his $100,000 grant was renewed annually for 33 years!
The metabolic specificity of metallo-redox cycling is indicated by the fact that copper efflux from the vacuole requires a Cu(II) → Cu(I) conversion catalyzed by another vacuolar reductase, Fre7 (86); these two close paralogs do not complement each other in their respective metallo-mobilizing functions (86, 100).
A. J. Terzulli and D. J. Kosman, unpublished data.
- Cp
- ceruloplasmin
- MCO
- multicopper oxidase
- hCp
- human Cp
- Tf
- transferrin
- Fpn
- ferroportin
- Hp
- hephaestin
- CNS
- central nervous system
- PM
- plasma membrane
- ROS
- reactive oxygen species
- GPI
- glycosylphosphatidylinositol
- Ft
- ferritin
- FRET
- fluorescence resonance energy transfer.
REFERENCES
- 1.Sumner J. B. (1926) J. Biol. Chem. 69, 435–441 [Google Scholar]
- 2.Deleted in proof
- 3.Deleted in proof
- 4.Deleted in proof
- 5.Mann T., Keilin D. (1938) Proc. R. Soc. Lond. B Biol. Sci. 126, 303–315 [Google Scholar]
- 6.Holmberg C. G., Laurell C. B. (1948) Acta Chem. Scand. 2, 550–556 [DOI] [PubMed] [Google Scholar]
- 7.Solomon E. I., Sundaram U. M., Machonkin T. E. (1996) Chem. Rev. 96, 2563–2606 [DOI] [PubMed] [Google Scholar]
- 8.Kosman D. J. (2002) Adv. Protein Chem. 60, 221–269 [DOI] [PubMed] [Google Scholar]
- 9.Stoj C. S., Kosman D. J. (2005) in Encyclopedia of Inorganic Chemistry (King R. B. ed) 2nd Ed., Vol. II, pp. 1134–1159, John Wiley & Sons, Inc., New York [Google Scholar]
- 10.Gaiger S. H. (1917) J. Comp. Pathol. 30, 185 [Google Scholar]
- 11.Bennetts H. W., Chapman F. E. (1937) Aust. Vet. J. 13, 138–149 [Google Scholar]
- 12.Barlow R. M. (1958) Proc. R. Soc. Med. 51, 748–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gubler C. J., Lahey M. E., Chase M. S., Cartwright G. E., Wintrobe M. M. (1952) Blood 7, 1075–1092 [PubMed] [Google Scholar]
- 14.Lahey M. E., Gubler C. J., Chase M. S., Cartwright G. E., Wintrobe M. M. (1952) Blood 7, 1053–1074 [PubMed] [Google Scholar]
- 15.Markowitz H., Gubler C. J., Mahoney J. P., Cartwright G. E., Wintrobe M. M. (1955) J. Clin. Invest. 34, 1498–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schleper B., Stuerenburg H. J. (2001) J. Neurol. 248, 705–706 [DOI] [PubMed] [Google Scholar]
- 17.Kumar N. (2006) Mayo Clin. Proc. 81, 1371–1384 [DOI] [PubMed] [Google Scholar]
- 18.Chase M. S., Gubler C. J., Cartwright G. E., Wintrobe M. M. (1952) J. Biol. Chem. 199, 757–763 [PubMed] [Google Scholar]
- 19.Osaki S., Johnson D. A., Frieden E. (1966) J. Biol. Chem. 241, 2746–2751 [PubMed] [Google Scholar]
- 20.Curzon G. (1961) Biochem. J. 79, 656–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osaki S. (1966) J. Biol. Chem. 241, 5053–5059 [PubMed] [Google Scholar]
- 22.Osaki S., Johnson D. A. (1969) J. Biol. Chem. 244, 5757–5758 [PubMed] [Google Scholar]
- 23.Osaki S., Johnson D. A., Frieden E. (1971) J. Biol. Chem. 246, 3018–3023 [PubMed] [Google Scholar]
- 24.Chidambaram M. V., Barnes G., Frieden E. (1983) FEBS Lett. 159, 137–140 [DOI] [PubMed] [Google Scholar]
- 25.De Domenico I., Ward D. M., di Patti M. C., Jeong S. Y., David S., Musci G., Kaplan J. (2007) EMBO J. 26, 2823–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vulpe C. D., Kuo Y. M., Murphy T. L., Cowley L., Askwith C., Libina N., Gitschier J., Anderson G. J. (1999) Nat. Genet. 21, 195–199 [DOI] [PubMed] [Google Scholar]
- 27.Frazer D. M., Vulpe C. D., McKie A. T., Wilkins S. J., Trinder D., Cleghorn G. J., Anderson G. J. (2001) Am. J. Physiol. Gastrointest. Liver Physiol. 281, G931–G939 [DOI] [PubMed] [Google Scholar]
- 28.Syed B. A., Beaumont N. J., Patel A., Naylor C. E., Bayele H. K., Joannou C. L., Rowe P. S., Evans R. W., Srai S. K. (2002) Protein Eng. 15, 205–214 [DOI] [PubMed] [Google Scholar]
- 29.Han O., Kim E. Y. (2007) J. Cell. Biochem. 101, 1000–1010 [DOI] [PubMed] [Google Scholar]
- 30.Chelly J., Tümer Z., Tønnesen T., Petterson A., Ishikawa-Brush Y., Tommerup N., Horn N., Monaco A. P. (1993) Nat. Genet. 3, 14–19 [DOI] [PubMed] [Google Scholar]
- 31.Mercer J. F., Livingston J., Hall B., Paynter J. A., Begy C., Chandrasekharappa S., Lockhart P., Grimes A., Bhave M., Siemieniak D., Glover T. W. (1993) Nat. Genet. 3, 20–25 [DOI] [PubMed] [Google Scholar]
- 32.Vulpe C., Levinson B., Whitney S., Packman S., Gitschier J. (1993) Nat. Genet. 3, 7–13 [DOI] [PubMed] [Google Scholar]
- 33.Bull P. C., Thomas G. R., Rommens J. M., Forbes J. R., Cox D. W. (1993) Nat. Genet. 5, 327–337 [DOI] [PubMed] [Google Scholar]
- 34.Petrukhin K., Fischer S. G., Pirastu M., Tanzi R. E., Chernov I., Devoto M., Brzustowicz L. M., Cayanis E., Vitale E., Russo J. J., et al. (1993) Nat. Genet. 5, 338–343 [DOI] [PubMed] [Google Scholar]
- 35.Tanzi R. E., Petrukhin K., Chernov I., Pellequer J. L., Wasco W., Ross B., Romano D. M., Parano E., Pavone L., Brzustowicz L. M., et al. (1993) Nat. Genet. 5, 344–350 [DOI] [PubMed] [Google Scholar]
- 36.La Fontaine S., Firth S. D., Lockhart P. J., Brooks H., Parton R. G., Camakaris J., Mercer J. F. (1998) Hum. Mol. Genet. 7, 1293–1300 [DOI] [PubMed] [Google Scholar]
- 37.Terada K., Schilsky M. L., Miura N., Sugiyama T. (1998) Int. J. Biochem. Cell Biol. 30, 1063–1067 [DOI] [PubMed] [Google Scholar]
- 38.Huffman D. L., O'Halloran T. V. (2001) Annu. Rev. Biochem. 70, 677–701 [DOI] [PubMed] [Google Scholar]
- 39.Lutsenko S., Efremov R. G., Tsivkovskii R., Walker J. M. (2002) J. Bioenerg. Biomembr. 34, 351–362 [DOI] [PubMed] [Google Scholar]
- 40.Tsivkovskii R., Eisses J. F., Kaplan J. H., Lutsenko S. (2002) J. Biol. Chem. 277, 976–983 [DOI] [PubMed] [Google Scholar]
- 41.Klomp L. W., Lin S. J., Yuan D. S., Klausner R. D., Culotta V. C., Gitlin J. D. (1997) J. Biol. Chem. 272, 9221–9226 [DOI] [PubMed] [Google Scholar]
- 42.Lin S. J., Pufahl R. A., Dancis A., O'Halloran T. V., Culotta V. C. (1997) J. Biol. Chem. 272, 9215–9220 [PubMed] [Google Scholar]
- 43.Askwith C., Eide D., Van Ho A., Bernard P. S., Li L., Davis-Kaplan S., Sipe D. M., Kaplan J. (1994) Cell 76, 403–410 [DOI] [PubMed] [Google Scholar]
- 44.Dancis A., Yuan D. S., Haile D., Askwith C., Eide D., Moehle C., Kaplan J., Klausner R. D. (1994) Cell 76, 393–402 [DOI] [PubMed] [Google Scholar]
- 45.De Silva D. M., Askwith C. C., Eide D., Kaplan J. (1995) J. Biol. Chem. 270, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 46.Yuan D. S., Stearman R., Dancis A., Dunn T., Beeler T., Klausner R. D. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 2632–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stearman R., Yuan D. S., Yamaguchi-Iwai Y., Klausner R. D., Dancis A. (1996) Science 271, 1552–1557 [DOI] [PubMed] [Google Scholar]
- 48.Miyajima H., Nishimura Y., Mizoguchi K., Sakamoto M., Shimizu T., Honda N. (1987) Neurology 37, 761–767 [DOI] [PubMed] [Google Scholar]
- 49.Au Y. P., Schilling R. F. (1986) Biochim. Biophys. Acta 883, 242–246 [DOI] [PubMed] [Google Scholar]
- 50.Kono S., Miyajima H. (2006) Biol. Res. 39, 15–23 [DOI] [PubMed] [Google Scholar]
- 51.Miyajima H. (2003) Neuropathology 23, 345–350 [DOI] [PubMed] [Google Scholar]
- 52.Miyajima H., Takahashi Y., Kono S. (2003) Biometals 16, 205–213 [DOI] [PubMed] [Google Scholar]
- 53.Chen H., Attieh Z. K., Su T., Syed B. A., Gao H., Alaeddine R. M., Fox T. C., Usta J., Naylor C. E., Evans R. W., McKie A. T., Anderson G. J., Vulpe C. D. (2004) Blood 103, 3933–3939 [DOI] [PubMed] [Google Scholar]
- 54.Kuo Y. M., Su T., Chen H., Attieh Z., Syed B. A., McKie A. T., Anderson G. J., Gitschier J., Vulpe C. D. (2004) Gut 53, 201–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dancis A., Roman D. G., Anderson G. J., Hinnebusch A. G., Klausner R. D. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 3869–3873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anderson G. J., Dancis A., Roman D. G., Klausner R. D. (1994) Adv. Exp. Med. Biol. 356, 81–89 [DOI] [PubMed] [Google Scholar]
- 57.Frieden E., Osaki S. (1974) Adv. Exp. Med. Biol. 48, 235–265 [DOI] [PubMed] [Google Scholar]
- 58.Hassett R. F., Yuan D. S., Kosman D. J. (1998) J. Biol. Chem. 273, 23274–23282 [DOI] [PubMed] [Google Scholar]
- 59.Taylor A. B., Stoj C. S., Ziegler L., Kosman D. J., Hart P. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15459–15464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singh A., Severance S., Kaur N., Wiltsie W., Kosman D. J. (2006) J. Biol. Chem. 281, 13355–13564 [DOI] [PubMed] [Google Scholar]
- 61.Chen H., Attieh Z. K., Dang T., Huang G., van der Hee R. M., Vulpe C. (2009) J. Cell. Biochem. 107, 803–808 [DOI] [PubMed] [Google Scholar]
- 62.Yeh K. Y., Yeh M., Mims L., Glass J. (2009) Am. J. Physiol. Gastrointest. Liver Physiol. 296, G55–G65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kwok E. Y., Severance S., Kosman D. J. (2006) Biochemistry 45, 6317–6327 [DOI] [PubMed] [Google Scholar]
- 64.Patel B. N., Dunn R. J., David S. (2000) J. Biol. Chem. 275, 4305–4310 [DOI] [PubMed] [Google Scholar]
- 65.Jeong S. Y., David S. (2003) J. Biol. Chem. 278, 27144–27148 [DOI] [PubMed] [Google Scholar]
- 66.Patel B. N., Dunn R. J., Jeong S. Y., Zhu Q., Julien J. P., David S. (2002) J. Neurosci. 22, 6578–6586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rathore K. I., Kerr B. J., Redensek A., López-Vales R., Jeong S. Y., Ponka P., David S. (2008) J. Neurosci. 28, 12736–12747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Finegold A. A., Shatwell K. P., Segal A. W., Klausner R. D., Dancis A. (1996) J. Biol. Chem. 271, 31021–31024 [DOI] [PubMed] [Google Scholar]
- 69.Shatwell K. P., Dancis A., Cross A. R., Klausner R. D., Segal A. W. (1996) J. Biol. Chem. 271, 14240–14244 [DOI] [PubMed] [Google Scholar]
- 70.McKie A. T., Barrow D., Latunde-Dada G. O., Rolfs A., Sager G., Mudaly E., Mudaly M., Richardson C., Barlow D., Bomford A., Peters T. J., Raja K. B., Shirali S., Hediger M. A., Farzaneh F., Simpson R. J. (2001) Science 291, 1755–1759 [DOI] [PubMed] [Google Scholar]
- 71.Latunde-Dada G. O., Van der Westhuizen J., Vulpe C. D., Anderson G. J., Simpson R. J., McKie A. T. (2002) Blood Cells Mol. Dis. 29, 356–360 [DOI] [PubMed] [Google Scholar]
- 72.McKie A. T., Latunde-Dada G. O., Miret S., McGregor J. A., Anderson G. J., Vulpe C. D., Wrigglesworth J. M., Simpson R. J. (2002) Biochem. Soc. Trans. 30, 722–724 [DOI] [PubMed] [Google Scholar]
- 73.Anderson G. J., Frazer D. M., McKie A. T., Vulpe C. D. (2002) Blood Cells Mol. Dis. 29, 367–375 [DOI] [PubMed] [Google Scholar]
- 74.Chen H., Huang G., Su T., Gao H., Attieh Z. K., McKie A. T., Anderson G. J., Vulpe C. D. (2006) J. Nutr. 136, 1236–1241 [DOI] [PubMed] [Google Scholar]
- 75.Harrison P. M., Bauminger E. R., Hechel D., Hodson N. W., Nowik I., Treffry A., Yewdall S. J. (1994) Adv. Exp. Med. Biol. 356, 1–12 [DOI] [PubMed] [Google Scholar]
- 76.Stillman T. J., Hempstead P. D., Artymiuk P. J., Andrews S. C., Hudson A. J., Treffry A., Guest J. R., Harrison P. M. (2001) J. Mol. Biol. 307, 587–603 [DOI] [PubMed] [Google Scholar]
- 77.Ohgami R. S., Campagna D. R., Antiochos B., Wood E. B., Sharp J. J., Barker J. E., Fleming M. D. (2005) Blood 106, 3625–3631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ohgami R. S., Campagna D. R., Greer E. L., Antiochos B., McDonald A., Chen J., Sharp J. J., Fujiwara Y., Barker J. E., Fleming M. D. (2005) Nat. Genet. 37, 1264–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ohgami R. S., Campagna D. R., McDonald A., Fleming M. D. (2006) Blood 108, 1388–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Knutson M. D. (2007) Nutr. Rev. 65, 335–340 [DOI] [PubMed] [Google Scholar]
- 81.Martins L. J., Jensen L. T., Simon J. R., Keller G. L., Winge D. R. (1998) J. Biol. Chem. 273, 23716–23721 [DOI] [PubMed] [Google Scholar]
- 82.Schafer F. Q., Buettner G. R. (2001) Free Radic. Biol. Med. 30, 1191–1212 [DOI] [PubMed] [Google Scholar]
- 83.Li L., Chen O. S., McVey Ward D., Kaplan J. (2001) J. Biol. Chem. 276, 29515–29519 [DOI] [PubMed] [Google Scholar]
- 84.Raguzzi F., Lesuisse E., Crichton R. R. (1988) FEBS Lett. 231, 253–258 [DOI] [PubMed] [Google Scholar]
- 85.Bode H. P., Dumschat M., Garotti S., Fuhrmann G. F. (1995) Eur. J. Biochem. 228, 337–342 [PubMed] [Google Scholar]
- 86.Singh A., Kaur N., Kosman D. J. (2007) J. Biol. Chem. 282, 28619–28626 [DOI] [PubMed] [Google Scholar]
- 87.Urbanowski J. L., Piper R. C. (1999) J. Biol. Chem. 274, 38061–38070 [DOI] [PubMed] [Google Scholar]
- 88.Herbik A., Bölling C., Buckhout T. J. (2002) Plant Physiol. 130, 2039–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.La Fontaine S., Quinn J. M., Nakamoto S. S., Page M. D., Göhre V., Moseley J. L., Kropat J., Merchant S. (2002) Eukaryot. Cell 1, 736–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Terzulli A. J., Kosman D. J. (2009) J. Biol. Inor.g Chem. 14, 315–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Williams R. J., Fraústo Da Silva J. J. (2003) J. Theor. Biol. 220, 323–343 [DOI] [PubMed] [Google Scholar]
- 92.Williams R. J. (2007) J. R. Soc. Interface 4, 1049–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Crichton R. R., Pierre J. L. (2001) Biometals 14, 99–112 [DOI] [PubMed] [Google Scholar]
- 94.Pierre J. L., Fontecave M., Crichton R. R. (2002) Biometals 15, 341–346 [DOI] [PubMed] [Google Scholar]
- 95.Rasmussen B., Fletcher I. R., Brocks J. J., Kilburn M. R. (2008) Nature 455, 1101–1104 [DOI] [PubMed] [Google Scholar]
- 96.Blank C. E., Sánchez-Baracaldo P. (2010) Geobiology 8, 1–23 [DOI] [PubMed] [Google Scholar]
- 97.White C., Kambe T., Fulcher Y. G., Sachdev S. W., Bush A. I., Fritsche K., Lee J., Quinn T. P., Petris M. J. (2009) J. Cell Sci. 122, 1315–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sarkar J., Seshadri V., Tripoulas N. A., Ketterer M. E., Fox P. L. (2003) J. Biol. Chem. 278, 44018–44024 [DOI] [PubMed] [Google Scholar]
- 99.Shi X., Stoj C., Romeo A., Kosman D. J., Zhu Z. (2003) J. Biol. Chem. 278, 50309–50315 [DOI] [PubMed] [Google Scholar]
- 100.Rees E. M., Thiele D. J. (2007) J. Biol. Chem. 282, 21629–21638 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.