Abstract
Iron is an essential element for diverse biological functions. In mammals, the majority of iron is enclosed within a single prosthetic group: heme. In metazoans, heme is synthesized via a highly conserved and coordinated pathway within the mitochondria. However, iron is acquired from the environment and subsequently assimilated into various cellular pathways, including heme synthesis. Both iron and heme are toxic but essential cofactors. How is iron transported from the extracellular milieu to the mitochondria? How are heme and heme intermediates coordinated with iron transport? Although recent studies have answered some questions, several pieces of this intriguing puzzle remain unsolved.
Keywords: Cell Metabolism, Heme, Iron Metabolism, Trafficking, Transport Metals
Introduction
Cellular Iron Transport
Iron is essential to most living organisms, which have different sophisticated ways to obtain this element from the environment. Uptake and regulation of iron in bacteria and yeast have been described in detail (1, 2). Here, we will highlight iron processing in higher eukaryotes, particularly mammals. The daily requirements for iron in mammals are high, exerted mainly by red blood cells (RBCs),4 which generate vast amounts of hemoglobin. The majority of this iron is efficiently recycled from senescent RBCs by macrophages (MΦ), but a small portion needs to be extracted from the diet (3). Dietary iron is taken up by the enterocytes in the proximal region of the small intestine. The proteins involved in intestinal iron absorption and transport have been reviewed in detail (4, 5). After a series of reduction and oxidation steps during its passage through the enterocyte, iron is released into the bloodstream by the basolateral transporter FPN1 (ferroportin 1) and then oxidized to ferric iron (Fe3+) by the ferroxidases hephaestin and ceruloplasmin. Ferric iron is bound by transferrin (Tf), which has high affinity for ferric but not ferrous (Fe2+) iron. Under normal circumstances, serum iron is bound to Tf and transported to all cells of the body.
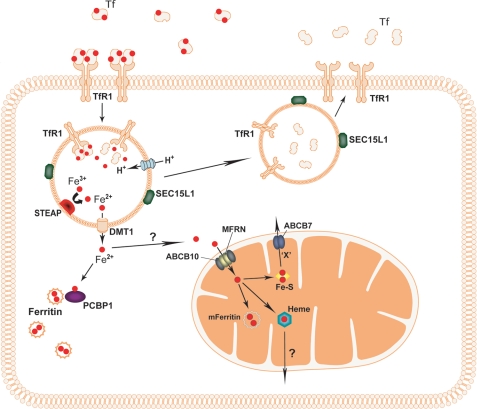
The best characterized mechanism for cellular iron uptake is uptake through Tf receptor-1 (TfR1; CD71) (Fig. 1). TfR1 binds two Tf molecules and has the highest affinity for diferric Tf. The TfR1-Tf complexes are concentrated in clathrin-coated pits on the cell surface and endocytosed. Once the complexes are internalized, a membrane-associated ATPase proton pump lowers the pH in the endosome to create an environment where ferric iron can be released from Tf. To be transported out of the endosome, iron must be reduced. In developing RBCs, this step is performed by the STEAP3 (six-transmembrane epithelial antigen of prostate 3) protein (6). Other members of the STEAP family are thought to be endosomal ferrireductases in non-erythroid cells (7). The reduced iron is exported out of the endosome by the divalent metal transporter (DMT1/SLC11A2/NRAMP2) in erythroid and non-erythroid cells (8–10). TfR1 is returned to the plasma membrane, where it can participate in another round of Tf-mediated iron uptake. A protein involved in TfR1 recycling in RBCs, Sec15l1, was identified from the hemoglobin-deficit (hbd) mouse, which shows microcytic hypochromic anemia (11). It was demonstrated that Mon1a plays a role in trafficking FPN1 to the cell surface of MΦ in mice (12).
FIGURE 1.
Cellular iron processing. Ferric iron (Fe3+) bound to Tf is taken up by cells through TfR1. The TfR1-Tf complex is internalized through endocytosis. The endosomal matrix is acidified by an ATPase proton pump, which allows dissociation of Tf and Fe3+ from TfR1. The ferric iron is reduced to Fe2+ by the STEAP family of proteins, and it can now be exported to the cytosol by DMT1. Ferrous iron can be delivered to ferritin for storage by the iron chaperone PCBP1. The larger part of iron will be transported to the mitochondrion for heme and FeS cluster synthesis. How iron is delivered to mitochondria is largely unknown. In erythrocytes, mitochondrial iron import is mediated by MFRN1 complexed with ABCB10. The MFRN1 paralog, MFRN2, is thought to import iron into mitochondria of non-erythroid cells, whether or not complexed with ABCB transporters. Factors involved in mitochondrial export of heme remain to be elucidated, whereas ABCB7 is thought to export a yet undefined component “X” needed for cytosolic FeS cluster assembly. After delivery of iron, TfR1 and Tf are recycled back to the cell surface, a pathway in which SEC15L1 plays an important role. mFerritin, mitochondrial ferritin.
The steps in cellular iron transport following release from the endosome are poorly understood. After its release, some ferrous iron will be stored in ferritin, a ubiquitously expressed iron storage protein that regulates intracellular iron availability (13). Storing iron in ferritin prevents free iron from generating toxic radicals and allows the regulated release of iron. Recent work identified a cytosolic iron chaperone, PCBP1 (poly(rC)-binding protein 1), that transports iron to ferritin (14). The human cell lines used in this study were non-erythroid; it remains to be investigated if PCBP1 or any orthologs serve this role in erythroid cells.
Mitochondrial Iron Uptake and Processing
The majority of cellular iron is utilized in the mitochondria for the biosynthesis of both heme and FeS clusters (15, 16). This makes the mitochondrion an important organelle in iron trafficking. Some cytosolic iron may be used for extramitochondrial FeS cluster synthesis (17–19). How iron is transferred from the endosome, the cytosol, or ferritin to the mitochondria is unknown. In developing RBCs, iron may be delivered by docking of the endosome to the mitochondrion (20). This “kiss-and-run” mechanism would provide an efficient means for delivering iron to mitochondria in RBCs, which generate vast amounts of heme. Recent evidence showed that mitochondria fuse with each other and with the endoplasmic reticulum (21, 22). Small organic compounds or some proteins may also bind free cellular iron (23). If and how these are involved in the trafficking of iron to the mitochondrion are unknown.
The vertebrate mitochondrial iron importer was discovered through studies with the anemic zebrafish mutant frascati (24). The gene responsible for the anemic phenotype, Mfrn1 (mitoferrin-1)/SLC25A37, belongs to the SLC25 (solute carrier 25) family of proteins, which are located primarily in the mitochondrial inner membrane (IM). The mouse Mfrn1 knock-out in early erythroid cells shows impaired mitochondrial iron import and reduced heme synthesis upon terminal differentiation (24). To import iron and regulate heme synthesis in RBCs, Mfrn1 must interact with the mitochondrial ATP-binding cassette transporter Abcb10 (25). The Mfrn1 paralog, Mfrn2 (Slc25a28), is ubiquitously expressed and was proposed to be the non-erythroid mitochondrial iron importer (24, 26). How iron is delivered to Mfrn1/2 from the cytosol for mitochondrial import remains unclear.
Once delivered to the mitochondria, iron is either stored in mitochondrial ferritin or utilized for the synthesis of heme and FeS clusters. FeS clusters are prosthetic groups in proteins that play an essential role in cell metabolism (15). In developing RBCs, iron is utilized primarily for the synthesis of large quantities of heme. Non-erythroid cells also make heme, but their iron requirements are much lower. Defects in heme synthesis genes result in human hematological disorders characterized by defective cellular iron homeostasis (27). The mechanisms that regulate vertebrate heme synthesis and the accompanying iron flux are beginning to be illuminated.
Heme biogenesis is closely linked to the availability of FeS clusters. The first gene in erythroid heme synthesis, ALAS2 (aminolevulinic acid synthase-2), is regulated by the FeS cluster-binding protein IRP1 (iron regulatory protein 1) (3). A pioneering study showed that, in the anemic zebrafish mutant shiraz, defective FeS cluster synthesis resulted in IRP1 binding constitutively to the iron regulatory element (IRE) in the 5′-untranslated region (UTR) of the ALAS2 mRNA. Consequently, translation of ALAS2 and synthesis of heme were blocked (28). Mammalian ferrochelatase (FECH), the terminal enzyme in heme synthesis, is a [2Fe-2S]-containing protein (29, 30). Deletion of the [2Fe-2S]-binding region at the C terminus of FECH abolished both the binding of the cluster and the enzyme activity (29). Similarly, inactivation of FeS clusters by nitric oxide significantly inhibited the activity of FECH (31). A recent study utilized a systems biology approach to analyze ≈35,000 cDNA microarrays and to identify mitochondrial genes that were tightly coexpressed and coregulated with the eight heme biosynthesis enzymes. Five candidate genes with putative roles in heme synthesis were selected for studies in the zebrafish. Two genes were known to be involved in FeS cluster synthesis, whereas the other three were mitochondrial transporters. Morpholino knockdown of all five genes resulted in profound anemia; silencing of the transporter SLC25A39 in differentiating mouse erythroleukemia (MEL) cells strongly reduced heme synthesis (32). These newly identified genes may play important roles in mammalian mitochondrial iron homeostasis.
In addition to heme biosynthesis, a significant portion of iron in the mitochondrion is utilized for the assembly of FeS clusters. This pathway is highly conserved among species, and studies in yeast have guided research on the mammalian components of the FeS cluster pathway, although additional proteins have been identified (33). Genetic defects in several genes involved in FeS cluster assembly are associated with human diseases in which defective mitochondrial iron homeostasis is a hallmark. Patients with mutations in GRX5 (glutaredoxin-5) suffer from microcytic anemia with accumulation of iron in mitochondria (34). Likewise, patients with Friedreich ataxia have genetic defects in the mitochondrial FeS cluster iron chaperone frataxin, which results in mitochondrial iron overload (35). In addition, defects in ABCB7, a protein implicated in export of mitochondrial FeS clusters, lead to anemia accompanied by mitochondrial iron deposits (36). These examples clearly indicate that the FeS cluster pathway plays an essential role in regulating mammalian mitochondrial and cellular iron homeostasis.
Cellular Iron Homeostasis: The IRE/IRP Regulatory Mechanism
Because both iron overload and iron deficiency are incompatible with normal body physiology, mammals regulate their iron levels at both the systemic and cellular levels. Excellent reviews detailing advances made in the past 2 decades have been published (37–39). This minireview focuses on the regulation of cellular iron homeostasis.
Many of the genes that are involved in iron transport or utilization contain one or more IREs in their mRNA. These elements are conserved RNA stem-loop structures that are recognized by IRP1 and IRP2. Depending on where the IRE is located in the mRNA, binding of an IRP leads to blocking translation or to stabilizing mRNA (reviewed in Ref. 40). Generally, mRNAs of genes such as FPN1, ALAS2, and ferritin that reduce cellular iron levels or the availability of iron have an IRE in the 5′-UTR of their mRNA; the IRE in the mRNA of genes like TfR1 and DMT1 that increase the availability of iron is located in the 3′-UTR. Therefore, binding of IRPs to IREs results in increased iron uptake and availability and reduced iron utilization and storage. This mechanism prevents a cell from becoming iron-deficient or iron-overloaded and is essential for cellular iron homeostasis.
The binding of IRP1 and IRP2 to their target mRNAs is differentially regulated. IRP1 (also known as aconitase 1) is a dual function protein that can convert citrate to isocitrate or bind IREs. IRP1 binds an FeS cluster and acts as an aconitase under iron-replete conditions when FeS cluster synthesis is normal. When cellular iron levels are low, there is a corresponding reduction in FeS cluster synthesis. Consequently, IRP1 loses its aconitase activity, binds IREs, and alters iron uptake and utilization. IRP2 does not bind an FeS cluster, and its activity is regulated by degradation by a proteasomal complex (41, 42). The key component of this complex is the cytosolic protein FBXL5 (F-box and leucine-rich repeat protein 5). The FBXL5 protein contains a domain that can bind iron. When iron levels are high, FBXL5 binds iron and targets IRP2 for degradation; iron-depleted conditions result in FBXL5 degradation. IRP2 appears to be regulated directly by cytosolic iron levels, whereas IRP1 binding to IREs depends on iron utilization by the mitochondrial FeS cluster assembly machinery. Vashisht et al. (42) showed that FBXL5 also binds IRP1 but does not target it for degradation under iron-replete conditions. The relevance of this interaction and how IRP1 escapes degradation remain to be elucidated.
The IRE/IRP mechanism is particularly important in tissues that regulate iron homeostasis or have high iron demands. IRPs play an essential role in regulating iron uptake in the intestine (43). The IRE/IRP system is also important in regulating iron processing by developing RBCs, as deletion of IRP2 results in microcytic anemia (44–46). The high demand for iron of immature erythroid cells may require, however, adaptations in the IRE/IRP network (47). In addition to being essential for cellular iron homeostasis, the IRE/IRP mechanism is also important in the regulation of systemic body iron levels (40).
Transport of Heme Synthesis Intermediates
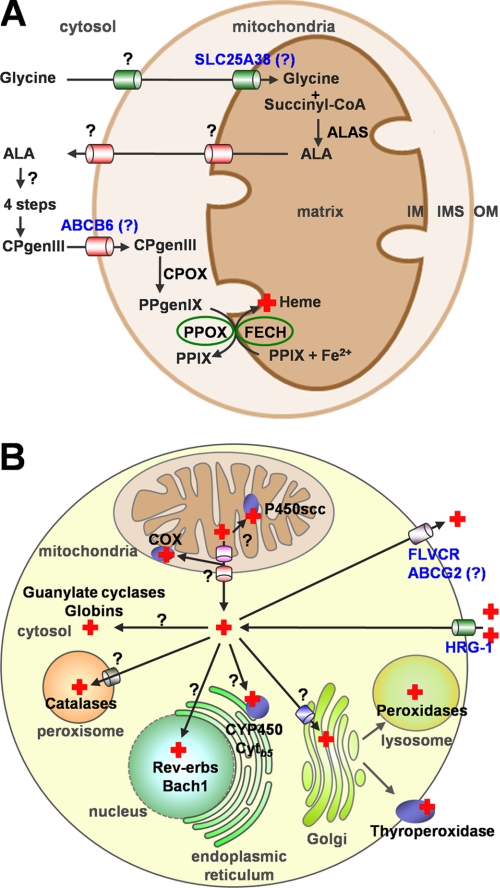
With few exceptions, metazoans synthesize heme via eight conserved, enzyme-catalyzed steps using glycine, succinyl-CoA, and ferrous iron as substrates (Fig. 2A). In the heme synthesis pathway, the first and the last three conversions take place in the mitochondria, whereas all remaining steps occur in the cytosol. The intermediates must therefore cross mitochondrial membranes for heme synthesis to progress.
FIGURE 2.
Transport of heme synthesis intermediates and heme in metazoans. A, heme is synthesized via a conserved eight-step pathway involving both mitochondrial and cytoplasmic enzymes. The intermediates ALA, CPgenIII, and PPIX and the substrate glycine need to be transported across mitochondrial membranes for the subsequent reactions. The solute carrier protein SLC25A38 may be involved in translocating glycine into mitochondria. The ATP-binding cassette transporter ABCB6 and the peripheral benzodiazepine receptor (PBR) were proposed to facilitate the import of CPgenIII into the mitochondria, whereas the 2-oxoglutarate carrier (OGC) and the adenine nucleotide translocator (ANT) may play a role in PPIX transport. The mechanisms for the export of ALA and the shuttling of heme precursors among the cytosolic enzymes are unknown. ALAS, aminolevulinic acid synthase; CPOX, coproporphyrinogen oxidase. B, the last step of heme biosynthesis occurs in the mitochondrial matrix. The nascent heme moiety must be translocated across membranes to multiple subcellular compartments where target hemoproteins reside. Heme can also be exported out of the cell or imported into the cell. The cell-surface FLVCR and the ABC transporter ABCG2 have been implicated in heme export in erythroid cells, whereas HRG-1 was identified as a heme importer. The question marks represent the presumptive heme trafficking pathways that are currently unclear. COX, cytochrome c oxidase; Cytb5, cytochrome b5.
Aminolevulinic acid synthase catalyzes the first reaction, which is the formation of δ-aminolevulinic acid (ALA) from glycine and succinyl-CoA. For this step, glycine needs to be imported into the mitochondrial matrix; the product ALA needs to be exported to the cytosol. A mitochondrial carrier family protein, SLC25A38, was proposed recently to facilitate glycine import or to exchange glycine for ALA across the IM (48). Patients with mutations in this gene manifest a form of nonsyndromic congenital sideroblastic anemia. Yeast lacking the SLC25A38 ortholog displayed decreased levels of ALA, possibly because of the reduced glycine levels in mitochondria. In addition, two mammalian oligopeptide transporters, PEPT1 and PEPT2, were found to transport ALA across the plasma membrane in a pH-dependent manner (49). Expression of either gene in Pichia pastoris yeast cells or Xenopus laevis oocytes significantly increased the influx of ALA. Further studies may help identify molecules on the mitochondrial membrane that transport ALA.
In the cytosol, ALA is converted to coproporphyrinogen III (CPgenIII) in four enzyme-catalyzed reactions. Because the accumulation of heme precursors is toxic to cells and usually causes porphyrias, the product of each reaction has to be quickly and efficiently delivered to the next enzyme. It is unknown how this is achieved.
The sixth step in heme synthesis is the oxidative decarboxylation of CPgenIII to generate protoporphyrinogen IX. This reaction is catalyzed by coproporphyrinogen oxidase. The majority of coproporphyrinogen oxidase is present in the mitochondrial intermembrane space (IMS), whereas a small fraction may be loosely attached to the IM (50). In either case, CPgenIII must be transported from the cytosol across the mitochondrial outer membrane (OM). This translocation is suggested to be mediated by the OM ATP-binding cassette transporter ABCB6 (51). ABCB6 was initially identified as a mammalian ortholog of the yeast mitochondrial iron transporter Atm1p (52). The expression profile of ABCB6 strongly correlates with that of heme synthesis genes (32). Overexpression of ABCB6 in K562 cells significantly increased the uptake of 55Fe-heme by mitochondria in an energy-dependent manner (51). A non-physiological CPgenIII-related compound, coproporphyrin III, inhibited both the heme-binding and heme uptake activities of ABCB6. It needs to be confirmed if CPgenIII is a bona fide ABCB6 substrate.
The final two steps are the conversion of protoporphyrinogen IX into protoporphyrin IX (PPIX) and the insertion of ferrous iron. The enzymes responsible for these reactions, protoporphyrinogen oxidase (PPOX) and FECH, are associated with the IM. However, the active site of PPOX faces the IMS, whereas the majority of FECH resides in the matrix (Fig. 2A) (53, 54). This poses the challenge of delivering the highly reactive PPIX from PPOX to FECH across the IM. A model of substrate channeling between PPOX and FECH has been proposed (53, 55). More recently, a co-immunoprecipitation experiment showed that PPOX and FECH physically interact in the cyanobacterium Thermosynechococcus elongates (56). Thus, the newly produced PPIX may be rapidly transferred from PPOX to FECH through protein-protein interactions.
Possible Mechanisms for Heme Transport from Mitochondria to Other Organelles
Free heme has inherent peroxidase activity and can intercalate and disrupt lipid bilayers of cell membranes, resulting in cytotoxicity. How then is heme delivered to the target hemoproteins once it is synthesized in the mitochondrial matrix (Fig. 2B)? A portion of heme may be shuttled from FECH to those hemoproteins in close proximity. For example, a heme-containing enzyme, cytochrome P450scc (P450 cholesterol side-chain cleavage), exhibits a similar localization pattern to FECH in mitochondria (57). Therefore, P450scc may acquire heme directly from FECH through protein-protein interaction. Hemoproteins such as cytochrome c oxidase, globins, guanylate cyclases, catalases, peroxidases, and certain transcription factors are found in distinct intracellular organelles, including the IMS, cytosol, peroxisomes, lysosomes, nucleus, and the secretory pathway (Fig. 2B) (58). In each of these cases, heme must traffic across at least one intracellular membrane to be incorporated into hemoproteins.
Metallochaperones have been demonstrated to be essential for intracellular trafficking of copper (59). It is likely that chaperones play a role in delivering heme from the site of synthesis to the sites of utilization. Heme chaperones that are important for cytochrome c biogenesis have been discovered in bacteria and plants (60, 61). Although no heme chaperone has been identified in mammals, several known cytosolic heme-binding proteins may facilitate intracellular heme transfer. For example, glutathione S-transferases (GSTs) in human RBCs and rat liver were shown to bind heme (62, 63). In fact, GSTs were first identified in mammalian liver as “ligandins” that could selectively bind steroids, bilirubin, and organic anions (64). A mixture of GSTs purified from rat liver cytosol increased the heme transport from mitochondria into apocytochrome b5 (65). Two other cytosolic proteins, p22HBP and HBP23, were also found to have high affinity for heme (66, 67). In MEL cells, p22HBP was induced during erythroid differentiation; knockdown of the gene resulted in reduced heme content (67). However, GSTs, p22HBP, and HBP23 are not dedicated heme-binding proteins because they also bind other tetrapyrrole compounds such as PPIX. Therefore, these molecules may not be dedicated intracellular heme transporters.
The endoplasmic reticulum (ER) may play an important role in intracellular heme allocation. It is likely that hemoproteins in the secretory pathway acquire heme in the ER. When the Golgi is disrupted by brefeldin A, the lysosomal heme-containing enzyme myeloperoxidase still receives its heme moiety, suggesting that heme incorporation occurs in the ER (68). It has been hypothesized that a portion of the heme may be transferred from the mitochondria directly to the ER through contact sites between the OM and ER, or mitochondria-associated membranes (MAMs). This interorganellar association was first reported 50 years ago in cells of the pseudobranch gland using electron microscopy (69). MAM has been implicated in non-vesicular transport of phospholipids and Ca2+ transmission from the ER to the mitochondria (70, 71). It was recently discovered that a dynamin-related protein, mitofusin 2, tethers the ER to mitochondria (72). The close contact between the ER and OM may provide a local route for efficient interorganellar heme delivery. This concept is supported by two earlier studies in rats. Both experiments showed that radiolabeled heme preferentially accumulated in the MAM fractions compared with the conventional microsomal fractions of the hepatic ER within 4 min after the rats were injected with [3H]ALA or [14C]ALA (73, 74).
Molecules Involved in Influx or Efflux of Heme
Heme transport machinery also exists on the plasma membrane. Dietary heme in the form of hemoglobin, myoglobin, and cytochromes is readily absorbed by the intestine. A nutritional study showed that humans absorb heme iron approximately four times more efficiently than inorganic iron (75). Iron supplementation significantly reduced the absorption of non-heme iron but not heme iron (75). The existence of heme receptors has been reported on the microvilli of pig small intestine and in cultured human enterocytes (76, 77). HCP1 (heme carrier protein 1) and HRG-1 (heme-responsive gene 1) are two newly identified molecules that may function to import heme into cells (78, 79).
Hcp1 was identified in a screen using hypotransferrinemic mice (79). Expression of HCP1 in Xenopus oocytes resulted in a 2–3-fold increase in heme uptake. However, Qiu et al. (80) showed that HCP1 was a high affinity folate/proton symporter and renamed it PCFT/HCP1. Expression of PCFT/HCP1 in Xenopus oocytes increased folate uptake by >200-fold. The high affinity folate transport activity suggested that folate may be the physiological ligand for PCFT/HCP1. It is unclear whether the low affinity heme transport activity of PCFT/HCP1 has physiological relevance.
HRG-1, the sole identified member of the SLC48 family, was initially discovered in a Caenorhabditis elegans microarray experiment (78). Humans have a single copy of HRG-1; worms have four hrg-1 paralogs. Worms may have evolved redundant heme acquisition pathways because they lack the ability to synthesize heme (81). Expression of human HRG-1 in MEL cells significantly increased the uptake of a fluorescent heme analog. Knockdown of zebrafish hrg-1 resulted in severe anemia, hydrocephalus, and a curved body with shortened yolk tube. Xenopus oocytes expressing hrg-1 revealed significant heme-induced inward currents, indicating heme uptake. These results suggest that HRG-1 is a bona fide heme transporter and is essential for normal metazoan development (Fig. 2B). Whether HRG-1 is involved in intestinal heme absorption remains to be determined.
Heme efflux may be a main mechanism of heme detoxification and could facilitate intercellular heme transport. The cell-surface receptor for feline leukemia virus subtype C (FLVCR) and the ABC transporter ABCG2 have been implicated in heme export in RBCs (Fig. 2B). Suppression of the FLVCR, a major facilitator superfamily protein, by feline leukemia virus C in feline embryonic fibroblasts significantly increased the cellular heme content, whereas ectopic expression of the FLVCR in renal epithelial cells reduced the intracellular heme levels (82). This result was supported by heme export assays using a fluorescent heme analog and 55Fe-heme in renal epithelial and K562 cells. The FLVCR is highly expressed in hematopoietic cells, and heme efflux mediated by the FLVCR is essential for erythroid differentiation (82, 83). No erythropoiesis is observed in FLVCR knock-out mice, which die at midgestation (84).
ABCG2, also named BCRP, was identified as a drug resistance protein in breast cancer cells. ABCG2 is highly expressed in hematopoietic progenitor cells (85); under hypoxic conditions, its expression is up-regulated by the transcription factor HIF-1 (86). Hemin-agarose pulldown assays showed that heme interacts with ABCG2 (86). Additionally, PPIX levels were 10 times higher in the RBCs of Abcg2-null mice than in wild-type mice (87), suggesting that ABCG2 may export porphyrin compounds. However, evidence to directly demonstrate that ABCG2 exports heme is still lacking.
Recycling of Heme and Heme Iron
In the human body, 65–75% of the total iron is present as heme iron in RBCs (88). After a life span of ≈120 days, senescent RBCs are phagocytosed by MΦ of the reticuloendothelial system and removed from the circulation. Once RBCs are lysed in MΦ, the heme moiety is released into the lumen of the phagolysosome. HO-1 (heme oxygenase-1) degrades heme into biliverdin, carbon monoxide, and iron. The iron generated during this process is either stored in ferritin or exported out of MΦ by FPN1 (89). Overexpression of FPN1 increased the levels of iron released from MΦ after erythrophagocytosis (90). Expression of both FPN1 and HO-1 was dramatically induced within 4 h after erythrophagocytosis in MΦ (91, 92). Whether heme is degraded within the phagolysosome or in the cytosol is unclear. NRAMP1 (natural resistance-associated macrophage protein 1), a divalent metal transporter localized to late endosomal and phagolysosomal membranes (93), is suggested to export iron out of phagolysosomes after erythrophagocytosis (94, 95). However, because HO-1 is associated with the ER, it is more likely that heme is first transported out of the phagolysosomes before being degraded.
The argument for heme translocation across phagolysosomal membranes is further supported by the observation that some heme is exported out of MΦ as an intact molecule following erythrophagocytosis. After J774 mouse MΦ phagocytosed 59Fe-labeled RBCs, 25–30% of the total 59Fe was released as 59Fe-heme (90). The FLVCR played a critical role in heme export from MΦ during erythrophagocytosis (84). After ingesting opsonized RBCs, FLVCR-deleted bone marrow-derived MΦ had higher ferritin levels in both the absence and presence of hepcidin, a negative regulator of iron export. Therefore, when the senescent RBCs are removed from circulation, intact heme, in addition to iron, may also be recycled.
Some heme and hemoglobin are released into the plasma during the destruction of senescent RBCs and enucleation of erythroblasts. Haptoglobin and hemopexin are plasma proteins responsible for recycling this portion of heme. Haptoglobin forms soluble complexes in an equimolar ratio with hemoglobin dimers. Haptoglobin-hemoglobin complexes bind to the CD163 receptor on the surface of monocytes and MΦ, and these complexes are subsequently endocytosed (96). The receptor for the haptoglobin-hemoglobin complexes also exists in hepatic parenchymal cells (97). Hemopexin is a heme-binding plasma protein that binds heme with high affinity. Low density lipoprotein receptor-related protein (CD91) is the receptor for hemopexin-heme complexes and is present in several cell types, including hepatocytes, MΦ, neurons, and syncytiotrophoblasts (98). After internalization, the haptoglobin-hemoglobin complexes are degraded in lysosomes (99). In contrast, the apohemopexin is recycled to the circulation after releasing heme into the cells (100). Following the endocytosis of haptoglobin-hemoglobin or hemopexin-heme complexes, heme is recycled as heme or heme iron.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DK74797 and R01 DK85035 (to I. H.) and Grants R01 DK70838, P01 HL032262, and P30 DK072437 (to B. H. P.). This work was also supported by grants from the Netherlands Organization for Scientific Research (NWO; to I. J. S.), the March of Dimes Foundation (to B. H. P. and I. H.), and the Roche Foundation for Anemia Research (to I. H.). This is the fourth article in the Thematic Minireview Series on Metals in Biology 2010. This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
- RBC
- red blood cell
- MΦ
- macrophage(s)
- Tf
- transferrin
- TfR1
- Tf receptor-1
- IM
- inner membrane
- IRE
- iron regulatory element
- UTR
- untranslated region
- FECH
- ferrochelatase
- MEL
- mouse erythroleukemia
- IRP
- iron regulatory protein
- ALA
- δ-aminolevulinic acid
- CPgenIII
- coproporphyrinogen III
- IMS
- intermembrane space
- OM
- outer membrane
- PPIX
- protoporphyrin IX
- PPOX
- protoporphyrinogen oxidase
- GST
- glutathione S-transferase
- ER
- endoplasmic reticulum
- MAM
- mitochondria-associated membrane
- FLVCR
- feline leukemia virus subtype C receptor.
REFERENCES
- 1.Krewulak K. D., Vogel H. J. (2008) Biochim. Biophys. Acta 1778, 1781–1804 [DOI] [PubMed] [Google Scholar]
- 2.Philpott C. C., Protchenko O. (2008) Eukaryot. Cell 7, 20–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hentze M. W., Muckenthaler M. U., Andrews N. C. (2004) Cell 117, 285–297 [DOI] [PubMed] [Google Scholar]
- 4.De Domenico I., McVey Ward D., Kaplan J. (2008) Nat. Rev. Mol. Cell Biol. 9, 72–81 [DOI] [PubMed] [Google Scholar]
- 5.Simpson R. J., McKie A. T. (2009) Cell Metab. 10, 84–87 [DOI] [PubMed] [Google Scholar]
- 6.Ohgami R. S., Campagna D. R., Greer E. L., Antiochos B., McDonald A., Chen J., Sharp J. J., Fujiwara Y., Barker J. E., Fleming M. D. (2005) Nat. Genet. 37, 1264–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohgami R. S., Campagna D. R., McDonald A., Fleming M. D. (2006) Blood 108, 1388–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fleming M. D., Romano M. A., Su M. A., Garrick L. M., Garrick M. D., Andrews N. C. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 1148–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fleming M. D., Trenor C. C., 3rd, Su M. A., Foernzler D., Beier D. R., Dietrich W. F., Andrews N. C. (1997) Nat. Genet. 16, 383–386 [DOI] [PubMed] [Google Scholar]
- 10.Gunshin H., Mackenzie B., Berger U. V., Gunshin Y., Romero M. F., Boron W. F., Nussberger S., Gollan J. L., Hediger M. A. (1997) Nature 388, 482–488 [DOI] [PubMed] [Google Scholar]
- 11.Lim J. E., Jin O., Bennett C., Morgan K., Wang F., Trenor C. C., 3rd, Fleming M. D., Andrews N. C. (2005) Nat. Genet. 37, 1270–1273 [DOI] [PubMed] [Google Scholar]
- 12.Wang F., Paradkar P. N., Custodio A. O., McVey Ward D., Fleming M. D., Campagna D., Roberts K. A., Boyartchuk V., Dietrich W. F., Kaplan J., Andrews N. C. (2007) Nat. Genet. 39, 1025–1032 [DOI] [PubMed] [Google Scholar]
- 13.Torti F. M., Torti S. V. (2002) Blood 99, 3505–3516 [DOI] [PubMed] [Google Scholar]
- 14.Shi H., Bencze K. Z., Stemmler T. L., Philpott C. C. (2008) Science 320, 1207–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lill R. (2009) Nature 460, 831–838 [DOI] [PubMed] [Google Scholar]
- 16.Ajioka R. S., Phillips J. D., Kushner J. P. (2006) Biochim. Biophys. Acta 1763, 723–736 [DOI] [PubMed] [Google Scholar]
- 17.Tong W. H., Jameson G. N., Huynh B. H., Rouault T. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9762–9767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tong W. H., Rouault T. (2000) EMBO J. 19, 5692–5700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tong W. H., Rouault T. A. (2006) Cell Metab. 3, 199–210 [DOI] [PubMed] [Google Scholar]
- 20.Sheftel A. D., Zhang A. S., Brown C., Shirihai O. S., Ponka P. (2007) Blood 110, 125–132 [DOI] [PubMed] [Google Scholar]
- 21.Liu X., Weaver D., Shirihai O., Hajnóczky G. (2009) EMBO J. 28, 3074–3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kornmann B., Currie E., Collins S. R., Schuldiner M., Nunnari J., Weissman J. S., Walter P. (2009) Science 325, 477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kruszewski M. (2003) Mutat. Res. 531, 81–92 [DOI] [PubMed] [Google Scholar]
- 24.Shaw G. C., Cope J. J., Li L., Corson K., Hersey C., Ackermann G. E., Gwynn B., Lambert A. J., Wingert R. A., Traver D., Trede N. S., Barut B. A., Zhou Y., Minet E., Donovan A., Brownlie A., Balzan R., Weiss M. J., Peters L. L., Kaplan J., Zon L. I., Paw B. H. (2006) Nature 440, 96–100 [DOI] [PubMed] [Google Scholar]
- 25.Chen W., Paradkar P. N., Li L., Pierce E. L., Langer N. B., Takahashi-Makise N., Hyde B. B., Shirihai O. S., Ward D. M., Kaplan J., Paw B. H. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16263–16268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paradkar P. N., Zumbrennen K. B., Paw B. H., Ward D. M., Kaplan J. (2009) Mol. Cell. Biol. 29, 1007–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sassa S. (2006) Br. J. Haematol. 135, 281–292 [DOI] [PubMed] [Google Scholar]
- 28.Wingert R. A., Galloway J. L., Barut B., Foott H., Fraenkel P., Axe J. L., Weber G. J., Dooley K., Davidson A. J., Schmid B., Schmidt B., Paw B. H., Shaw G. C., Kingsley P., Palis J., Schubert H., Chen O., Kaplan J., Zon L. I. (2005) Nature 436, 1035–1039 [DOI] [PubMed] [Google Scholar]
- 29.Dailey H. A., Finnegan M. G., Johnson M. K. (1994) Biochemistry 33, 403–407 [DOI] [PubMed] [Google Scholar]
- 30.Wu C. K., Dailey H. A., Rose J. P., Burden A., Sellers V. M., Wang B. C. (2001) Nat. Struct. Biol. 8, 156–160 [DOI] [PubMed] [Google Scholar]
- 31.Sellers V. M., Johnson M. K., Dailey H. A. (1996) Biochemistry 35, 2699–2704 [DOI] [PubMed] [Google Scholar]
- 32.Nilsson R., Schultz I. J., Pierce E. L., Soltis K. A., Naranuntarat A., Ward D. M., Baughman J. M., Paradkar P. N., Kingsley P. D., Culotta V. C., Kaplan J., Palis J., Paw B. H., Mootha V. K. (2009) Cell Metab. 10, 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rouault T. A., Tong W. H. (2008) Trends Genet. 24, 398–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Camaschella C., Campanella A., De Falco L., Boschetto L., Merlini R., Silvestri L., Levi S., Iolascon A. (2007) Blood 110, 1353–1358 [DOI] [PubMed] [Google Scholar]
- 35.Rötig A., de Lonlay P., Chretien D., Foury F., Koenig M., Sidi D., Munnich A., Rustin P. (1997) Nat. Genet. 17, 215–217 [DOI] [PubMed] [Google Scholar]
- 36.Bekri S., Kispal G., Lange H., Fitzsimons E., Tolmie J., Lill R., Bishop D. F. (2000) Blood 96, 3256–3264 [PubMed] [Google Scholar]
- 37.Nemeth E., Ganz T. (2006) Annu. Rev. Nutr. 26, 323–342 [DOI] [PubMed] [Google Scholar]
- 38.Andrews N. C. (2008) Blood 112, 219–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang A. S., Enns C. A. (2009) J. Biol. Chem. 284, 711–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muckenthaler M. U., Galy B., Hentze M. W. (2008) Annu. Rev. Nutr. 28, 197–213 [DOI] [PubMed] [Google Scholar]
- 41.Salahudeen A. A., Thompson J. W., Ruiz J. C., Ma H. W., Kinch L. N., Li Q., Grishin N. V., Bruick R. K. (2009) Science 326, 722–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vashisht A. A., Zumbrennen K. B., Huang X., Powers D. N., Durazo A., Sun D., Bhaskaran N., Persson A., Uhlen M., Sangfelt O., Spruck C., Leibold E. A., Wohlschlegel J. A. (2009) Science 326, 718–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galy B., Ferring-Appel D., Kaden S., Gröne H. J., Hentze M. W. (2008) Cell Metab. 7, 79–85 [DOI] [PubMed] [Google Scholar]
- 44.Cooperman S. S., Meyron-Holtz E. G., Olivierre-Wilson H., Ghosh M. C., McConnell J. P., Rouault T. A. (2005) Blood 106, 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galy B., Ferring D., Minana B., Bell O., Janser H. G., Muckenthaler M., Schümann K., Hentze M. W. (2005) Blood 106, 2580–2589 [DOI] [PubMed] [Google Scholar]
- 46.Meyron-Holtz E. G., Ghosh M. C., Iwai K., LaVaute T., Brazzolotto X., Berger U. V., Land W., Ollivierre-Wilson H., Grinberg A., Love P., Rouault T. A. (2004) EMBO J. 23, 386–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schranzhofer M., Schifrer M., Cabrera J. A., Kopp S., Chiba P., Beug H., Müllner E. W. (2006) Blood 107, 4159–4167 [DOI] [PubMed] [Google Scholar]
- 48.Guernsey D. L., Jiang H., Campagna D. R., Evans S. C., Ferguson M., Kellogg M. D., Lachance M., Matsuoka M., Nightingale M., Rideout A., Saint-Amant L., Schmidt P. J., Orr A., Bottomley S. S., Fleming M. D., Ludman M., Dyack S., Fernandez C. V., Samuels M. E. (2009) Nat. Genet. 41, 651–653 [DOI] [PubMed] [Google Scholar]
- 49.Döring F., Walter J., Will J., Föcking M., Boll M., Amasheh S., Clauss W., Daniel H. (1998) J. Clin. Invest. 101, 2761–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grandchamp B., Phung N., Nordmann Y. (1978) Biochem. J. 176, 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krishnamurthy P. C., Du G., Fukuda Y., Sun D., Sampath J., Mercer K. E., Wang J., Sosa-Pineda B., Murti K. G., Schuetz J. D. (2006) Nature 443, 586–589 [DOI] [PubMed] [Google Scholar]
- 52.Mitsuhashi N., Miki T., Senbongi H., Yokoi N., Yano H., Miyazaki M., Nakajima N., Iwanaga T., Yokoyama Y., Shibata T., Seino S. (2000) J. Biol. Chem. 275, 17536–17540 [DOI] [PubMed] [Google Scholar]
- 53.Ferreira G. C., Andrew T. L., Karr S. W., Dailey H. A. (1988) J. Biol. Chem. 263, 3835–3839 [PubMed] [Google Scholar]
- 54.Harbin B. M., Dailey H. A. (1985) Biochemistry 24, 366–370 [DOI] [PubMed] [Google Scholar]
- 55.Koch M., Breithaupt C., Kiefersauer R., Freigang J., Huber R., Messerschmidt A. (2004) EMBO J. 23, 1720–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Masoumi A., Heinemann I. U., Rohde M., Koch M., Jahn M., Jahn D. (2008) Microbiology 154, 3707–3714 [DOI] [PubMed] [Google Scholar]
- 57.Cherradi N., Rossier M. F., Vallotton M. B., Timberg R., Friedberg I., Orly J., Wang X. J., Stocco D. M., Capponi A. M. (1997) J. Biol. Chem. 272, 7899–7907 [DOI] [PubMed] [Google Scholar]
- 58.Severance S., Hamza I. (2009) Chem. Rev. 109, 4596–4616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O'Halloran T. V., Culotta V. C. (2000) J. Biol. Chem. 275, 25057–25060 [DOI] [PubMed] [Google Scholar]
- 60.Feissner R. E., Richard-Fogal C. L., Frawley E. R., Kranz R. G. (2006) Mol. Microbiol. 61, 219–231 [DOI] [PubMed] [Google Scholar]
- 61.Spielewoy N., Schulz H., Grienenberger J. M., Thony-Meyer L., Bonnard G. (2001) J. Biol. Chem. 276, 5491–5497 [DOI] [PubMed] [Google Scholar]
- 62.Harvey J. W., Beutler E. (1982) Blood 60, 1227–1230 [PubMed] [Google Scholar]
- 63.Ketterer B., Srai K. S., Christodoulides L. (1976) Biochim. Biophys. Acta 428, 683–689 [DOI] [PubMed] [Google Scholar]
- 64.Litwack G., Ketterer B., Arias I. M. (1971) Nature 234, 466–467 [DOI] [PubMed] [Google Scholar]
- 65.Senjo M., Ishibashi T., Imai Y. (1985) J. Biol. Chem. 260, 9191–9196 [PubMed] [Google Scholar]
- 66.Iwahara S., Satoh H., Song D. X., Webb J., Burlingame A. L., Nagae Y., Muller-Eberhard U. (1995) Biochemistry 34, 13398–13406 [DOI] [PubMed] [Google Scholar]
- 67.Taketani S., Adachi Y., Kohno H., Ikehara S., Tokunaga R., Ishii T. (1998) J. Biol. Chem. 273, 31388–31394 [DOI] [PubMed] [Google Scholar]
- 68.Nauseef W. M., McCormick S., Yi H. (1992) Blood 80, 2622–2633 [PubMed] [Google Scholar]
- 69.Copeland D. E., Dalton A. J. (1959) J. Biophys. Biochem. Cytol. 5, 393–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Achleitner G., Gaigg B., Krasser A., Kainersdorfer E., Kohlwein S. D., Perktold A., Zellnig G., Daum G. (1999) Eur. J. Biochem. 264, 545–553 [DOI] [PubMed] [Google Scholar]
- 71.Rizzuto R., Pinton P., Carrington W., Fay F. S., Fogarty K. E., Lifshitz L. M., Tuft R. A., Pozzan T. (1998) Science 280, 1763–1766 [DOI] [PubMed] [Google Scholar]
- 72.de Brito O. M., Scorrano L. (2008) Nature 456, 605–610 [DOI] [PubMed] [Google Scholar]
- 73.Asagami H., Hino Y., Kang D., Minakami S., Takeshige K. (1994) Biochim. Biophys. Acta 1193, 345–352 [DOI] [PubMed] [Google Scholar]
- 74.Meier P. J., Gasser R., Hauri H. P., Stieger B., Meyer U. A. (1984) J. Biol. Chem. 259, 10194–10200 [PubMed] [Google Scholar]
- 75.Roughead Z. K., Hunt J. R. (2000) Am. J. Clin. Nutr. 72, 982–989 [DOI] [PubMed] [Google Scholar]
- 76.Gräsbeck R., Majuri R., Kouvonen I., Tenhunen R. (1982) Biochim. Biophys. Acta 700, 137–142 [DOI] [PubMed] [Google Scholar]
- 77.Worthington M. T., Cohn S. M., Miller S. K., Luo R. Q., Berg C. L. (2001) Am. J. Physiol. Gastrointest. Liver Physiol. 280, G1172–G1177 [DOI] [PubMed] [Google Scholar]
- 78.Rajagopal A., Rao A. U., Amigo J., Tian M., Upadhyay S. K., Hall C., Uhm S., Mathew M. K., Fleming M. D., Paw B. H., Krause M., Hamza I. (2008) Nature 453, 1127–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shayeghi M., Latunde-Dada G. O., Oakhill J. S., Laftah A. H., Takeuchi K., Halliday N., Khan Y., Warley A., McCann F. E., Hider R. C., Frazer D. M., Anderson G. J., Vulpe C. D., Simpson R. J., McKie A. T. (2005) Cell 122, 789–801 [DOI] [PubMed] [Google Scholar]
- 80.Qiu A., Jansen M., Sakaris A., Min S. H., Chattopadhyay S., Tsai E., Sandoval C., Zhao R., Akabas M. H., Goldman I. D. (2006) Cell 127, 917–928 [DOI] [PubMed] [Google Scholar]
- 81.Rao A. U., Carta L. K., Lesuisse E., Hamza I. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4270–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Quigley J. G., Yang Z., Worthington M. T., Phillips J. D., Sabo K. M., Sabath D. E., Berg C. L., Sassa S., Wood B. L., Abkowitz J. L. (2004) Cell 118, 757–766 [DOI] [PubMed] [Google Scholar]
- 83.Quigley J. G., Burns C. C., Anderson M. M., Lynch E. D., Sabo K. M., Overbaugh J., Abkowitz J. L. (2000) Blood 95, 1093–1099 [PubMed] [Google Scholar]
- 84.Keel S. B., Doty R. T., Yang Z., Quigley J. G., Chen J., Knoblaugh S., Kingsley P. D., De Domenico I., Vaughn M. B., Kaplan J., Palis J., Abkowitz J. L. (2008) Science 319, 825–828 [DOI] [PubMed] [Google Scholar]
- 85.Zhou S., Schuetz J. D., Bunting K. D., Colapietro A. M., Sampath J., Morris J. J., Lagutina I., Grosveld G. C., Osawa M., Nakauchi H., Sorrentino B. P. (2001) Nat. Med. 7, 1028–1034 [DOI] [PubMed] [Google Scholar]
- 86.Krishnamurthy P., Ross D. D., Nakanishi T., Bailey-Dell K., Zhou S., Mercer K. E., Sarkadi B., Sorrentino B. P., Schuetz J. D. (2004) J. Biol. Chem. 279, 24218–24225 [DOI] [PubMed] [Google Scholar]
- 87.Jonker J. W., Buitelaar M., Wagenaar E., Van Der Valk M. A., Scheffer G. L., Scheper R. J., Plosch T., Kuipers F., Elferink R. P., Rosing H., Beijnen J. H., Schinkel A. H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 15649–15654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Andrews N. C. (2000) Nat. Rev. Genet. 1, 208–217 [DOI] [PubMed] [Google Scholar]
- 89.Donovan A., Lima C. A., Pinkus J. L., Pinkus G. S., Zon L. I., Robine S., Andrews N. C. (2005) Cell Metab. 1, 191–200 [DOI] [PubMed] [Google Scholar]
- 90.Knutson M. D., Oukka M., Koss L. M., Aydemir F., Wessling-Resnick M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1324–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Delaby C., Pilard N., Hetet G., Driss F., Grandchamp B., Beaumont C., Canonne-Hergaux F. (2005) Exp. Cell Res. 310, 43–53 [DOI] [PubMed] [Google Scholar]
- 92.Knutson M. D., Vafa M. R., Haile D. J., Wessling-Resnick M. (2003) Blood 102, 4191–4197 [DOI] [PubMed] [Google Scholar]
- 93.Gruenheid S., Pinner E., Desjardins M., Gros P. (1997) J. Exp. Med. 185, 717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Soe-Lin S., Apte S. S., Andriopoulos B., Jr., Andrews M. C., Schranzhofer M., Kahawita T., Garcia-Santos D., Ponka P. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 5960–5965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Soe-Lin S., Sheftel A. D., Wasyluk B., Ponka P. (2008) Exp. Hematol. 36, 929–937 [DOI] [PubMed] [Google Scholar]
- 96.Kristiansen M., Graversen J. H., Jacobsen C., Sonne O., Hoffman H. J., Law S. K., Moestrup S. K. (2001) Nature 409, 198–201 [DOI] [PubMed] [Google Scholar]
- 97.Kino K., Tsunoo H., Higa Y., Takami M., Hamaguchi H., Nakajima H. (1980) J. Biol. Chem. 255, 9616–9620 [PubMed] [Google Scholar]
- 98.Hvidberg V., Maniecki M. B., Jacobsen C., Højrup P., Møller H. J., Moestrup S. K. (2005) Blood 106, 2572–2579 [DOI] [PubMed] [Google Scholar]
- 99.Schaer C. A., Schoedon G., Imhof A., Kurrer M. O., Schaer D. J. (2006) Circ. Res. 99, 943–950 [DOI] [PubMed] [Google Scholar]
- 100.Smith A., Hunt R. C. (1990) Eur. J. Cell Biol. 53, 234–245 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.