Abstract
Friedreich ataxia is an inherited neurodegenerative disease caused by frataxin deficiency. Frataxin is a conserved mitochondrial protein that plays a role in FeS cluster assembly in mitochondria. FeS clusters are modular cofactors that perform essential functions throughout the cell. They are synthesized by a multistep and multisubunit mitochondrial machinery that includes the scaffold protein Isu for assembling a protein-bound FeS cluster intermediate. Frataxin interacts with Isu, iron, and the cysteine desulfurase Nfs1, which supplies sulfide, thus placing it at the center of mitochondrial FeS cluster biosynthesis.
Keywords: Iron, Iron-Sulfur Protein, Metals, Mitochondria, Neurodegeneration, Frataxin
Introduction
Friedreich ataxia is an inherited disease that is characterized by progressive symptoms of ataxia and sensory loss, often leading to gait impairment and the need for a wheelchair. A progressive and sometimes lethal cardiomyopathy is another feature, and in some cases, diabetes mellitus is associated. At the pathological level, degenerative changes affect certain large sensory neurons, heart cells, and islet cells, thus involving a unique target tissue distribution (1). The gene was identified by genome mapping of affected kindreds, and the encoded protein, frataxin, is decreased in affected individuals most often due to expansion of a GAA repeat in the first intron of the gene (2). The protein is found primarily in mitochondria (3). Human frataxin is synthesized on cytoplasmic ribosomes with a mitochondrial targeting sequence that mediates organelle targeting and is subsequently removed by proteolytic processing steps in the mitochondrial matrix (4–7). Deficiencies of aconitase and other mitochondrial FeS proteins occur as first noted in cardiac biopsies of affected individuals (8). FeS clusters are modular cofactors consisting of iron and sulfur, most often anchored by bonds joining cysteine sulfur atoms in the polypeptide chain of a protein and iron atoms of the cluster. They perform essential and diverse biochemical functions (electron transfer in cellular respiration, substrate interaction, and biological signal transduction among other functions), and their biogenesis is catalyzed by a multisubunit machinery (9). Frataxin has been a subject of intense study with contributions from many disciplines: structural biology, cell biology, genetics of model organisms, evolutionary biology, and medicine. A picture is emerging of a direct role of frataxin in the complex and highly conserved machinery of FeS cluster biogenesis in mitochondria. Additional functions may be mediated by direct frataxin-iron effects or by other protein-protein interactions.
Frataxin Evolution
Frataxin is highly conserved throughout evolution, being present in humans, plants, flies, worms, and bacteria (proteobacteria but not archaebacteria) (10). Some hints about frataxin function can be gleaned from the evolutionary record. Three different bacterial operons capable of mediating FeS cluster assembly have been identified: nif, suf, and isc. The nif operon is specialized for high volume biosynthesis needed to support nitrogenase supply during diazotrophic growth of some organisms (e.g. Azotobacter vinelandii). The suf operon is specially adapted for FeS cluster synthesis under conditions of iron starvation and oxidative stress. Finally, the isc operon handles housekeeping FeS cluster biogenesis (9). Although not found on any of these operons, frataxin is strongly associated with the isc operon. It appeared in proteobacteria about the time that the specialized chaperones hscA and hscB appeared on the isc operon. A frataxin ortholog was subsequently lost in this phylogenetic lineage on two separate occasions, coinciding with loss of hscA and hscB. The appearance of frataxin in eukaryotes occurred about the time of the endosymbiotic event creating mitochondria from the purple bacterial ancestor, and it was probably acquired by mitochondria together with other components of the isc operon (10). This notion of co-evolution of frataxin and FeS assembly components of the isc operon is supported by studies of primitive eukaryotes that lack typical mitochondria. In Trichomonas vaginalis, a unicellular parasite, a modified and stripped-down organelle called the hydrogenosome performs FeS cluster assembly and contains a frataxin ortholog able to complement the phenotypic defects of a yeast mutant lacking its own frataxin (11). An even more severe case of organelle simplification is presented by mitosomes. These are rudimentary organelles of obligate intracellular parasites called microsporidia, and they lack respiratory complexes or mitochondrial DNA. Virtually all that remain are a few components recognizable for their homology to FeS cluster assembly components (e.g. Isu, Nfs1, and Hsp70 homologs) and frataxin (12). The co-retention of frataxin with these very few other components involved in FeS cluster assembly in the mitosome emphasizes their close functional relationship.
Mitochondrial Iron Accumulation
In Saccharomyces cerevisiae, Yfh1 is the yeast frataxin homolog, and the initial studies of the Δyfh1 deletion strain described a pleiotropic phenotype. The mutant exhibited constitutive up-regulation of the cellular iron uptake system and tremendous accumulation of iron in mitochondria (13). The accumulated iron appeared as dense aggregates in unstained electron micrographs. Chemical analyses revealed that the accumulated iron was in the form of ferric phosphate nanoparticles (14). Many additional studies have established a close association of the mitochondrial iron accumulation phenotype and deficiency of FeS clusters. Cellular depletion studies, in which Yfh1 was put under the control of a regulated promoter and turned off, recapitulated the phenotypes of deficient FeS cluster proteins and mitochondrial iron accumulation. If the promoter was turned on again, FeS clusters were restored, and the accumulated iron returned to a normal distribution (15). Other mutants of the mitochondrial FeS cluster assembly pathway were similarly associated with this iron homeostatic phenotype (ssq1, jac1, nfs1, etc.). In human cells, mitochondrial iron accumulation has also been observed both in tissues from Friedreich patients (e.g. heart and dorsal root ganglia) and in cultured cells engineered to exhibit stable frataxin deficiency (16). Thus, low level frataxin is associated with mitochondrial iron accumulation and FeS cluster deficiency in both human and yeast cells. However, an FeS cluster protein capable of mediating these effects and conserved between yeast and humans has not been identified. The best candidate protein for this function, human IRP1, is not conserved with yeast, and the yeast iron regulatory machinery, including Aft1/2, is not found in humans (17).
FeS Cluster Assembly in Mitochondria
Frataxin has been found in mitochondria of virtually all eukaryotes using biochemical and microscopic tools to ascertain its subcellular location. Similarly, orthologs of many of the other Isc FeS cluster assembly components have been found in mitochondria (18). The steps and components involved in the biogenesis of FeS cluster proteins in mitochondria are quite similar to those in bacteria. The process can be understood in terms of the central role of scaffolds as first shown by Dennis Dean and co-workers (19) and now well established. An FeS cluster intermediate is formed on the scaffold protein Isu and then transferred to apoproteins (Fig. 1). Many steps must occur in a coordinated fashion for proper FeS cluster synthesis on Isu. The sulfide for FeS clusters originates from cysteine via the action of Nfs1, a pyridoxal phosphate-containing cysteine desulfurase. In eukaryotes, Nfs1 must be assembled with Isd11, a small accessory subunit of unknown function (20, 21). The sulfide is probably transferred to Isu from the Nfs1 active site as a persulfide. The source of the iron for the Isu intermediate is unclear, although a role for mitochondrial carrier proteins Mrs3 and Mrs4 in yeast (mitoferrin in humans) has been proposed (22). Frataxin may play a role here and has been shown to interact genetically with the mitochondrial carrier proteins. This was shown by the exacerbated slow growth and mitochondrial iron starvation in the combined Δmrs3Δmrs4Δyfh1 mutant (23). Other components implicated at this stage are Arh1 and Yah1 (adrenodoxin reductase and adrenodoxin in humans), which are needed to provide reducing equivalents in an electron transfer chain, although the substrate requiring reduction has not been defined (24). The Isu protein appears to be conserved in the context of primary sequence and function, and structural information exists for a few bacterial orthologs. Key features are three critical cysteines and one aspartate that bind the cluster intermediate probably on each subunit of the Isu dimer (25).
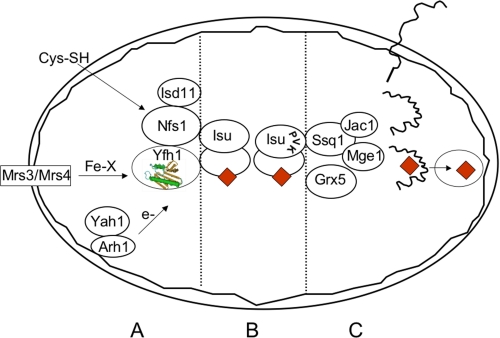
FIGURE 1.
Scheme showing role of yeast frataxin (Yfh1) in mitochondrial FeS cluster assembly. The mitochondrion is shown as an oval bounded by a double membrane. Yfh1 is shown as a helix-sheet sandwich (green and tan). A, mitochondrial carriers Mrs3 and Mrs4 (box) play a role in transfer of iron in some form (Fe-X) across the mitochondrial inner membrane. Cysteine (Cys-SH) enters mitochondria and is acted on by the enzyme Nfs1 and its accessory protein Isd11 to provide sulfur for FeS clusters. Electrons or reducing equivalents are provided by the reductase Arh1 in the membrane and the associated ferredoxin Yah1. Yfh1 physically interacts with Nfs1, Isd11, and Isu. B, the FeS cluster intermediate (red diamonds) is assembled on the scaffold Isu, which interacts with Nfs1, Isd11, and Yfh1. Another complex is formed by Isu with Ssq1, Jac1, and Mge1, with the binding site being provided by the PVK tripeptide on Isu1. Grx5 acts at this step. C, the precursor proteins (squiggly black lines) are nuclear encoded, translated on cytoplasmic ribosomes, targeted to mitochondria, and imported in an unfolded state. After proteolytic processing, they are folded and acquire the FeS cluster cofactor (red diamonds) by action of the chaperones and glutaredoxin.
Most mitochondrial proteins, including FeS cluster proteins, are translated on cytoplasmic ribosomes as precursors with mitochondrial targeting sequences. Upon import into mitochondria, the unfolded precursors are subjected to various processing steps that remove the targeting sequences (26). At this point, the processed apoproteins become substrates for a machinery dedicated to transferring the Isu cluster intermediate to recipient proteins. The scaffold Isu displays an interaction site consisting of the tripeptide sequence PVK, and this mediates binding to the Hsp70 chaperone (Ssq1 in yeast and HSPA9 in humans) (27, 28). This interaction is modulated by the ATP hydrolysis cycle in conjunction with accessory proteins Jac1 and Mge1 (29). A monothiol glutaredoxin, Grx5, is also implicated here (30). Through concerted action of the chaperones, glutaredoxin, and accessory proteins, the Isu FeS cluster intermediate is transferred to apoproteins.
A compartment problem exists for eukaryotic cells in that many FeS cluster proteins reside in the cytoplasm and in the nucleus. The biogenesis of these FeS clusters requires both mitochondrial and extramitochondrial machineries (31, 32). In this regard, it is notable that small amounts of frataxin have been found outside mitochondria, and a role in synthesis of extramitochondrial FeS clusters has been proposed (7).
Placement of Yfh1 in the Pathway of Mitochondrial FeS Cluster Assembly
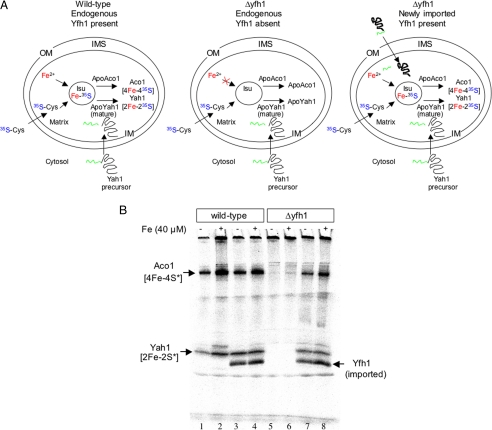
An experiment is described here that illustrates the role of frataxin in promoting FeS cluster assembly in mitochondria. Mitochondria were isolated from wild-type or Δyfh1 yeast strains. New synthesis of FeS clusters in the isolated organelles was examined by providing [35S]cysteine as a source of sulfide. In the wild-type strain, radioactive sulfide was incorporated into a pool of apoaconitase by the mitochondrial FeS cluster machinery. Synthesis of new clusters was also detected by labeling of imported ferredoxin, and these could be distinguished by signals on a native gel (Fig. 2, upper panel, left; and lower panel, lanes 1 and 2). In the mutant, however, no such labeling occurred, indicating failure to make FeS clusters (Fig. 2, upper panel, middle; and lower panel, lanes 5 and 6). Subsequently, frataxin was imported into the mutant Δyfh1 mitochondria, and the mitochondria were recovered by centrifugation. Now, when a similar [35S]cysteine labeling experiment was performed, the mutant mitochondria demonstrated restored ability to make FeS clusters on both aconitase and imported ferredoxin (Fig. 2, upper panel, right; and lower panel, lanes 7 and 8). Thus, a very small amount of frataxin (estimated at <1 pmol) imported into isolated Δyfh1 mitochondria even without iron addition was able to rapidly (in <10 min) restore FeS cluster-synthesizing activity. This effect in isolated mitochondria shows that everything is present in mitochondria for FeS cluster formation and that a small quantity of frataxin is capable of activating or mediating the process (23).
FIGURE 2.
Imported frataxin (Yfh1) restores FeS cluster assembly in isolated yeast mitochondria. Upper panel, wild-type mitochondria (left) synthesize FeS clusters on endogenous apoaconitase and imported apoYah1. Mutant Δyfh1 mitochondria (middle) fail to synthesize clusters. Following Yfh1 precursor import, mutant Δyfh1 mitochondria (right) now synthesize clusters. OM, outer membrane; IMS, intermembrane space; IM, inner membrane. Lower panel, mitochondria were isolated from wild-type or Δyfh1 strains. Full-length yeast frataxin (produced and radiolabeled on methionine and cysteine in the polypeptide) was imported (18 °C, 10 min) (lanes 3, 4, 7, and 8), and mitochondria were recovered by centrifugation. In the second stage, [35S]cysteine and ferredoxin precursor Yah1 were imported (30 °C, 10 min) with or without added iron. The mitochondria were recovered, and the matrix fraction was analyzed on a native gel and by autoradiography (lanes 1–8). The imported Yfh1 labeled on the polypeptide is visible on the gel. The imported ferredoxin labeled on the sulfur of the FeS is visible in Δyfh1 mitochondria only following Yfh1 import (lanes 7 and 8). Similarly, endogenous aconitase labeled on its FeS cluster is visible in Δyfh1 only following Yfh1 import (lanes 7 and 8) (23).
To begin to address where frataxin might act to promote FeS cluster assembly, it was important to determine whether frataxin acts before or after Isu. In one approach to this problem, a form of metabolic labeling was devised in which radioactive iron was added to growing yeast cells, and the radiolabeled Isu FeS cluster intermediate was recovered by immunoprecipitation (33). This approach was combined with promoter swaps to produce cellular depletion of one or another of the assembly component(s) prior to iron labeling and Isu recovery. A reduction in iron labeling of Isu would indicate decreased formation of FeS cluster intermediates. In that case, the depleted component was placed upstream of Isu because it was necessary for intermediate formation. In the case of increased iron labeling of Isu, the component was placed downstream of Isu because it was needed for transfer of the Isu clusters to recipient apoproteins. Using this assay design, Nfs1, Arh1, and Yah1 were placed upstream, and Ssq1, Jac1, and Grx5 were placed downstream of Isu in the FeS cluster assembly process. Frataxin was placed upstream because frataxin-depleted cells failed to efficiently form the radiolabeled Isu intermediate (34).
A completely independent set of observations led to a similar conclusion. The manganese superoxide dismutase (Mn-SOD),2 the dismutase of the mitochondrial matrix, was found to incorporate iron instead of manganese in some settings, with concomitant inactivation of the enzyme. For example, some of the FeS cluster assembly mutants with a mitochondrial iron accumulation phenotype were associated with Mn-SOD inactivation secondary to iron incorporation. Specifically, these mutants, such as ssq1 and grx5, belonged to Isu downstream events in transfer of FeS cluster intermediates to apoproteins (35). By contrast, although frataxin mutants (Δyfh1) dramatically accumulated iron in mitochondria, Mn-SOD was active because the iron was not available for insertion (36). Similarly, Isu mutants (depletion or dominant-negative) were also not associated with Mn-SOD inactivation (35). These data suggest that frataxin should be grouped with Isu in the upstream part of the FeS cluster biogenesis pathway and distinguished from Ssq1 and Grx5. Although mismetallation is an aberrant situation, these observations may point to a physiological role of frataxin in modulating bioavailable iron pools in mitochondria.
Frataxin Structure, Isu Interaction, and Iron Interaction
Structures have been obtained for yeast (37), human (38), and bacterial (39) frataxins. They are strikingly alike, characterized by an αβ-sandwich motif creating the two protein planes. The α-helical plane consists of two parallel (N- and C-terminal) α-helices, and the β-sheet plane consists of five antiparallel β-strands; a sixth and possibly seventh strand intersect the planes depending on the species (40). The structure and length of the respective N termini vary among orthologs, whereas the unstructured C termini, also of variable length, help control protein stability (41). A negatively charged surface is created by clustering of contiguous acidic residues contributed by portions of helix 1 and β-strand 1. Although the individual residues are not always conserved, all known frataxins are characterized by the presence of such an acidic surface. A second conserved structural feature is a neutral flat area on the β-sheet surface, which is more hydrophobic toward the center and more hydrophilic toward the periphery. This region contains a number of perfectly conserved residues and appears to be well suited to mediate an interaction with a protein partner.
Several lines of evidence point to Isu as a frataxin protein partner. A functional relationship of frataxin and Isu was initially suggested by genetic experiments showing synthetic lethality of Δyfh1 (deletion of the YFH1 gene) and Δisu1 (deletion of one of two homologous ISU genes, causing lowered levels of Isu) (42). Physical interactions between frataxin and Isu were subsequently demonstrated in pulldown experiments from mitochondrial lysates. Interestingly, the pulldowns of Yfh1 by Isu, or reciprocally of Isu by Yfh1, were dependent on iron addition to the buffer and were inhibited by the presence of metal chelators (43). Iron dependence of a frataxin-Isu interaction was also shown for the purified human proteins (44). The clearest demonstration of the importance of frataxin-Isu interaction in FeS cluster assembly was provided by careful analyses of frataxin proteins with substitutions of surface-exposed amino acids in the interaction interface (Fig. 3A). Alteration of conserved surface-exposed residues of β-strand 3 (N122A/K123T/Q124A) was associated with severely impaired frataxin-Isu interaction (pulldown of Isu from a lysate on immobilized Yfh1-His6). The same mutant frataxin expressed in cells was associated with decreased FeS cluster protein enzyme activities. Other substituted forms of frataxin with alterations in exposed areas of the β-sheets (Q129A in strand 4, W131A in strand 4, and R141A in strand 5) (Fig. 3A) were studied in vitro and in vivo. These various amino acid changes were each associated with decreased frataxin-Isu interaction and deficient activities of FeS cluster proteins (45). These studies with minimally altered frataxins were especially informative because they avoided many of the secondary phenotypes associated with yfh1 deletion. The conclusion from these studies is that the interaction between frataxin and Isu in mitochondria is required for FeS cluster assembly.
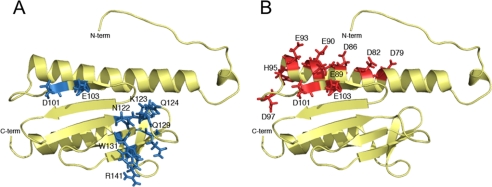
FIGURE 3.
A, solution structure of monomeric apo-Yfh1 (Protein Data Bank code 2GA5) with residues involved in Isu binding identified in blue. Yfh1 amino acid substitutions with effects on Isu interactions are as follows: N122A/K123T/Q124A, diminished Isu interaction, low aconitase and succinate dehydrogenase, and iron accumulation (58); single amino acid changes Q129A, W131A, and R141A, diminished Isu interaction and low aconitase (45); and D101A/E103A, no interaction with Isu and decreased aconitase (50). B, solution structure of monomeric apo-Yfh1 with residues involved in iron binding identified in red (37). Published data on frataxins with substitutions of acidic amino acids are as follows: D86A/E90A/E93A/D101A/E103A, decreased iron-binding affinity, low aconitase, and slow growth (50); D86A/E89A/E101A/E103A, decreased iron-dependent Isu interaction, low aconitase, and iron accumulation (51); D86A/E90A/E93A, no oligomerization in response to iron exposure in vitro, normal Isu interaction, no FeS deficit, and no iron accumulation (53); D79A/D82A, low ferroxidase, oxidant sensitivity, normal aconitase, and no iron accumulation (69); and E93A/D97A/E103A, oxidant sensitivity, normal aconitase, and no iron accumulation (69). C-term and N-term, C and N termini.
Frataxins interact with iron in vitro, although not in the manner of typical iron-binding proteins. Binding is relatively low affinity, occurs on the protein surface rather than in a cavity, and is mediated primarily by carboxylic amino acids rather than cysteines and histidines (46). Analysis of iron interactions with yeast frataxin monomers showed two ferrous iron atoms binding with an affinity of 2.5–5 μm as high spin ferrous iron. Chemical shifts and line broadening induced by exposure of 15N-labeled yeast frataxin to paramagnetic iron (ferrous iron under anaerobic conditions) further defined iron-binding residues as primarily carboxylate-containing amino acids in the helix 1-strand 1 region (37, 47). Studies with other frataxin orthologs gave similar results, although with some variation in iron-binding stoichiometry and affinity: the human ortholog bound six atoms with Kd = 12–55 μm (44), the Escherichia coli ortholog bound two atoms with Kd = 4 μm (48), and the Drosophila ortholog bound one atom with Kd = 6 μm (49).
Mutagenesis studies performed with the objective of correlating in vitro iron binding and in vivo function have been problematic in part because of the redundant nature of the iron-binding site(s) distributed over a surface and mediated by multiple iron-binding amino acids. Alterations of single acidic residues showed minimal effects. However, frataxins with multiple acidic residues changed to alanine were compromised in terms of iron binding and in terms of supporting FeS cluster formation. For example, an altered frataxin with Asp86, Glu90, Glu93, Asp101, and Glu103 all changed to alanine was associated with decreased iron binding by tryptophan fluorescence titration and decreased FeS cluster enzyme activities (50). Similarly, frataxin with Asp86, Glu89, Asp101, and Glu103 changed to alanine (Fig. 3B) was associated with decreased aconitase activity, iron accumulation in mitochondria, and abrogation of iron-dependent Isu interaction in a mitochondrial lysate (51). Some frataxins oligomerize when exposed to iron. Exposure of yeast or E. coli frataxins to a 20-fold molar excess of elemental iron in the absence of competing cations induced oligomerization, but this did not occur with the human protein (52). Mutation of three carboxylates (Asp86, Glu90, and Glu93) to alanine (Fig. 3B) completely abrogated iron-dependent oligomerization of the yeast protein. When tested in vivo, this triple mutant showed no deleterious effects on FeS cluster protein activities and iron homeostasis, even when expressed at low levels (53); and thus, frataxin oligomerization is probably not required for FeS cluster synthesis.
How Frataxin Might Work
In summary, frataxin interacts with the FeS cluster assembly scaffold protein Isu using the frataxin β-sheet surface. It also interacts with iron via surface carboxylates of the α-helix 1/β-strand 1 area. Furthermore, the frataxin-Isu interaction is iron-dependent. Various in vitro assays have demonstrated these interactions, and in vivo experiments with mutant alleles have shown the importance of these interactions for FeS cluster formation. Biochemical data show that frataxin facilitates iron use for FeS cluster formation on Isu (44, 49). Frataxin might work as a metallochaperone, similar to copper metallochaperones. Copper chaperones function by high affinity interactions with copper and with recipient proteins. Copper is bound to the chaperone, then liganded between the chaperone and target protein, and finally handed off to the target protein. The copper is thus never free in solution and is targeted to its correct destination in the cell by the protein-protein interactions between the chaperone and target (54). Frataxin might work in a similar fashion by transiently binding iron in mitochondria, interacting with Isu in an iron-dependent fashion, transferring protein-bound iron to Isu, and finally disengaging from Isu after metal delivery. However, the affinity of frataxin for iron is relatively low and coordination is incomplete, with the carboxylate-binding sites being completed by water molecules. The driving force and mechanism of metal transfer to recipients also might be different from those for the copper chaperones. A prediction of this model is that the iron-liganding sites should overlap or be contiguous with the Isu-binding sites. This may be the case especially for the iron-binding site on the strand 1 sheet consisting of the conserved Asp101 and Glu103 residues (Fig. 3, A and B). Other functions in mitochondrial iron trafficking are also possible. In line with the iron chaperone idea, frataxin might bind iron and bind to other proteins, thereby targeting the metal for delivery to aconitase (55), ferrochelatase (14, 37), and succinate dehydrogenase (56). Iron-specific activities of frataxin have also been suggested, and some bypass of frataxin mutant phenotypes, including FeS cluster deficiencies, has been observed as a result of forced expression of mitochondrial ferritin (57).
Alternatively, the function of frataxin in FeS cluster biogenesis might be primarily a regulatory one. The iron-dependent frataxin-Isu interaction might signal formation of a protein complex for creation of the FeS intermediate on Isu. Of note, pulldown experiments with mitochondria using yeast frataxin as bait yielded not only Isu but also Nfs1 (43) and Isd11 (58), the functional cysteine desulfurase complex responsible for providing sulfide for FeS. Perhaps the data are pointing to the existence of a multisubunit complex consisting of frataxin, Nfs1/Isd11, and Isu. Such a complex would be well situated to mediate formation of the Isu-bound FeS cluster intermediate. Iron and sulfide are toxic intermediates, and their insertion into FeS clusters must be controlled to avoid toxicity. Frataxin might regulate the flux of these key intermediates, permitting delivery of enough for physiological FeS cluster formation and not so much as to create iron and/or sulfide toxicity. The source of iron and its biological form are not well understood. The sulfide generated by the action of Nfs1/Isd11 on cysteine may be provided to Isu as persulfide by direct protein interaction as observed for some bacterial components. A role for frataxin in regulating sulfide transfer has been proposed in bacteria (59, 60). For eukaryotes, regulation of sulfide delivery for FeS has not been defined, and structural details of the frataxin-Nfs1/Isd11 interaction remain to be clarified. The details of the frataxin-Isu interaction from the Isu side also have not been solved. These interactions might compete with Isu-chaperone interactions that mediate transfer of the cluster intermediate to apoproteins.
Is Friedreich Ataxia a Disease of FeS Cluster Assembly?
The molecular basis of Friedreich ataxia is deficiency of frataxin protein. The question then arises as to how frataxin deficiency causes the disease. As far as is known, neither the cellular phenotype nor the human disease phenotype can be explained by deficiency of a single FeS cluster protein. Perhaps it is the combined effects from decreased activities of many FeS cluster proteins that are pathogenic. Also the degree of frataxin deficiency may be important for producing the unique cellular disease phenotypes. A complete lack of frataxin is lethal for growing mammalian cells, and somewhere between 20 and 30% is needed for normal growth. In cells with decreased levels of frataxin, some FeS cluster proteins are deficient, iron accumulates in mitochondria, and oxidant sensitivity is observed as in the human disease (16). The causal links among these effects are not well defined, and an FeS cluster protein mediating the mitochondrial iron accumulation or oxidant sensitivity has not been identified. Conversely, several lines of evidence indicate that mitochondrial iron accumulation (61, 62) and oxidative stress (63) worsen the cellular phenotypes, perhaps mediated by detrimental effects on FeS clusters or perhaps mediated independently. Interventions that limit cellular iron have improved growth of yeast mutants in some cases and have had some efficacy in treatment of the human disease (64). However, opposite effects have also been observed (i.e. worsening of phenotypes associated with iron starvation) (65, 66). One final issue merits mention, i.e. the tissue specificity of the disease. FeS cluster biogenesis is required for every cell in every tissue, as many essential processes require FeS cluster proteins. Frataxin is ubiquitously expressed in all cell types. Therefore, it is unclear why some tissues are compromised by frataxin deficiency and some are spared. The target tissue distribution for the disease includes dorsal root ganglia, cerebellum, and heart muscle and does not involve skeletal muscle or blood, for example. By contrast, an inherited disease ascribed to Isu deficiency, also characterized by deficient FeS cluster proteins, affects primarily skeletal muscle and spares nervous tissues (67, 68). Thus, Isu deficiency and frataxin deficiency in humans bear some resemblance, but the tissue specificities are distinct with the differences still unexplained.
This work was supported, in whole or in part, by National Institutes of Health Grants DK068139 (to T. L. S.), AG030504 (to D. P.), and R37DK053953 (to A. D.). This work was also supported by an Association Francaise de l'Ataxie de Friedreich grant (to E. L.) and American Heart Association Grant 09GRNT2260364 (to D. P.). This is the second article in the Thematic Minireview Series on Metals in Biology 2010. This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
- Mn-SOD
- manganese superoxide dismutase.
REFERENCES
- 1.Pandolfo M. (2009) J. Neurol. 256, Suppl. 1, 3–8 [DOI] [PubMed] [Google Scholar]
- 2.Campuzano V., Montermini L., Moltò M. D., Pianese L., Cossée M., Cavalcanti F., Monros E., Rodius F., Duclos F., Monticelli A., Zara F., Cañizares J., Koutnikova H., Bidichandani S. I., Gellera C., Brice A., Trouillas P., De Michele G., Filla A., De Frutos R., Palau F., Patel P. I., Di Donato S., Mandel J. L., Cocozza S., Koenig M., Pandolfo M. (1996) Science 271, 1423–1427 [DOI] [PubMed] [Google Scholar]
- 3.Wilson R. B., Roof D. M. (1997) Nat. Genet. 16, 352–357 [DOI] [PubMed] [Google Scholar]
- 4.Campuzano V., Montermini L., Lutz Y., Cova L., Hindelang C., Jiralerspong S., Trottier Y., Kish S. J., Faucheux B., Trouillas P., Authier F. J., Dürr A., Mandel J. L., Vescovi A., Pandolfo M., Koenig M. (1997) Hum. Mol. Genet. 6, 1771–1780 [DOI] [PubMed] [Google Scholar]
- 5.Gordon D. M., Shi Q., Dancis A., Pain D. (1999) Hum. Mol. Genet. 8, 2255–2262 [DOI] [PubMed] [Google Scholar]
- 6.Branda S. S., Cavadini P., Adamec J., Kalousek F., Taroni F., Isaya G. (1999) J. Biol. Chem. 274, 22763–22769 [DOI] [PubMed] [Google Scholar]
- 7.Condo I., Malisan F., Guccini I., Serio D., Rufini A., Testi R. (2010) Hum. Mol. Genet. 19, 1221–1229 [DOI] [PubMed] [Google Scholar]
- 8.Rötig A., de Lonlay P., Chretien D., Foury F., Koenig M., Sidi D., Munnich A., Rustin P. (1997) Nat. Genet. 17, 215–217 [DOI] [PubMed] [Google Scholar]
- 9.Johnson D. C., Dean D. R., Smith A. D., Johnson M. K. (2005) Annu. Rev. Biochem. 74, 247–281 [DOI] [PubMed] [Google Scholar]
- 10.Gibson T. J., Koonin E. V., Musco G., Pastore A., Bork P. (1996) Trends Neurosci. 19, 465–468 [DOI] [PubMed] [Google Scholar]
- 11.Dolezal P., Dancis A., Lesuisse E., Sutak R., Hrdý I., Embley T. M., Tachezy J. (2007) Eukaryot. Cell 6, 1431–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldberg A. V., Molik S., Tsaousis A. D., Neumann K., Kuhnke G., Delbac F., Vivares C. P., Hirt R. P., Lill R., Embley T. M. (2008) Nature 452, 624–628 [DOI] [PubMed] [Google Scholar]
- 13.Babcock M., de Silva D., Oaks R., Davis-Kaplan S., Jiralerspong S., Montermini L., Pandolfo M., Kaplan J. (1997) Science 276, 1709–1712 [DOI] [PubMed] [Google Scholar]
- 14.Lesuisse E., Santos R., Matzanke B. F., Knight S. A., Camadro J. M., Dancis A. (2003) Hum. Mol. Genet. 12, 879–889 [DOI] [PubMed] [Google Scholar]
- 15.Chen O. S., Hemenway S., Kaplan J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12321–12326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calmels N., Schmucker S., Wattenhofer-Donzé M., Martelli A., Vaucamps N., Reutenauer L., Messaddeq N., Bouton C., Koenig M., Puccio H. (2009) PLoS One 4, e6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rouault T. A., Tong W. H. (2005) Nat. Rev. Mol. Cell Biol. 6, 345–351 [DOI] [PubMed] [Google Scholar]
- 18.Lill R., Dutkiewicz R., Elsässer H. P., Hausmann A., Netz D. J., Pierik A. J., Stehling O., Urzica E., Mühlenhoff U. (2006) Biochim. Biophys. Acta 1763, 652–667 [DOI] [PubMed] [Google Scholar]
- 19.Frazzon J., Fick J. R., Dean D. R. (2002) Biochem. Soc. Trans. 30, 680–685 [DOI] [PubMed] [Google Scholar]
- 20.Wiedemann N., Urzica E., Guiard B., Müller H., Lohaus C., Meyer H. E., Ryan M. T., Meisinger C., Mühlenhoff U., Lill R., Pfanner N. (2006) EMBO J. 25, 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adam A. C., Bornhövd C., Prokisch H., Neupert W., Hell K. (2006) EMBO J. 25, 174–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mühlenhoff U., Stadler J. A., Richhardt N., Seubert A., Eickhorst T., Schweyen R. J., Lill R., Wiesenberger G. (2003) J. Biol. Chem. 278, 40612–40620 [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y., Lyver E. R., Knight S. A., Pain D., Lesuisse E., Dancis A. (2006) J. Biol. Chem. 281, 22493–22502 [DOI] [PubMed] [Google Scholar]
- 24.Lange H., Kaut A., Kispal G., Lill R. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 1050–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bandyopadhyay S., Chandramouli K., Johnson M. K. (2008) Biochem. Soc. Trans. 36, 1112–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neupert W., Herrmann J. M. (2007) Annu. Rev. Biochem. 76, 723–749 [DOI] [PubMed] [Google Scholar]
- 27.Dutkiewicz R., Schilke B., Cheng S., Knieszner H., Craig E. A., Marszalek J. (2004) J. Biol. Chem. 279, 29167–29174 [DOI] [PubMed] [Google Scholar]
- 28.Schilke B., Williams B., Knieszner H., Pukszta S., D'Silva P., Craig E. A., Marszalek J. (2006) Curr. Biol. 16, 1660–1665 [DOI] [PubMed] [Google Scholar]
- 29.Dutkiewicz R., Schilke B., Knieszner H., Walter W., Craig E. A., Marszalek J. (2003) J. Biol. Chem. 278, 29719–29727 [DOI] [PubMed] [Google Scholar]
- 30.Rodríguez-Manzaneque M. T., Tamarit J., Bellí G., Ros J., Herrero E. (2002) Mol. Biol. Cell 13, 1109–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rouault T. A., Tong W. H. (2008) Trends Genet. 24, 398–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lill R., Mühlenhoff U. (2008) Annu. Rev. Biochem. 77, 669–700 [DOI] [PubMed] [Google Scholar]
- 33.Pierik A. J., Netz D. J., Lill R. (2009) Nat. Protoc. 4, 753–766 [DOI] [PubMed] [Google Scholar]
- 34.Mühlenhoff U., Gerber J., Richhardt N., Lill R. (2003) EMBO J. 22, 4815–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naranuntarat A., Jensen L. T., Pazicni S., Penner-Hahn J. E., Culotta V. C. (2009) J. Biol. Chem. 284, 22633–22640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang M., Cobine P. A., Molik S., Naranuntarat A., Lill R., Winge D. R., Culotta V. C. (2006) EMBO J. 25, 1775–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He Y., Alam S. L., Proteasa S. V., Zhang Y., Lesuisse E., Dancis A., Stemmler T. L. (2004) Biochemistry 43, 16254–16262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dhe-Paganon S., Shigeta R., Chi Y. I., Ristow M., Shoelson S. E. (2000) J. Biol. Chem. 275, 30753–30756 [DOI] [PubMed] [Google Scholar]
- 39.Nair M., Adinolfi S., Pastore C., Kelly G., Temussi P., Pastore A. (2004) Structure 12, 2037–2048 [DOI] [PubMed] [Google Scholar]
- 40.Bencze K. Z., Kondapalli K. C., Cook J. D., McMahon S., Millán-Pacheco C., Pastor N., Stemmler T. L. (2006) Crit. Rev. Biochem. Mol. Biol. 41, 269–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adinolfi S., Nair M., Politou A., Bayer E., Martin S., Temussi P., Pastore A. (2004) Biochemistry 43, 6511–6518 [DOI] [PubMed] [Google Scholar]
- 42.Ramazzotti A., Vanmansart V., Foury F. (2004) FEBS Lett. 557, 215–220 [DOI] [PubMed] [Google Scholar]
- 43.Gerber J., Mühlenhoff U., Lill R. (2003) EMBO Rep. 4, 906–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoon T., Cowan J. A. (2003) J. Am. Chem. Soc. 125, 6078–6084 [DOI] [PubMed] [Google Scholar]
- 45.Leidgens S., De Smet S., Foury F. (2010) Hum. Mol. Genet. 19, 276–286 [DOI] [PubMed] [Google Scholar]
- 46.Pastore C., Franzese M., Sica F., Temussi P., Pastore A. (2007) FEBS J. 274, 4199–4210 [DOI] [PubMed] [Google Scholar]
- 47.Cook J. D., Bencze K. Z., Jankovic A. D., Crater A. K., Busch C. N., Bradley P. B., Stemmler A. J., Spaller M. R., Stemmler T. L. (2006) Biochemistry 45, 7767–7777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bou-Abdallah F., Adinolfi S., Pastore A., Laue T. M., Dennis Chasteen N. (2004) J. Mol. Biol. 341, 605–615 [DOI] [PubMed] [Google Scholar]
- 49.Kondapalli K. C., Kok N. M., Dancis A., Stemmler T. L. (2008) Biochemistry 47, 6917–6927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Correia A. R., Wang T., Craig E. A., Gomes C. M. (2010) Biochem. J. 426, 197–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Foury F., Pastore A., Trincal M. (2007) EMBO Rep. 8, 194–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adinolfi S., Trifuoggi M., Politou A. S., Martin S., Pastore A. (2002) Hum. Mol. Genet. 11, 1865–1877 [DOI] [PubMed] [Google Scholar]
- 53.Aloria K., Schilke B., Andrew A., Craig E. A. (2004) EMBO Rep. 5, 1096–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O'Halloran T. V., Culotta V. C. (2000) J. Biol. Chem. 275, 25057–25060 [DOI] [PubMed] [Google Scholar]
- 55.Bulteau A. L., O'Neill H. A., Kennedy M. C., Ikeda-Saito M., Isaya G., Szweda L. I. (2004) Science 305, 242–245 [DOI] [PubMed] [Google Scholar]
- 56.González-Cabo P., Vázquez-Manrique R. P., García-Gimeno M. A., Sanz P., Palau F. (2005) Hum. Mol. Genet. 14, 2091–2098 [DOI] [PubMed] [Google Scholar]
- 57.Campanella A., Isaya G., O'Neill H. A., Santambrogio P., Cozzi A., Arosio P., Levi S. (2004) Hum. Mol. Genet. 13, 2279–2288 [DOI] [PubMed] [Google Scholar]
- 58.Wang T., Craig E. A. (2008) J. Biol. Chem. 283, 12674–12679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Layer G., Ollagnier-de Choudens S., Sanakis Y., Fontecave M. (2006) J. Biol. Chem. 281, 16256–16263 [DOI] [PubMed] [Google Scholar]
- 60.Adinolfi S., Iannuzzi C., Prischi F., Pastore C., Iametti S., Martin S. R., Bonomi F., Pastore A. (2009) Nat. Struct. Mol. Biol. 16, 390–396 [DOI] [PubMed] [Google Scholar]
- 61.Foury F. (1999) FEBS Lett. 456, 281–284 [DOI] [PubMed] [Google Scholar]
- 62.Radisky D. C., Babcock M. C., Kaplan J. (1999) J. Biol. Chem. 274, 4497–4499 [DOI] [PubMed] [Google Scholar]
- 63.Bulteau A. L., Dancis A., Gareil M., Montagne J. J., Camadro J. M., Lesuisse E. (2007) Free Radic. Biol. Med. 42, 1561–1570 [DOI] [PubMed] [Google Scholar]
- 64.Kakhlon O., Manning H., Breuer W., Melamed-Book N., Lu C., Cortopassi G., Munnich A., Cabantchik Z. I. (2008) Blood 112, 5219–5227 [DOI] [PubMed] [Google Scholar]
- 65.Goncalves S., Paupe V., Dassa E. P., Rustin P. (2008) BMC Neurol. 8, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li K., Besse E. K., Ha D., Kovtunovych G., Rouault T. A. (2008) Hum. Mol. Genet. 17, 2265–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mochel F., Knight M. A., Tong W. H., Hernandez D., Ayyad K., Taivassalo T., Andersen P. M., Singleton A., Rouault T. A., Fischbeck K. H., Haller R. G. (2008) Am. J. Hum. Genet. 82, 652–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Olsson A., Lind L., Thornell L. E., Holmberg M. (2008) Hum. Mol. Genet. 17, 1666–1672 [DOI] [PubMed] [Google Scholar]
- 69.Gakh O., Park S., Liu G., Macomber L., Imlay J. A., Ferreira G. C., Isaya G. (2006) Hum. Mol. Genet. 15, 467–479 [DOI] [PubMed] [Google Scholar]