Abstract
FeS cluster biogenesis is an essential process in virtually all forms of life. Complex protein machineries that are conserved from bacteria through higher eukaryotes facilitate assembly of the FeS cofactor in proteins. In the last several years, significant strides have been made in our understanding of FeS cluster assembly and the functional overlap of this process with cellular iron homeostasis. This minireview summarizes the present understanding of the cytosolic iron-sulfur cluster assembly (CIA) system in eukaryotes, with a focus on information gained from studies in budding yeast and mammalian systems.
Keywords: Iron, Iron Metabolism, Iron-Sulfur Protein, Metalloproteins, Metals
Introduction
The chemical and structural versatility of FeS clusters makes these cofactors uniquely suited to participate in a diverse set of cellular processes (1). FeS clusters are used as part of catalytic centers, for chemical sensing, to stabilize protein structure, to transfer electrons, to generate radicals, and to determine protein function (1, 2). Organisms rely on these inorganic cofactors for critical roles in cellular metabolism and regulation (1). Comprehensive reviews on FeS protein biogenesis have been published recently (3–7), and we will not attempt to replicate them here. The focus of this minireview is the maturation of FeS proteins in the eukaryotic cytosol, with emphasis on the emerging CIA2 system in Saccharomyces cerevisiae and animals and how this process is linked to cellular iron homeostasis.
Interest in FeS cluster biogenesis in the eukaryotic cytosol intensified with the discovery that mammalian IRP1 is an FeS protein whose activity as a RNA-binding gene regulator is controlled through cluster assembly and disassembly (8). Cellular iron status determines the extent of FeS cluster assembly in IRP1 and thereby regulates expression of genes for iron storage, transport, and utilization (8). FeS proteins are now recognized to contribute to processes covering virtually all areas of cell biology, including DNA metabolism, protein synthesis, transcription, and iron metabolism itself (Table 1), making the biogenesis of the FeS cofactor a centrally important, essential process.
TABLE 1.
Yeast and mammalian extramitochondrial FeS proteins
h, human; SAM, S-adenosylmethionine; SNARE, soluble N-ethylmaleimide-sensitive factor attachment protein receptor.
| Yeast | Mammalian | Localization | Function | Ref. |
|---|---|---|---|---|
| Genome integrity and expression | ||||
| Ntg2 | hNTH1 | Nucleus | DNA N-glycosylase | 81 |
| Rad3 | XPD | Nucleus | DNA helicase, TFIIH transcription complex | 82 |
| Chl1 | FancJ | Nucleus | DNA helicase | 82 |
| Dna2 | Dna2 | Nucleus | ATP-dependant nuclease, helicase | 83 |
| Pri2 | PRIM2 | Nucleus | Subunit of DNA primase | 84 |
| Elp3 | Elp3 | Cytosol, nucleus | Subunit of elongator complex, transcription | 31 |
| MUTYH | Nucleus | DNA glycosylase | 85 | |
| Protein synthesis | ||||
| Rli1 | ABCE1 | Cytosol, nucleus | Ribosome maturation, translation | 86 |
| Tyw1 | TYW1 | Endoplasmic reticulum | Synthesis of wybutosine, tRNA modification | 87 |
| Leu1 | Cytosol | α-Isopropylmalate isomerase, leucine biosynthesis | 88 | |
| Ecm17 | Cytosol | Sulfite reductase, methionine biosynthesis | ||
| Iron metabolism | ||||
| Cfd1 | Nubp2 | Cytosol | FeS scaffold | 25 |
| Nbp35 | Nubp1 | Cytosol, nucleus | FeS scaffold | 25 |
| Nar1 | IOP1 | Cytosol, nucleus | FeS biogenesis | 28 |
| Dre2 | Ciapin1 | Cytosol | FeS biogenesis | 31 |
| Grx3/4 | PICOT | Cytosol | Iron regulation, monothiol glutaredoxin | 74 |
| Fra2 | Cytosol | Negative regulator of Aft1 | 74 | |
| IRP1 | Cytosol | Cellular iron homeostasis, cytosolic aconitase | 80 | |
| Intermediary metabolism | ||||
| Grx6 | Endoplasmic reticulum | Monothiol glutaredoxin | 89 | |
| Viperin | Endoplasmic reticulum | Radical SAM enzyme | 90 | |
| CMAH | Cytosol | Monooxygenase, hydroxylase | 91 | |
| DPD | Cytosol | Dihydropyrimidine dehydrogenase | 92 | |
| XDH | Cytosol | Xanthine dehydrogenase | 93 | |
| XOR | Cytosol | Xanthine oxidoreductase | 93 | |
| Sprouty2 | Cytosol | Inhibitor of receptor tyrosine kinase signaling | 94 | |
| Miner1 | Endoplasmic reticulum | Unknown | 95 | |
| SNAP-25 | Plasma membrane | Component of SNARE complex | 96 | |
| IOP2 | Cytosol | Nar1 homolog, unknown function | 42 | |
FeS Cluster Biogenesis in Non-photosynthetic Eukaryotes
Early studies of nitrogenase in nitrogen-fixing bacteria were instrumental in revealing the need for specialized proteins for assembly of FeS clusters in proteins (2). Genome sequencing combined with a high degree of conservation of genes involved in FeS cluster biogenesis across species accelerated identification of systems for cluster biogenesis, including the ISC system in mitochondria and bacteria and the SUF (sulfur formation) system in bacteria, archaea, and plant chloroplasts (2–6). These protein-assisted FeS cluster assembly systems follow a common strategy, which may represent the fundamental steps in all FeS cluster assembly systems. Specifically, sulfide is generated from cysteine via a cysteine desulfurase (9). The enzyme captures and donates the sulfide through an enzyme-linked persulfide (10, 11). Iron also appears to enter the process bound to protein (12). Both elements are preassembled into a labile FeS cluster on a scaffold protein, from which the cluster is transferred to apo-FeS proteins. Although FeS cluster transfer to apo-targets by entropy-driven ligand exchange has been demonstrated (13), the mechanism of transfer in the cell is likely more complex.
The ISC system of mitochondria is composed of at least 15 proteins (6). Nfs1 and Isu1/2 are the cysteine desulfurase and major FeS scaffold proteins for this system. Nfs1 forms a complex with Isd11 in mitochondria, which modulates cysteine desulfurase activity (14). Initial FeS cluster assembly on the Isu1/2 scaffold requires sulfide from Nfs1/Isd11, iron from Yfh1 (frataxin), and electrons from Yah1/Arh1 (14, 15). Ssq1, Jac1, and Mge1, protein chaperon and co-chaperons, along with the glutaredoxin Grx5 facilitate transfer of the cluster from the Isu1/2 scaffold to apo-targets (16–18). Together, these factors compose the essential core activities for all FeS cluster biogenesis in the eukaryotic cell. Homologous proteins/activities make up the ISC systems in bacteria (Table 2) and archaea (3, 4, 6, 7). Maturation of subsets of FeS proteins in mitochondria requires additional assembly factors, including Ind1, Isa1/2, and Iba57 (19–21).
TABLE 2.
CIA and ISC FeS cluster assembly factors in yeast, mammals, and bacteria
hu, human.
| Cytoplasm (CIA): yeast (mammal) | Mitochondria (ISC and ISC export): yeast (mammal) | Bacteria | Proposed function |
|---|---|---|---|
| Cfd1, Nbp35 (NUBP2, NUBP1) | Ind1 (huIND1) | ApbC | P-loop NTPase, FeS scaffold |
| Nar1 (IOP1) | Electron donor, FeS cluster transfer | ||
| Cia1 (CIAO1) | CIA protein complex formation | ||
| Dre2 (CIAPIN1) | Electron transfer, ISC/CIA link | ||
| Nfs1 (NFS1) | IscS | Cysteine desulfurase | |
| Isd11 (LYRM4) | Sulfur transfer | ||
| Isu1, Isu2 (ISCU1, ISCU2) | IscU | FeS scaffold | |
| Nfu1 (NFU) | NfuA | FeS scaffold | |
| Isa1, Isa2 (ISCA1, ISCA2) | IscA | FeS scaffold | |
| Iba57 (C1orf69) | FeS cluster assembly factor for biotin synthase and Aco1-like proteins | ||
| Yah1 (FDX1) | Fdx | Ferredoxin | |
| Arh1 (Fdxr/ADR) | Ferredoxin reductase | ||
| Yfh1 (frataxin/FXN) | CyaY | Iron donor | |
| Ssq1 (mortalin/HSPA9) | HscA | Protein chaperone | |
| Jac1 (HSCB) | HscB | Co-chaperone | |
| Mge1 | GrpE | Co-chaperone, nucleotide exchange factor | |
| Grx5 (GLRX5) | Glutaredoxin | ||
| Atm1 (ABCB7) | ABC transporter, ISC export system | ||
| Erv1 (GFER) | Sulfhydryl oxidase, ISC export system |
Various explanations have been given for the requirement of the mitochondrial ISC system for extramitochondrial FeS cluster biogenesis. It was posited early on that all FeS cluster biogenesis occurred in mitochondria, with maturation of cytosolic FeS proteins depending on export of preformed clusters. This idea grew out of findings that, in addition to the ISC system, a putative ISC export system consisting of the inner mitochondrial membrane ABC transporter Atm1 and the intermembrane space protein Erv1 and glutathione were uniquely required for extramitochondrial FeS cluster biogenesis (22–24). The orientation of Atm1 indicated a role in export from the mitochondrial matrix, prompting the suggestion of export of FeS clusters (22). More recent evidence suggests that a form of sulfur generated via the ISC system may be the exported substance (25). Consistent with this view, peptides rich in sulfur-containing amino acids stimulated the ATPase activity of Atm1 (26).
The first exclusively cytosolic protein specific for extramitochondrial FeS protein maturation, Cfd1 (cytosolic FeS cluster-deficient 1), was identified in a yeast genetic screen based on the FeS cluster-dependent conversion of mammalian IRP1 to cytosolic aconitase in the yeast cytosol (27). Discovery of Cfd1 suggested a unique cytosolic FeS cluster assembly (CIA) system. This was confirmed when three additional protein components of the CIA system (Nar1, a member of the iron-only hydrogenase family (28); Nbp35, a P-loop NTPase homologous to Cfd1 (29); and Cia1, a WD40 protein (30)) were found. Depletion of any one of these proteins resulted in defective cytosolic and nuclear FeS protein maturation. A fifth CIA factor, Dre2, was identified in a search for mutations that were synthetic-lethal with deletion of the mitochondrial iron transport genes MRS3 and MRS4 (31). Loss of Mrs3 and Mrs4 impairs cluster assembly via the ISC system (32, 33). Thus, Dre2 may link the ISC and CIA systems for cytosolic FeS cluster assembly.
With the exception of Nfs1, which is needed in the nucleus for tRNA modification and maturation (34, 35), ISC factors in budding yeast are restricted to the mitochondria. However, in animal cells, some ISC factors are found in the cytosol, leading to the suggestion that these proteins function directly in cytosolic FeS protein maturation (36–39). Although the notion of ISC function in the cytosol has remained controversial and unresolved, recent observations support a specific role for ISC factors in the cytosol of mammalian cells. For example, a cytosolic isoform of frataxin restored cytosolic aconitase and IRE-binding activity of IRP1 to normal levels in frataxin-deficient lymphoblasts derived from a Friedreich ataxia patient (36). Mitochondrial aconitase activity was unaltered, indicating that the effect of this frataxin isoform was specific to the cytosol. A physical interaction between IRP1 and frataxin was also detected (36).
The mammalian Nar1 homolog IOP1 (iron-only hydrogenase-like protein 1) was shown to interact with a cytosolic isoform of Isa1 (40), raising the possibility of extramitochondrial cooperation between CIA and ISC. Although cytosolic isoforms of the ISC factors Nfs1, Isu1, and frataxin have been reported to be important for cytosolic FeS cluster biogenesis (36, 38), the possibility of functional cooperation between the ISC and CIA systems is an exciting avenue to be explored.
The CIA System
CIA proteins are defined by having a primary location in the cytoplasm and a requirement for their function in cytosolic and not mitochondrial FeS protein maturation. This distinguishes CIA proteins from ISC factors that are important for both mitochondrial and cytosolic FeS cluster biogenesis and ISC export proteins that are required for cytosolic cluster biogenesis but are located exclusively within the mitochondria. The number and nature of the FeS proteins dependent on cytosolic cluster biogenesis suggest a critical role for CIA in cell biology (Table 1). Consistent with this view, each of the CIA factor genes is essential in yeast (27–31), and their depletion slows growth of animal cells (37, 41, 42). To date, only [4Fe-4S] proteins have been shown to require the CIA system for maturation.
Cfd1 and Nbp35
The current thinking is that Cfd1 and Nbp35 are the scaffolds for initial FeS cluster assembly in the CIA system. These P-loop NTPases bear high sequence similarity (∼49% identity) but are not redundant (25). An important difference between the two proteins is at the N terminus, where Nbp35 has an extension of ∼50 amino acids that contains four conserved cysteine residues implicated in binding a [4Fe-4S] cluster (25). Deletion of the first 52 residues or mutation of the two central N-terminal cysteines in yeast Nbp35 was lethal, consistent with an essential role for the putative FeS cluster within this region (25, 43).
The identification of Cfd1 as an essential factor for extramitochondrial FeS protein maturation and the demonstration in bacteria of a role for the homologous protein ApbC in cluster biogenesis revealed a new family of proteins involved in FeS cluster assembly (27, 44). Cfd1, Nbp35, and their homologs throughout nature belong to a class of deviant P-loop NTPases that includes the bacterial cell division protein MinD, the iron protein of nitrogenase NifH, and the arsenic resistance ATPase ArsA (27, 29, 45–49). This class of NTPases typically forms homodimers in which a signature lysine (Lys26 in Cfd1 and Lys81 in Nbp35) within the Walker A (nucleotide-binding) motif of one monomer extends into the nucleotidase active site of the other monomer and plays a role in ATP binding and/or hydrolysis (49).
Cfd1 and Nbp35 belong to a subfamily of deviant P-loop NTPases often referred to as the MRP/Nbp35 subfamily (49). MRP/Nbp35 members are distinguished by a conserved -ENMS- sequence, followed by a metal-coordinating motif, CX2C (at Cfd1 residues 194–197 and 201–204, respectively). Mutation of either cysteine in the CX2C sequence of Cfd1 or Nbp35 is lethal, consistent with this motif being essential to function (25, 27). The asparagine in the -ENMS- sequence is predicted to contact the adenosine ring of bound ATP (49). MRP/Nbp35 subfamily members appear to function in FeS cluster biogenesis in all kingdoms (25, 27, 41, 47, 50, 51).
The putative metal-binding CX2C motif maps to the molecular surface of MRP/Nbp35 family members. Structural information for these proteins comes from the x-ray crystal structure of Af2382, a homolog of unknown function in Archaeoglobus fulgidus. In the crystal structure of Af2382, the CX2C sequence coordinates a single zinc atom between monomers. It is imagined that, in MRP/Nbp35 the homodimer, the CX2C motif would be oriented to bind a bridging FeS cluster coordinated by the cysteine residues from each monomer. The proximity to the putative nucleotide-binding asparagine in the adjacent -ENMS- sequence raises the possibility that ATP binding and/or hydrolysis invokes a conformational change that alters the stability (kinetic lability) of a coordinated FeS cluster, facilitating transfer to apo-FeS proteins. Whether Cfd1 and Nbp35 bind nucleotide triphosphates or respond to nucleotide binding and/or hydrolysis in a manner similar to other deviant P-loop NTPases, such as NifH (52), has yet to be shown.
Nar1
The finding that eukaryotes possess a protein with high similarity to bacterial hydrogenases aroused curiosity about its role before it was shown to function in cytosolic FeS cluster biogenesis (28, 53, 54). Animal cells express two orthologs of Nar1, IOP1 and IOP2 (42). Depletion of IOP1, but not IOP2, by RNA interference resulted in defective cytosolic FeS cluster assembly (42). The position of conserved cysteine residues, structural modeling based on hydrogenases, and mutagenesis and 55Fe binding studies suggest that Nar1 contains two FeS clusters (54, 55). The homology of Nar1 to hydrogenases has fueled speculation that it acts as an electron donor for cytosolic FeS cluster maturation and/or transfer (6).
Cia1
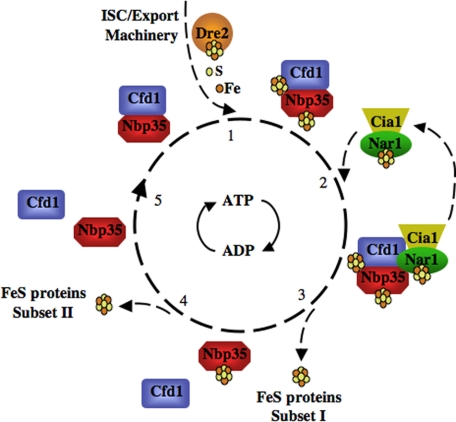
Cia1 is a seven-bladed propeller typical of the WD40 repeat family (56). WD40 proteins mediate protein-protein interactions (57). Depletion of Cia1 impaired maturation of cytosolic and nuclear FeS proteins but did not reduce 55Fe binding to Nbp35 or Nar1, prompting the suggestion that Cia1 acts in the transfer of FeS from Nbp35 to target proteins (30). CFD1 and CIA1 are fused in Schizosaccharomyces pombe (30), consistent with a role in mediating protein complex formation with Cfd1. Because Cfd1 interacts transiently with Nbp35 and Nar1 (25),3 we envision formation of a transient complex of CIA factors for cluster transfer to apo-targets (Fig. 1).
FIGURE 1.
Model for FeS cluster assembly via the CIA system. A nucleotide-dependent cycle for FeS cluster assembly on a Cfd1-Nbp35 scaffold and transfer to apo-targets is depicted. ATP binding, hydrolysis, and release of ADP by Cfd1 and Nbp35 are proposed to occur based on the high homology of these proteins to known deviant P-loop ATPases (49). The precise steps within the process affected by nucleotide binding and hydrolysis are not suggested here. FeS cluster assembly on the apo-Cfd1-Nbp35 scaffold complex is proposed to depend on the mitochondrial ISC and export systems, the CIA factor Dre2, and a source of iron and sulfur (Step 1). Nar1 and Cia1 interact with the Cfd1-Nbp35 complex, facilitating cluster transfer to Subset I of cytosolic and nuclear FeS proteins (Steps 2 and 3). It is proposed here that dissociation of Nar1, Cia1, and Cfd1 frees Nbp35 to support FeS cluster assembly in Subset II of cytosolic and nuclear FeS proteins (Steps 3 and 4). Apo-Cfd1 and apo-Nbp35 reform the heterocomplex to restart the process (Step 5).
Dre2
Recombinant Dre2 contains a [4Fe-4S] cluster and a [2Fe-2S] cluster (31). These clusters are stable even after prolonged exposure to air, suggesting that they play structural and/or catalytic roles in the protein. Depletion of Dre2 impaired cytosolic but not mitochondrial FeS cluster biogenesis, although Dre2 was found partially localized in the mitochondrial intermembrane space (31). It is speculated that Dre2 is involved in an early step in cytosolic FeS cluster biogenesis, possibly working in concert with the ISC export system to deliver a substrate necessary for FeS cluster formation on Cfd1 and Nbp35 (Fig. 1). Dre2 was reported to function in a complex with Tah18, a protein with FAD- and NAD-binding motifs that raise the possibility of an electron transfer function (58).
Mechanism of FeS Cluster Biogenesis via the CIA System
A model for FeS cluster assembly through the CIA system is beginning to emerge as studies provide information on the characteristics of individual CIA proteins. Fig. 1 illustrates our current thinking on CIA-mediated FeS cluster assembly. P-loop ATPases typically cycle in and out of protein interactions, driven by nucleotide binding, hydrolysis, and release (49). The model presented is centered on such a hypothetical cycle for Cfd1 and Nbp35. Although evidence for nucleotide binding by these proteins is lacking, the effect of mutation at predicted nucleotide-binding residues on protein function supports a nucleotide-directed process.3 The model posits that Cfd1 and Nbp35 cycle in and out of a heterocomplex, transiently binding [4Fe-4S] clusters for transfer to apo-targets.
Several lines of evidence support the view that Cfd1 and Nbp35 function in FeS cluster biogenesis as a complex. When coexpressed in Escherichia coli, Cfd1 and Nbp35 were isolated in a heterotetramer complex (25). Notably, this complex bound multiple [4Fe-4S] clusters upon cluster reconstitution. Cfd1 and Nbp35 co-immunoprecipitated from yeast extracts, indicating that they physically interact in their natural environment (25). Iron binding by Cfd1 and Nbp35, as well as complex formation, was disrupted by mutation of the CX2C motif in either protein.4
The model in Fig. 1 shows initial assembly of cytosolic FeS clusters occurring on the Cfd1-Nbp35 complex. The loading of iron onto Cfd1 and Nbp35 requires the mitochondrial ISC and ISC export systems (25, 29), consistent with a role in assembly of the initial clusters on the Cfd1-Nbp35 complex (Fig. 1). Because of its dual localization in mitochondria and the cytoplasm (31), Dre2 is shown here to participate at this initial step.
The source of sulfur and iron for cytosolic FeS cluster assembly has not yet been resolved. To date, Nfs1 is the only cysteine desulfurase known to be required for FeS cluster biogenesis in non-photosynthetic eukaryotes. In yeast, Nfs1 supplies this function in the mitochondria; restricting Nfs1 to the cytosol did not rescue the cytosolic FeS cluster defect associated with deficiency in the mitochondria (34). However, in S. cerevisiae, Icp55 clips off three amino acids from the N terminus of Nfs1, promoting its translocation from mitochondria to the nucleus (59). It seems plausible that a portion of this modified Nfs1 also functions in the cytoplasm, providing sulfide to the CIA system. In animals, a cytoplasmic splice variant of Nfs1 may function directly in cytosolic cluster assembly (38). Clearly, additional work is necessary to fully resolve this issue.
Reconstituted Cfd1 or Nbp35 or the Cfd1-Nbp35 heterotetramer transferred FeS clusters to apo-Leu1 in vitro (25). This transfer reaction occurred much faster than chemical reconstitution of Leu1 and was insensitive to iron chelator, suggesting direct cluster transfer to Leu1. The efficiency of activation of Leu1 by Cfd1, Nbp35, and the heterotetramer complex implicates all three entities as potential FeS scaffolds. Indeed, all of the MRP/Nbp35 family members tested show similar abilities, indicating an evolutionarily conserved function (25, 47, 60).
The CIA factors Nar1 and Cia1 are believed to act downstream of FeS cluster loading on Cfd1 and Nbp35. Nar1 deficiency had little effect on iron binding to Cfd1 or Nbp35, and depletion of Cia1 did not affect iron binding to Cfd1, Nbp35, or Nar1 (25, 30). These observations place Nar1 and Cia1 at the transfer of FeS clusters from the Cfd1-Nbp35 complex (or Nbp35 alone; see below) to apo-targets. Nar1 and Cia1 likely interact transiently with Cfd1 or Nbp35 (and/or the heterocomplex), with Nar1 potentially altering the electrochemical state of the nascent cluster, making it competent for transfer, and with Cia1 acting as an adapter protein for specific targeting of the labile FeS clusters to apo-targets.
Approximately half of Cfd1 and Nbp35 are in the heterocomplex in yeast cells.5 However, ∼80% of Nbp35-bound iron was found associated with the protein that was free of Cfd1; only 20% was associated with the Cfd1-Nbp35 complex, and none was detected with Cfd1 alone. This raises the question of whether the heterocomplex and free Nbp35 serve different subsets of FeS proteins. The fact that cells harbor FeS proteins of many types in the cytoplasm, nucleus, and other subcellular locations (Table 1) points to a potential need for specialized systems for each compartment or for subsets of apo-FeS proteins. The MRP/Nbp35 family member Ind1 is required for maturation of only a subset of mitochondrial FeS proteins, for example (51). Therefore, we depict the Cfd1-Nbp35 complex delivering FeS to one set of proteins and free holo-Nbp35 delivering FeS to a different set of apo-targets (Fig. 1). The argument against such a view is that deficiency in any of the CIA factors impaired FeS cluster assembly in all extramitochondrial proteins tested. On the other hand, the essential nature of the CIA system may lead to wide-ranging and pleiotropic effects that are not indicative of direct action. The answers to these questions await further investigation.
FeS Cluster Biogenesis and Cellular Iron Regulation
Organisms have evolved very sophisticated and complex processes to regulate iron at various levels (3, 7, 8, 61, 62). Regulation of iron acquisition is generally balanced with utilization and storage through regulation of synthesis of proteins that perform these functions. Considering the number of FeS proteins within cells (e.g. Table 1), the biogenesis of this cofactor is a major use of iron. It is therefore not surprising that sensitive iron regulatory mechanisms have evolved to detect the need for iron in FeS cluster biogenesis.
Link in Cellular Iron Regulation and FeS Cluster Biogenesis in Animals: IRP1
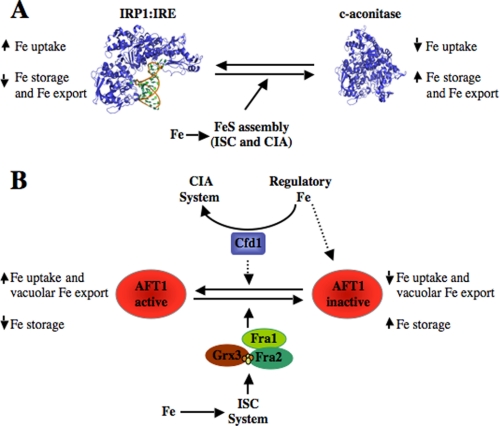
Cellular iron regulation in animals occurs mainly through the action of two IRPs. IRP1 and IRP2 are RNA-binding proteins whose activity is regulated by iron (8, 62). IRPs bind to conserved stem-loop structures located within the 5′- or 3′-UTRs of mRNAs that encode proteins for cellular iron transport, storage, and utilization, as well as proteins for energy and oxygen metabolism (63). The first linkage of FeS cluster biogenesis with iron homeostasis in eukaryotes came with the discovery that IRP1 and cytosolic aconitase were one and the same protein and that interconversion between the RNA-binding protein and enzyme was through assembly and disassembly of a [4Fe-4S] cluster (Fig. 2A) (8, 62).
FIGURE 2.
Intersection of FeS cluster assembly and cellular iron regulation. A, IRP1 is one of two post-transcriptional regulators of iron-related gene expression in animal cells. FeS cluster assembly and disassembly regulate IRP1 structure and activity. Both the ISC and CIA machineries are required for cluster assembly in IRP1, resulting in its conversion to cytosolic aconitase (c-aconitase). Changes in cellular iron affect efficiency of FeS cluster assembly in IRP1. This alters iron-related mRNA metabolism and iron transport, storage, and export as shown. The structure models for the IRP1-IRE complex and cytosolic aconitase are from Refs. 65 and 80, respectively. B, Aft1 is the primary transcriptional regulator of iron-related gene expression in S. cerevisiae. A complex that includes Fra1, Fra2, and Grx3 (or Grx4) controls Aft1 activity and links ISC-mediated FeS cluster biogenesis to Aft1 signaling. Overexpressed Cfd1 leads to activation of Aft1, which could be due either to increased consumption of iron from a regulatory iron pool or to direct action of Cfd1 on Aft1.
During iron starvation, IRP1 and IRP2 bind to conserved stem-loop structures, called IREs, found within the 5′-UTRs of mRNAs encoding the iron storage protein ferritin, the erythroid isoform of the heme biosynthetic enzyme δ-aminolevulinate synthase, mitochondrial aconitase, and the iron efflux transporter ferroportin, inhibiting translation initiation on these mRNAs (8, 62). IRPs also bind IREs located within the 3′-UTRs of transferrin receptor 1 and DMT1 (divalent metal ion transporter 1) mRNAs, stabilizing these transcripts. When the cellular iron level is sufficient, IRP1 acquires an FeS cluster, converting it to cytosolic aconitase and inhibiting its IRE-binding activity (64, 65). The consequence of IRP activity is that cellular iron storage and export are suppressed and iron uptake is stimulated when iron is limited, whereas loss of IRP activity when iron is in excess has the reciprocal effect (Fig. 2A).
FeS cluster assembly in IRP1 depends on the CIA system (7). Nbp35 depletion in human cells by RNA interference impaired cytosolic FeS cluster biogenesis and conversion of IRP1 to cytosolic aconitase (41). Likewise, depletion of Nar1 in cultured animal cells caused an increase in the IRE-binding activity of IRP1 (42). Although these manipulations of the CIA system affected IRP1 activity, little effect on IRP2 was seen. This is to be expected because IRP2 does not bind an FeS cluster (64). The effects in mammalian cells of CIA system deficiency on overall cellular iron metabolism appear to be solely through effects on efficiency of FeS cluster assembly in IRP1.
It is of note that an increase in cellular iron results in an increase in conversion of IRP1 to cytosolic aconitase, indicating that iron excess stimulates cytosolic FeS cluster biogenesis (Fig. 2A). Therefore, it is reasonable to conclude that animal cells have an excess capacity for cluster assembly, the process being limited by iron availability. It seems also likely that IRP1 is not an ideal target for cluster assembly because it can adopt conformations that cannot readily accept an FeS cluster, such as when bound IRE occupies the FeS cluster-binding site (65). Combined, these features make IRP1 an effective sentinel for cellular iron status in general.
Link in Cellular Iron Regulation and FeS Cluster Biogenesis in Budding Yeast: Aft1
The budding yeast S. cerevisiae achieves iron homeostasis through transcriptional and post-transcriptional regulation of iron-related genes, called the iron regulon (66). The combined effects of transcription factor Aft1 (activator of ferrous transport 1), its paralog Aft2, and the RNA-binding protein Cth2 balance expression of the iron regulon and give yeast the ability to coordinate iron uptake, storage, and utilization with availability (67).
FeS cluster biogenesis is tied to iron homeostasis in yeast. Yeast cells bearing defects in mitochondrial FeS cluster biogenesis or in the ISC export system increase expression of the iron regulon and cellular iron accumulation mainly targeted to mitochondria (22, 68–70). A compelling set of experiments implicated a cytosolic FeS protein in the signaling pathway that communicates mitochondrial FeS metabolism to Aft1 (70). This signaling pathway includes the cytosolic proteins Fra1 and Fra2 and either of the cytosolic glutaredoxins Grx3 and Grx4 (71). Fra1 and Fra2 form a complex with Grx3/4 (71–73). Deletion of either FRA1 or FRA2 resulted in hyperexpression of the iron regulon through activation of Aft1, suggesting that the regulatory complex they are part of normally acts to repress Aft1 activity (Fig. 2B).
A Fra2-Grx3 complex was found to coordinate a [2Fe-2S] cluster (74). Serving as a scaffold for [2Fe-2S] cluster assembly may be a conserved role of cytosolic glutaredoxins. GrxC1, a cytosolic glutaredoxin in plants, binds a bridging [2Fe-2S] cluster between monomers and has been implicated in FeS cluster biogenesis (75). These observations raise the intriguing possibility that Aft1 senses the [2Fe-2S] cluster on the Fra2-Grx3 complex. The model in Fig. 2B posits that the ISC system and export machinery are required for assembly of the [2Fe-2S] cluster on this complex. Failure to assemble this cluster would lead to activation of Aft1.
Given that Aft1 appears to sense FeS status in the cytoplasm, a surprising finding was that deficiency in CIA did not stimulate Aft1-responsive gene expression (69). Although this seems at odds with the notion of a cytosolic FeS protein serving to signal Aft1, it is possible that such an FeS protein utilizes a pathway other than CIA for assembly of cluster. It is not yet known whether the CIA system supports assembly of [2Fe-2S] clusters.
The critical functions provided by cytosolic and nuclear FeS proteins dependent on the CIA system for cluster assembly make this system a significant pathway for iron utilization in non-photosynthetic eukaryotes (Table 1). Given the demand for iron by the pathway, it was expected that the CIA system in yeast would intersect with the cellular iron regulatory system. In preliminary studies, we found that overexpression of Cfd1 caused a 3–4-fold stimulation of Aft1-responsive gene expression.6 Overexpression of other CIA factors did not stimulate Aft1, suggesting that the effect was unique to Cfd1.
A plausible model is that CIA consumes iron from a pool monitored by the Aft1 regulatory system (Fig. 2B). Cfd1 may play the critical role of promoting iron entry into the CIA pathway, competing with other pathways for available iron. An alternative view is that apo-Cfd1, which would be more abundant when iron is limiting or upon overexpression, serves to directly signal the status of cytosolic FeS cluster biogenesis to the Aft1 regulatory system (Fig. 2B). That there are Cfd1 mutants that fail to support FeS cluster assembly but stimulate Aft1-responsive gene expression argues for this latter view.6
Evolution of the CIA system for cytosolic FeS cluster assembly likely reflects the challenges associated with cluster biogenesis in the cytoplasm, such as exposure to oxygen and other reactive species potentially damaging to FeS clusters and their assembly. Consistent with this view, recent evidence suggests that an important role of Nar1 is related to oxygen metabolism and resistance to oxygen stress (76). The importance of oxygen to iron metabolism has been recognized for many years (77). The fact that IRP2 in animals and Aft1 in yeast respond as vigorously to changes in oxygen levels as to iron further strengthens this connection (78, 79). The challenges of the future will be to understand the interconnections between the various pathways for FeS cluster biogenesis, their relationship to other central metabolic pathways, and how organisms coordinate the activities inherent to achieve homeostasis.
This work was supported, in whole or in part, by National Institutes of Health Grant DK047281. This is the third article in the Thematic Minireview Series on Metals in Biology 2010. This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
A. K. Sharma, L. J. Pallesen, R. J. Spang, and W. E. Walden, unpublished data.
D. J. A. Netz, A. J. Pierik, M. Stumpfig, E. Bill, L. J. Pallesen, A. K. Sharma, W. E. Walden, and R. Lill, manuscript in preparation.
L. J. Pallesen, A. K. Sharma, N. Solodovnikova, and W. E. Walden, manuscript in preparation.
A. K. Sharma, unpublished data.
- CIA
- cytosolic iron-sulfur cluster assembly
- IRP
- iron regulatory protein
- IRE
- iron-responsive element
- UTR
- untranslated region
- ISC
- iron-sulfur cluster.
REFERENCES
- 1.Beinert H., Holm R. H., Münck E. (1997) Science 277, 653–659 [DOI] [PubMed] [Google Scholar]
- 2.Johnson D. C., Dean D. R., Smith A. D., Johnson M. K. (2005) Annu. Rev. Biochem. 74, 247–281 [DOI] [PubMed] [Google Scholar]
- 3.Ayala-Castro C., Saini A., Outten F. W. (2008) Microbiol. Mol. Biol. Rev. 72, 110–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fontecave M., Ollagnier-de-Choudens S. (2008) Arch. Biochem. Biophys. 474, 226–237 [DOI] [PubMed] [Google Scholar]
- 5.Pilon M., Abdel-Ghany S. E., Van Hoewyk D., Ye H., Pilon-Smits E. A. (2006) Genet. Eng. 27, 101–117 [DOI] [PubMed] [Google Scholar]
- 6.Lill R. (2009) Nature 460, 831–838 [DOI] [PubMed] [Google Scholar]
- 7.Rouault T. A., Tong W. H. (2008) Trends Genet. 24, 398–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hentze M. W., Muckenthaler M. U., Andrews N. C. (2004) Cell 117, 285–297 [DOI] [PubMed] [Google Scholar]
- 9.Zheng L., White R. H., Cash V. L., Jack R. F., Dean D. R. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 2754–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng L., White R. H., Cash V. L., Dean D. R. (1994) Biochemistry 33, 4714–4720 [DOI] [PubMed] [Google Scholar]
- 11.Shi R., Proteau A., Villarroya M., Moukadiri I., Zhang L., Trempe J. F., Matte A., Armengod M. E., Cygler M. (2010) PLoS Biol. 8, e1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoon T., Cowan J. A. (2003) J. Am. Chem. Soc. 125, 6078–6084 [DOI] [PubMed] [Google Scholar]
- 13.Bandyopadhyay S., Naik S. G., O'Carroll I. P., Huynh B. H., Dean D. R., Johnson M. K., Dos Santos P. C. (2008) J. Biol. Chem. 283, 14092–14099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adam A. C., Bornhövd C., Prokisch H., Neupert W., Hell K. (2006) EMBO J. 25, 174–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J., Saxena S., Pain D., Dancis A. (2001) J. Biol. Chem. 276, 1503–1509 [DOI] [PubMed] [Google Scholar]
- 16.Voisine C., Schilke B., Ohlson M., Beinert H., Marszalek J., Craig E. A. (2000) Mol. Cell. Biol. 20, 3677–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voisine C., Cheng Y. C., Ohlson M., Schilke B., Hoff K., Beinert H., Marszalek J., Craig E. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 1483–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miao B., Davis J. E., Craig E. A. (1997) J. Mol. Biol. 265, 541–552 [DOI] [PubMed] [Google Scholar]
- 19.Bych K., Kerscher S., Netz D. J., Pierik A. J., Zwicker K., Huynen M. A., Lill R., Brandt U., Balk J. (2008) EMBO J. 27, 1736–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mühlenhoff U., Gerl M. J., Flauger B., Pirner H. M., Balser S., Richhardt N., Lill R., Stolz J. (2007) Eukaryot. Cell 6, 495–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gelling C., Dawes I. W., Richhardt N., Lill R., Mühlenhoff U. (2008) Mol. Cell. Biol. 28, 1851–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kispal G., Csere P., Prohl C., Lill R. (1999) EMBO J. 18, 3981–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lange H., Lisowsky T., Gerber J., Mühlenhoff U., Kispal G., Lill R. (2001) EMBO Rep. 2, 715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sipos K., Lange H., Fekete Z., Ullmann P., Lill R., Kispal G. (2002) J. Biol. Chem. 277, 26944–26949 [DOI] [PubMed] [Google Scholar]
- 25.Netz D. J., Pierik A. J., Stümpfig M., Mühlenhoff U., Lill R. (2007) Nat. Chem. Biol. 3, 278–286 [DOI] [PubMed] [Google Scholar]
- 26.Kuhnke G., Neumann K., Mühlenhoff U., Lill R. (2006) Mol. Membr. Biol. 23, 173–184 [DOI] [PubMed] [Google Scholar]
- 27.Roy A., Solodovnikova N., Nicholson T., Antholine W., Walden W. E. (2003) EMBO J. 22, 4826–4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balk J., Pierik A. J., Netz D. J., Mühlenhoff U., Lill R. (2004) EMBO J. 23, 2105–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hausmann A., Aguilar Netz D. J., Balk J., Pierik A. J., Mühlenhoff U., Lill R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3266–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balk J., Aguilar Netz D. J., Tepper K., Pierik A. J., Lill R. (2005) Mol. Cell. Biol. 25, 10833–10841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y., Lyver E. R., Nakamaru-Ogiso E., Yoon H., Amutha B., Lee D. W., Bi E., Ohnishi T., Daldal F., Pain D., Dancis A. (2008) Mol. Cell. Biol. 28, 5569–5582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mühlenhoff U., Stadler J. A., Richhardt N., Seubert A., Eickhorst T., Schweyen R. J., Lill R., Wiesenberger G. (2003) J. Biol. Chem. 278, 40612–40620 [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y., Lyver E. R., Knight S. A., Pain D., Lesuisse E., Dancis A. (2006) J. Biol. Chem. 281, 22493–22502 [DOI] [PubMed] [Google Scholar]
- 34.Mühlenhoff U., Balk J., Richhardt N., Kaiser J. T., Sipos K., Kispal G., Lill R. (2004) J. Biol. Chem. 279, 36906–36915 [DOI] [PubMed] [Google Scholar]
- 35.Nakai Y., Umeda N., Suzuki T., Nakai M., Hayashi H., Watanabe K., Kagamiyama H. (2004) J. Biol. Chem. 279, 12363–12368 [DOI] [PubMed] [Google Scholar]
- 36.Condò I., Malisan F., Guccini I., Serio D., Rufini A., Testi R. (2010) Hum. Mol. Genet. 19, 1221–1229 [DOI] [PubMed] [Google Scholar]
- 37.Tong W. H., Rouault T. A. (2006) Cell Metab. 3, 199–210 [DOI] [PubMed] [Google Scholar]
- 38.Li K., Tong W. H., Hughes R. M., Rouault T. A. (2006) J. Biol. Chem. 281, 12344–12351 [DOI] [PubMed] [Google Scholar]
- 39.Land T., Rouault T. A. (1998) Mol. Cell 2, 807–815 [DOI] [PubMed] [Google Scholar]
- 40.Song D., Tu Z., Lee F. S. (2009) J. Biol. Chem. 284, 35297–35307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stehling O., Netz D. J., Niggemeyer B., Rösser R., Eisenstein R. S., Puccio H., Pierik A. J., Lill R. (2008) Mol. Cell. Biol. 28, 5517–5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song D., Lee F. S. (2008) J. Biol. Chem. 283, 9231–9238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vitale G., Fabre E., Hurt E. C. (1996) Gene 178, 97–106 [DOI] [PubMed] [Google Scholar]
- 44.Skovran E., Downs D. M. (2003) J. Bacteriol. 185, 98–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boyd J. M., Pierik A. J., Netz D. J., Lill R., Downs D. M. (2008) Biochemistry 47, 8195–8202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bych K., Netz D. J., Vigani G., Bill E., Lill R., Pierik A. J., Balk J. (2008) J. Biol. Chem. 283, 35797–35804 [DOI] [PubMed] [Google Scholar]
- 47.Kohbushi H., Nakai Y., Kikuchi S., Yabe T., Hori H., Nakai M. (2009) Biochem. Biophys. Res. Commun. 378, 810–815 [DOI] [PubMed] [Google Scholar]
- 48.Lezhneva L., Amann K., Meurer J. (2004) Plant J. 37, 174–185 [DOI] [PubMed] [Google Scholar]
- 49.Leipe D. D., Wolf Y. I., Koonin E. V., Aravind L. (2002) J. Mol. Biol. 317, 41–72 [DOI] [PubMed] [Google Scholar]
- 50.Schwenkert S., Netz D. J., Frazzon J., Pierik A. J., Bill E., Gross J., Lill R., Meurer J. (2010) Biochem. J. 425, 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sheftel A. D., Stehling O., Pierik A. J., Netz D. J., Kerscher S., Elsässer H. P., Wittig I., Balk J., Brandt U., Lill R. (2009) Mol. Cell. Biol. 29, 6059–6073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schindelin H., Kisker C., Schlessman J. L., Howard J. B., Rees D. C. (1997) Nature 387, 370–376 [DOI] [PubMed] [Google Scholar]
- 53.Barton R. M., Worman H. J. (1999) J. Biol. Chem. 274, 30008–30018 [DOI] [PubMed] [Google Scholar]
- 54.Nicolet Y., Cavazza C., Fontecilla-Camps J. C. (2002) J. Inorg. Biochem. 91, 1–8 [DOI] [PubMed] [Google Scholar]
- 55.Urzica E., Pierik A. J., Mühlenhoff U., Lill R. (2009) Biochemistry 48, 4946–4958 [DOI] [PubMed] [Google Scholar]
- 56.Srinivasan V., Netz D. J., Webert H., Mascarenhas J., Pierik A. J., Michel H., Lill R. (2007) Structure 15, 1246–1257 [DOI] [PubMed] [Google Scholar]
- 57.Li D., Roberts R. (2001) Cell. Mol. Life Sci. 58, 2085–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vernis L., Facca C., Delagoutte E., Soler N., Chanet R., Guiard B., Faye G., Baldacci G. (2009) PLoS One 4, e4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Naamati A., Regev-Rudzki N., Galperin S., Lill R., Pines O. (2009) J. Biol. Chem. 284, 30200–30208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boyd J. M., Drevland R. M., Downs D. M., Graham D. E. (2009) J. Bacteriol. 191, 1490–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bandyopadhyay S., Chandramouli K., Johnson M. K. (2008) Biochem. Soc. Trans. 36, 1112–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eisenstein R. S. (2000) Annu. Rev. Nutr. 20, 627–662 [DOI] [PubMed] [Google Scholar]
- 63.Theil E. C. (2000) Biochem. Pharmacol. 59, 87–93 [DOI] [PubMed] [Google Scholar]
- 64.Kühn L. C. (2009) Cell Metab. 10, 439–441 [DOI] [PubMed] [Google Scholar]
- 65.Walden W. E., Selezneva A. I., Dupuy J., Volbeda A., Fontecilla-Camps J. C., Theil E. C., Volz K. (2006) Science 314, 1903–1908 [DOI] [PubMed] [Google Scholar]
- 66.Yamaguchi-Iwai Y., Stearman R., Dancis A., Klausner R. D. (1996) EMBO J. 15, 3377–3384 [PMC free article] [PubMed] [Google Scholar]
- 67.Puig S., Vergara S. V., Thiele D. J. (2008) Cell Metab. 7, 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schilke B., Voisine C., Beinert H., Craig E. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 10206–10211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rutherford J. C., Ojeda L., Balk J., Mühlenhoff U., Lill R., Winge D. R. (2005) J. Biol. Chem. 280, 10135–10140 [DOI] [PubMed] [Google Scholar]
- 70.Chen O. S., Crisp R. J., Valachovic M., Bard M., Winge D. R., Kaplan J. (2004) J. Biol. Chem. 279, 29513–29518 [DOI] [PubMed] [Google Scholar]
- 71.Kumánovics A., Chen O. S., Li L., Bagley D., Adkins E. M., Lin H., Dingra N. N., Outten C. E., Keller G., Winge D., Ward D. M., Kaplan J. (2008) J. Biol. Chem. 283, 10276–10286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ojeda L., Keller G., Muhlenhoff U., Rutherford J. C., Lill R., Winge D. R. (2006) J. Biol. Chem. 281, 17661–17669 [DOI] [PubMed] [Google Scholar]
- 73.Pujol-Carrion N., Belli G., Herrero E., Nogues A., de la Torre-Ruiz M. A. (2006) J. Cell Sci. 119, 4554–4564 [DOI] [PubMed] [Google Scholar]
- 74.Li H., Mapolelo D. T., Dingra N. N., Naik S. G., Lees N. S., Hoffman B. M., Riggs-Gelasco P. J., Huynh B. H., Johnson M. K., Outten C. E. (2009) Biochemistry 48, 9569–9581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rouhier N., Unno H., Bandyopadhyay S., Masip L., Kim S. K., Hirasawa M., Gualberto J. M., Lattard V., Kusunoki M., Knaff D. B., Georgiou G., Hase T., Johnson M. K., Jacquot J. P. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7379–7384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fujii M., Adachi N., Shikatani K., Ayusawa D. (2009) Genes Cells 14, 457–468 [DOI] [PubMed] [Google Scholar]
- 77.Peyssonnaux C., Nizet V., Johnson R. S. (2008) Cell Cycle 7, 28–32 [DOI] [PubMed] [Google Scholar]
- 78.Meyron-Holtz E. G., Ghosh M. C., Rouault T. A. (2004) Science 306, 2087–2090 [DOI] [PubMed] [Google Scholar]
- 79.Hassett R. F., Romeo A. M., Kosman D. J. (1998) J. Biol. Chem. 273, 7628–7636 [DOI] [PubMed] [Google Scholar]
- 80.Dupuy J., Volbeda A., Carpentier P., Darnault C., Moulis J. M., Fontecilla-Camps J. C. (2006) Structure 14, 129–139 [DOI] [PubMed] [Google Scholar]
- 81.Alseth I., Eide L., Pirovano M., Rognes T., Seeberg E., Bjørås M. (1999) Mol. Cell. Biol. 19, 3779–3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rudolf J., Makrantoni V., Ingledew W. J., Stark M. J., White M. F. (2006) Mol. Cell 23, 801–808 [DOI] [PubMed] [Google Scholar]
- 83.Yeeles J. T., Cammack R., Dillingham M. S. (2009) J. Biol. Chem. 284, 7746–7755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Klinge S., Hirst J., Maman J. D., Krude T., Pellegrini L. (2007) Nat. Struct. Mol. Biol. 14, 875–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Boal A. K., Yavin E., Barton J. K. (2007) J. Inorg. Biochem. 101, 1913–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Barthelme D., Scheele U., Dinkelaker S., Janoschka A., Macmillan F., Albers S. V., Driessen A. J., Stagni M. S., Bill E., Meyer-Klaucke W., Schünemann V., Tampé R. (2007) J. Biol. Chem. 282, 14598–14607 [DOI] [PubMed] [Google Scholar]
- 87.Goto-Ito S., Ishii R., Ito T., Shibata R., Fusatomi E., Sekine S. I., Bessho Y., Yokoyama S. (2007) Acta Crystallogr. D Biol. Crystallogr. 63, 1059–1068 [DOI] [PubMed] [Google Scholar]
- 88.Bigelis R., Umbarger H. E. (1976) J. Biol. Chem. 251, 3545–3552 [PubMed] [Google Scholar]
- 89.Luo M., Jiang Y. L., Ma X. X., Tang Y. J., He Y. X., Yu J., Zhang R. G., Chen Y., Zhou C. Z. (2010) J. Mol. Biol. 398, 614–622 [DOI] [PubMed] [Google Scholar]
- 90.Duschene K. S., Broderick J. B. (2010) FEBS Lett. 584, 1263–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schlenzka W., Shaw L., Kelm S., Schmidt C. L., Bill E., Trautwein A. X., Lottspeich F., Schauer R. (1996) FEBS Lett. 385, 197–200 [DOI] [PubMed] [Google Scholar]
- 92.Schnackerz K. D., Dobritzsch D., Lindqvist Y., Cook P. F. (2004) Biochim. Biophys. Acta 1701, 61–74 [DOI] [PubMed] [Google Scholar]
- 93.Nishino T., Okamoto K. (2000) J. Inorg. Biochem. 82, 43–49 [DOI] [PubMed] [Google Scholar]
- 94.Wu X., Alexander P. B., He Y., Kikkawa M., Vogel P. D., McKnight S. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14058–14062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Conlan A. R., Axelrod H. L., Cohen A. E., Abresch E. C., Zuris J., Yee D., Nechushtai R., Jennings P. A., Paddock M. L. (2009) J. Mol. Biol. 392, 143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huang Q., Hong X., Hao Q. (2008) FEBS Lett. 582, 1431–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]