Abstract
During translation, aminoacyl-tRNAs are delivered to the ribosome by specialized GTPases called translation factors. Here, we report the tRNA binding to the P-site of 40 S ribosomes by a novel GTP-independent factor eIF2D isolated from mammalian cells. The binding of tRNAiMet occurs after the AUG codon finds its position in the P-site of 40 S ribosomes, the situation that takes place during initiation complex formation on the hepatitis C virus internal ribosome entry site or on some other specific RNAs (leaderless mRNA and A-rich mRNAs with relaxed scanning dependence). Its activity in tRNA binding with 40 S subunits does not require the presence of the aminoacyl moiety. Moreover, the factor possesses the unique ability to deliver non-Met (elongator) tRNAs into the P-site of the 40 S subunit. The corresponding gene is found in all eukaryotes and includes an SUI1 domain present also in translation initiation factor eIF1. The versatility of translation initiation strategies in eukaryotes is discussed.
Keywords: Ribosome Function, RNA, RNA Viruses, Translation Initiation Factors, Translation Regulation, Hepatitis C Virus, SUI1 Domain, eIF2A, eIF2D, Ligatin
Introduction
Translation initiation in eukaryotes is a complex process involving a number of canonical initiation factors and auxiliary trans-acting proteins. According to present knowledge, among the canonical initiation factors, the key role in recruitment of mRNA and delivery of the initiator tRNA onto ribosomes belongs to factors eIF4F and eIF2, respectively, which have no analogs in bacterial cells. Therefore, it is not surprising that they are the principal targets of translational regulation in eukaryotes, especially in mammalian cells (1). In the last few years, the focus of our interests has been concentrated on eIF2, a three-subunit factor that not only delivers the Met-tRNAiMet to the 40 S ribosome but presumably participates in the scanning of 5′-UTRs of eukaryotic mRNAs and selection of the start codon along with other factors (eIF1, eIF1A, and eIF5). The unique properties of eIF2 are underscored by the fact that another Met-tRNAiMet-binding factor of eukaryotes, eIF5B (homolog of bacterial IF2), is not capable of scanning and searching for the start codon in mRNAs and works at the later step, combining the 48 S preinitiation complex with the 60 S subunit (2, 3).
In mammalian cells, the eIF2 activity is severely suppressed by phosphorylation of its α-subunit by one of the following four specific kinases: HRI, GCN2, PKR, and PERK (4). They are activated under quite different stress conditions (hemin deprivation, amino acid starvation, viral infection, and unfolded protein response, respectively), but they all result in a strong inhibition of overall protein synthesis in mammalian cells. Curiously, some mRNAs, especially of viral origin, somehow escape inhibition of their translation initiation. As we have recently shown, for those IRES2-containing mRNAs that do not use scanning and deliver their start codons directly to the vicinity of the P-site of the 40 S subunit, the function of Met-tRNAiMet delivery may be accomplished by eIF5B in a way similar to that operating in bacteria (5). However, in the case of a majority of cellular mRNAs, such a mechanism is unlikely or can be excluded. This prompts us to look for other protein factors that could either modify the activity of eIF2 or replace this ubiquitous eukaryotic factor for some specific mRNAs in some specific cases.
Our attention was drawn by initiation factor IF-M1 whose activities were described by Merrick and co-workers in the 1970s (6). According to these data, partially purified IF-M1 possessed two principal activities as follows: 1) it stimulated poly(Phe) synthesis directed by poly(U); and 2) it stimulated GTP-independent binding of Met-tRNAiMet to 40 S subunits programmed with the AUG triplet (6). With the discovery of eIF2 as a true factor that delivers the Met-tRNAiMet to ribosomes in all eukaryotic organisms, the interest in IF-M1 temporarily waned until 2002 when the major 65-kDa protein in the fraction containing IF-M1 activity was sequenced and, according to current nomenclature, named “initiation factor eIF2A” (7). The knock-out of the respective gene in yeast resulted in totally viable cells with growth characteristics similar to the wild strain. However, yeast eIF2A was found to genetically interact with initiation factors eIF4E and eIF5B, suggesting that these proteins function in the same pathway (8). The double eIF2A/eIF4E-ts mutant strain displayed a severe slow growth phenotype, which correlated with the accumulation of 85% of the double mutant cells arrested at the G2/M border. It was also found that eIF2A functions as a specific suppressor of Ure2p internal ribosome entry site-mediated translation in yeast cells (8). These data point to the involvement of eIF2A in the translation initiation events in yeast, at least for some individual mRNAs. However, its functional role in mammalian cells remained poorly investigated.
Attempts to obtain eIF2A in a recombinant form have been unsuccessful. Therefore, it was not possible to verify the in vitro activities formerly attributed to IF-M1 (see above) with the preparation of eIF2A that was obviously free of any contamination with other mammalian activities. Taking into account these circumstances, we undertook here a new exhaustive analysis of the IF-M1 containing fraction from rabbit reticulocyte lysate and HeLa cells. As a result, we describe here a novel factor with properties unprecedented for characterized translation factors that are known to be involved in recruitment of aminoacyl-tRNAs to ribosomes. The novel factor promotes binding of not only the initiator tRNA with the AUG codon but also of some elongator tRNAs with the corresponding codons on the 40 S ribosomal subunit. The binding is directed into the P-site of the subunit and occurs in the absence of GTP. The factor has the same molecular mass of 65 kDa as eIF2A but migrates in the gel as a 70–75-kDa polypeptide. Earlier, this protein was erroneously identified as ligatin (LGTN), a membrane receptor of glycoproteins of eukaryotic cells. The factor, termed here eIF2D, has two functional domains, PUA and SUI1. The first one is the RNA-binding domain found in several families of enzymes, including those that post-transcriptionally modify transfer RNAs (9). The second one is known to constitute initiation factor eIF1, the factor that plays a crucial role in selection of the start codon in mRNAs (10).
EXPERIMENTAL PROCEDURES
Plasmid Constructs and in Vitro Transcription
Plasmids coded for mRNAs with β-globin (11) or HCV (12) 5′-UTRs, as well as construct pcIlacZ and its derivatives 5A-cI, 5G-cI, cI-GUG, and (CAA)19-cIlacZ (13), were described. pHCV-UUU-NS' was created from pHCV-NS' plasmid by replacing ATG triplet with TTT by PCR with primers 5′-TTTAGCACGAATCCTAAACC-3′ and 5′-GGTGCACGGTCTACGAGACC-3′. For in vitro transcription, plasmids were linearized at positions downstream to primer annealing sites, and the RiboMAX kit (Promega) was used. The resulting transcripts were precipitated with 2 m LiCl followed by G-50 spin column purification and additional LiCl precipitation to remove any residual amount of GTP and other nucleotides. For β-globin, ScriptCap m7G capping system (EPICENTRE® Biotechnologies) was used to obtain 100% capped transcript, whereas all other mRNAs were uncapped.
To obtain eIF2D expressing vectors, the LGTN coding region (corresponding to GenBankTM accession number NM_006893) was first amplified from total HEK293T cell RNA by RT-PCR with primers 5′-GCGCACCATGGTTGCCAAGGCCTTTCGGGTC-3′ and 5′-CATATGAATTCTTCTTCTTGCCAGGTTTGAGG-3′ followed by EcoRI and NcoI treatment and inserting into pET33b vector (Novagen). The identity and correctness of the LGTN sequence were confirmed by sequencing of the complete ORF. The corresponding His6-tagged protein was found to be unstable, so the second expressing plasmid was created based on the pGEX-6p1 vector (GE Healthcare). For this purpose, eIF2D ORF was amplified from the above plasmid with primers 5′-CCGAGATCTATGTTTGCCAAGGCCTTTC-3′ and 5′-GGGTTTACTTCTTCTTGCCAGGTTTG-3′, digested with BglII, and ligated into BamHI-SmaI sites of the pGEX-6p1. The plasmid for eIF2D expression in mammalian cells was prepared by insertion of the eIF2D coding region obtained by PCR from the above plasmid with primers ATCATGGTTTTTGCCAAGGCCTTTCG and CCGACTAGTTTTACTTCTTCTTGCCAGG and digested with AhlI into XbaI and blunted NheI sites of the pcDNA3.1(+) expression vector (Invitrogen).
Purification of Ribosome Subunits, Initiation Factors, and Met-tRNAiMet
eIF2, eIF3, and eIF4F were purified from RRL; 40 S was isolated from HeLa cell extract; eIF1, eIF1A, eIF4A, and eIF4B were expressed in Escherichia coli as described previously (14–17). Purified tRNAfMet, a kind gift from V. Makhno and Y. Semenkov, was used as initiator tRNA. For aminoacylation, recombinant methionyl-tRNA synthetase was used as described previously (13). For deacylation, the tRNA was incubated for 1.5 h at 37 °C in Tris-HCl buffer, pH 8.8, followed by ethanol precipitation.
Purification of native eIF2A and eIF2D from RRL and HeLa cell extract is described under “Results.” To obtain recombinant eIF2D, E. coli Rosetta strain bearing pET33b-eIF2D or pGEX-6p1-eIF2D plasmid was grown in standard LB medium supplemented with 0.1 mm PMSF during 6 h at 28 °C after induction with 0.3 mm IPTG. eIF2D-His6 was purified using nickel-nitrilotriacetic acid-agarose (Qiagen) according to the manufacturer's instruction (the buffers was supplemented with 500 mm KCl, 10% glycerol, and 0.5 mm PMSF) followed by phosphocellulose chromatography (cut 200–400 mm KCl). N-GST-tagged eIF2D was expressed in a similar way, followed by binding to glutathione-Sepharose 4B (GE Healthcare) and elution with PreScission protease (Amersham Biosciences). Untagged protein was then purified by FPLC on Mono S column with KCl gradient of 100–300 mm. Apparently, the homogeneous protein was eluted at 270 mm KCl.
Assembly and Analysis of Translation Initiation Complexes
48 S ribosomal complexes were assembled and analyzed by toeprinting or RelE-printing assays as described earlier (14, 15, 18). Briefly, 48 S complexes were assembled by incubating 1 pmol of mRNA for 10 min at 30 °C in a 20-μl reaction volume that contained the reconstitution buffer (20 mm Tris-HCl, pH 7.5, 110 mm KOAc, 1 mm Mg(OAc)2, 0.25 mm spermidine-HCl, 1 mm DTT), 0.4 mm GTP-Mg(OAc)2 and 1 mm ATP-Mg(OAc)2 where indicated, 10 pmol of Met-tRNAiMet or deacyl- tRNAiMet or tRNAPhe, and 2.5 pmol of 40 S ribosomal subunits and combination of factors (eIF1 (10 pmol), eIF1A (10 pmol), eIF2 (5 pmol), eIF2A (5 pmol), eIF2D (5 pmol), eIF3 (5 pmol), eIF4A (10 pmol), eIF4B (5 pmol), eIF4F (2 pmol)), as described in the text. For toeprinting, 32P-labeled oligonucleotides 5′-GGGATTTCTGATCTCGGCG-3′ (for HCV), 5′-TCACCACCAACTTCTTCCAC-3′ (for β-globin), or 5′-CCAGGGTTTTCCCAGTCACG-3′ (for cIlacZ derivatives) were used. Assembled complexes were analyzed directly by primer extension using avian myeloblastosis virus RT (Promega) essentially as described previously (11) or treated with recombinant RelE protein, followed by deproteinization, RNA precipitation, and primer extension analysis (for details, see Ref. 18).
Binding of [35S]Met-tRNA to 40 S Ribosomal Subunits and Nitrocellulose Filter Binding Assay
Enzymatic binding of [35S]Met-tRNA with 40 S subunits was a modification of the procedure described in Ref. 19. Incubation in a total volume of 25 μl in the buffer containing 20 mm Tris-HCl, pH 7.5, 100 mm KCl, 5 mm Mg(OAc)2, and 1 mm DTT was performed at 25 °C for 15 min. The reaction mixture contained 12 pmol of 40 S subunits, 5 pmol of [35S]Met-tRNAiMet (15,000 cpm/pmol), 0.1 A260 units of ApUpG ribonucleotide, and indicated amount of eIF2A or eIF2D. After incubation, the reaction was stopped by the addition of 75 μl of Wash buffer (20 mm Tris-HCl, pH 7.5, 100 mm KCl, 5 mm Mg(OAc)2, 1 mm dl-methionine), and bound tRNA was measured by retention on nitrocellulose filters (Millipore type HA, 0.45 μm), which were washed five times with 1 ml of the Wash buffer. The radioactivity was determined using standard toluene scintillator.
Transfection of Mammalian Cells and Immunofluorescent Microscopy
HeLa cells were grown and transfected with pcDNA3.1-eIF2D plasmid as described previously (20). 24 h after transfection, cells were fixed with 1% glutaraldehyde in PBS, treated with sodium borohydride and 0.1% Triton X-100, and immunostained with rabbit polyclonal anti-eIF3a antibody (21), mouse anti-eIF2D serum raised against recombinant eIF2D-His6 protein, and goat-derived IgG-specific secondary antibodies conjugated with FITC or TRITC (Jackson ImmunoResearch). Distribution of eIF2D and eIF3a was analyzed with 100× Plan-NEOFLUAR objectives using an Axiovert 2000 M (Carl Zeiss) inverted microscope equipped with a Hammamatsu Orca digital camera. Images were acquired and processed with AxioVision and Adobe Photoshop software.
Polysome Profile Analysis
72 h after transfection with pcDNA3.1-eIF2D vector, logarithmically growing HEK293T cells from two 10-cm dishes were washed with ice-cold PBS, harvested by scraping, and lysed in 200 μl of Polysome Extraction Buffer (15 mm Tris-HCl, pH 7.5, 150 mm NaCl, 15 mm MgCl2, 1 mg/ml heparin, 1% Triton X-100, 0.5 mm PMSF, 40 units/ml RiboLock (Fermentas)). After pelleting debris, the lysate was applied onto 11 ml of 10–50% sucrose gradient (prepared with the buffer containing 15 mm Tris-HCl, pH 7.5, 150 mm NaCl, 15 mm MgCl2, 1 mm DTT, and 0.5 mm PMSF) and centrifuged 3.5 h at 35,000 rpm in an SW40 rotor (Beckman). 440-ml fractions were collected, and the absorbance at 260 nm was measured. 50 μl of indicated fractions were analyzed by Western blotting with anti-eIF2D serum (see above) or anti-RPSA antibody (22).
RESULTS
Factor eIF2A Separates from the GTP-independent tRNAiMet Binding Activity in the Course of RSW Fractionation
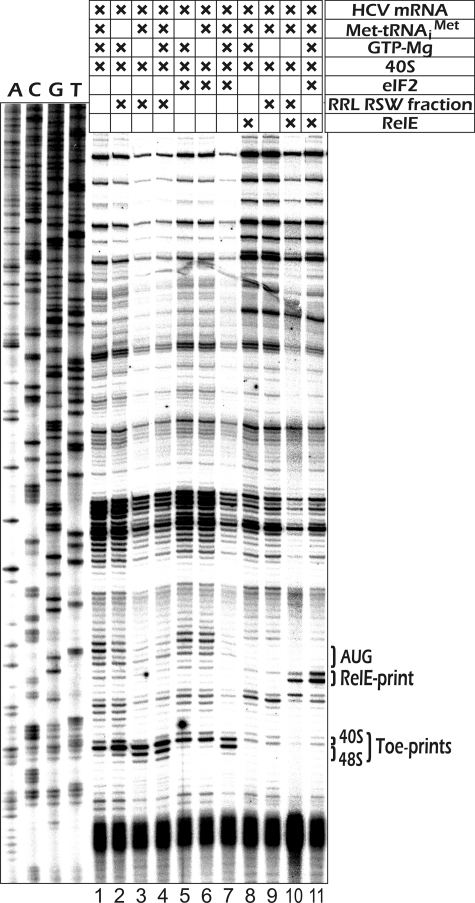
To identify the protein whose activity was attributed to factor IF-M1 and to check whether it is identical to eIF2A, we first looked for a more convenient and more natural test system alternative to membrane binding of the initiator tRNA with 40 S ribosomal subunits programmed with the AUG triplet. We reasoned that the HCV IRES element may be suitable for this purpose because it is known to place its AUG initiation triplet directly in the vicinity of the P-site of ribosomal 40 S subunits without scanning its nucleotide sequence (12). Indeed, the experiments demonstrated (Fig. 1) that the partially purified fraction of RSW from RRL, which stimulated AUG-dependent Met-tRNA binding with 40 S subunits on membranes (but does not contain eIF2), also produced the toe print signal characteristic of the HCV IRES. Authenticity of the complex was supported by its sensitivity to bacterial toxin RelE, which cleaves the mRNA at the nucleotide triplet accommodated in the A-site of the ribosome only when the P-site is occupied by peptidyl- or Met-tRNA (18). However, unlike the signal observed with the eIF2, the appearance of the toe-print signal in that case was not affected by the presence of GTP. Thereafter, subsequent experiments searching for the identification of the factor with the GTP independent activity in Met-tRNAiMet binding were performed predominantly by the toe-printing method with the HCV IRES RNA. As an additional tool to see whether this activity co-purifies with eIF2A in different fractions, we used eIF2A-specific antibodies raised against an internal peptide sequence, HEAKKAAKQEAR (amino acids 491–502).
FIGURE 1.
Reconstitution of 40 S preinitiation complexes on the HCV IRES using a partially purified RSW fraction of RRL containing eIF2A and GTP-independent Met-tRNA binding activity and detection of the complexes by toe-printing (lanes 1–7) or by RelE printing (lanes 8–11). Position of the AUG start codon is indicated. The bands originating from the binary HCV IRES × 40 S complex and those originating from the HCV IRES × 40 S × Met-tRNAiMet ×eIF2 (eIF2D) complexes are denoted on the right of the gel as 40 S and 48 S, respectively. Sequencing lanes obtained for the corresponding cDNA using the same primer are presented on the left.
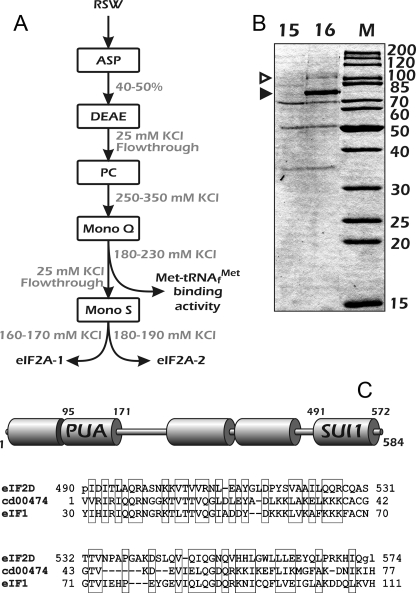
The scheme of purification is schematically presented in Fig. 2A. After a fine FPLC fractionation on Mono Q (gradient 25–300 mm KCl), it became clear that the protein with a molecular mass of 65 kDa, which reacted with eIF2A antibodies in Western blot assay, separated from the Met-tRNAiMet binding activity; eIF2A was not retained on the Mono Q column even at 25 mm KCl, whereas the initiator tRNA binding activity was eluted around 180 mm KCl (supplemental Fig. 1, A–C). Further analysis showed that in fact there were one or two more polypeptides with a molecular mass of 65 kDa that were retained on the Mono Q column. However, they were negative in the Western blot assay with eIF2A antibodies (supplemental Fig. 1B). The proteins not retained on Mono Q were further fractionated on Mono S. At least two polypeptides with a molecular mass of 65 kDa were eluted between 150 and 200 mm KCl; they were both positive in the Western blot assay, but one of them reacted much stronger with eIF2A antibodies than the other (supplemental Fig. 1, D and E). The subsequent analysis of these polypeptides by MS peptide fingerprinting unambiguously showed that they are both eIF2A and presumably differ in a post-translational modification(s). It should be noted that unlike the fractionation on Mono Q, we failed to resolve eIF2A and Met-tRNAiMet binding activity when the fraction 250–350 mm KCl from phosphocellulose was applied directly on the Mono S column; they eluted approximately in the same fractions (data not shown).
FIGURE 2.
Purification of the GTP-independent Met-tRNA binding activity. A, scheme of purification ((NH4)SO4 precipitation is abbreviated as ASP. B, Coomassie-stained gel that exemplifies two fractions (15 and 16) of HeLa RSW from the last step of purification of GTP-independent Met-tRNAiMet binding activity on Mono Q, of which fraction 16 had a much higher activity. Positions of two bands (∼70 and ∼100 kDa), with which this activity could be associated, are shown with filled and open arrowheads, respectively. M denotes position of standard molecular weight markers. C, upper part, conserved domains identified in the eIF2D sequence; bottom part, alignment of SUI1 domains of eIF2D and translation initiation factor eIF1. cd00474, consensus sequence of SUI1 domain from Conserved Domain Data base.
Neither purified eIF2A itself nor its post-translationally modified form(s), alone or in combination, possessed any Met-tRNAiMet binding activity (supplemental Fig. 2, A and B, also see below).The activity in the Met-tRNAiMet binding to the 40 S subunit programmed with the HCV IRES (estimated by the intensity of toe-print bands) best correlated with the presence and the amount of a protein with a molecular mass of ∼70 kDa (marked with arrow in supplemental Fig. 1A). Unfortunately, we failed to identify this protein with MS, presumably because the rabbit protein had some amino acid substitutions or covalent modifications as compared with its human or mouse homolog, as the rabbit genome has not been sequenced yet. Therefore, the entire procedure of purification of the Met-tRNAiMet binding factor was repeated for RSW from HeLa cells. The same band with a molecular mass of ∼70 kDa was found in the analogous fractions from the Mono Q separation (marked with the filled arrowhead in Fig. 2B). Again, the Met-tRNAiMet binding activity strongly correlated with the presence of this polypeptide (data not shown) and did not correlate with other weaker bands present also in the neighboring fraction, except probably for the weak band with a molecular mass of ∼100 kDa (indicated by open arrowhead in Fig. 2B). The latter band, although absent from the corresponding RRL fraction, was taken for MS analysis along with the 70-kDa protein.
Identification of Proteins, Candidates for Met-tRNAiMet Binding Activity, by MS Peptide Fingerprinting
The band that migrates in the gel as a 70–72-kDa protein was identified by MS as human LGTN (accession number NP_008824). Analysis of the LGTN CDS by the Phyre program (23) revealed that the protein likely consist of five structural domains (Fig. 2C) with unusually long disordered region between the second and the third ones. Judging by the conserved functional motifs identified within this sequence, the encoded protein looks like a translation factor rather than a receptor that binds and localizes phosphoglycoproteins at the cell surface (24, 25). It clearly does not contain any hydrophobic region, which might be attributed to a membrane-associated domain, and has a predicted molecular mass of 65 kDa (although LGTN monomer is a 10-kDa polypeptide, see under “Discussion”). In the identified protein, the N-terminal part harbors a PUA domain (named after PseudoUridine synthase and Archaeosine transglycosylase), which has been originally identified in silico as a putative RNA binding domain found in a wide range of proteins involved in RNA metabolism (9). It is implicated in many processes, including tRNA and rRNA modifications, translation initiation, and ribosome biogenesis (for review see Ref. 26). The C-terminal part of “LGTN” contains an SUI1 domain. Although the PUA motif was identified in several protein families, the SUI1 domain, apart from NP_008824 homologs, was found in eukaryotes only in translation initiation factor eIF1 and in another translation-associated protein DRP/DENR (27, 28). eIF1 is known to play an important role in accurate initiator codon recognition (for review, see Ref. 10). Therefore, we concluded that the sequence with accession number NP_008824 was attributed to LGTN by mistake, and hereafter throughout the text, the respective protein will be called eIF2D.
Another band with the mobility in the gel as 100 kDa (see above) was identified by MS as one of two eukaryotic RNases Z. This protein termed “ElaC homolog 2” (ELAC2) has a molecular mass of 92 kDa and participates in the endonuclease processing of pre-tRNAs and rRNAs (29). Other functions of ELAC2 are not related to translation. We suggest that eIF2D is a better candidate for the factor with GTP-independent Met-tRNAiMet binding activity than ElaC2. To support this suggestion, the recombinant eIF2D was prepared in E. coli, and its functions were analyzed.
Recombinant eIF2D Is Capable of Delivering Met-tRNAiMet to the Ribosomal 40 S Subunits in a GTP-independent Way
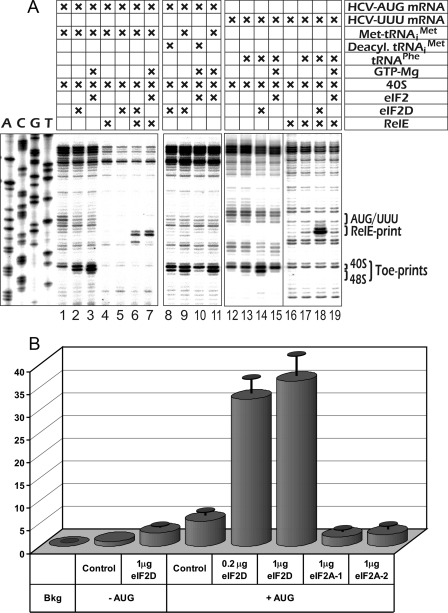
The cDNA for eIF2D was amplified from RNA isolated from HEK293 cells and cloned. The corresponding protein in GST fusion form was expressed in E. coli, cleaved from GST moiety, and purified to homogeneity (see “Experimental Procedures”). Fig. 3A shows that eIF2D is able to produce the toe-print signal with 40 S ribosomal subunits and the HCV IRES and that this signal is observed in the absence of GTP. To confirm that this activity is similar to that revealed for factor IF-M1, we also tested eIF2D in the membrane Met-tRNAiMet binding assay in the presence of the AUG triplet, the test initially used to characterize IF-M1 activities (6). The data of membrane binding assay (Fig. 3B) present strong evidence that the recombinant eIF2D does stimulate the delivery of Met-tRNAiMet onto 40 S ribosomes in the presence of AUG and is not capable of doing so in the absence of the nucleotide triplet. That the binding of the initiator tRNA occurs correctly, i.e. to the P-site of the 40 S subunit, was supported by the fact that the 40 S·HCV IRES·Met-tRNAiMet·eIF2D complex was also positive in RelE printing (Fig. 3A, lanes 4–7).
FIGURE 3.
Functional properties of eIF2D. A, lanes 1–7 show experiments analogous to those presented in Fig. 1A, but the purified recombinant factor was used instead of partially purified RSW fraction. Lanes 8–11 analyze the contribution of aminoacyl moiety of Met-tRNAiMet to its binding with 40 S ribosomal subunits. The HCV-UUU mRNA represents the HCV IRES where the initiation codon was mutated from AUG to UUU to test binding of cognate tRNAPhe in the presence of eIF2D (lanes 12–14). B, membrane binding assay. Recombinant eIF2D was tested for activity in [35S]Met-tRNAiMet binding with 40 S ribosomal subunits programmed with the AUG triplet. Bkg, background (no tRNA); control, no eIF2D added; eIF2A-1 and eIF2A-2 are two forms of eIF2A (see the text) purified to homogeneity.
Remarkably, unlike all known translation factors delivering aminoacyl-tRNAs to ribosomes, the aminoacyl moiety of Met-tRNAiMet is dispensable for the eIF2D promoted formation of its complex with 40 S subunits (Fig. 3A, lanes 8–11). At the same time, lanes 12–14 in Fig. 3 demonstrate that eIF2D is not strictly specific for the initiator tRNA and AUG codon; substitution of the AUG codon in the HCV IRES and the initiator tRNA for UUU triplet and tRNAPhe, respectively, resulted in a rather intensive toe print and RelE signals. Similar observations were once obtained for factor IF-M1; it also stimulated binding of Phe- and N-acetyl-Phe-tRNA with 40 S ribosomal subunits in the presence of UUU codon (6). On the other hand, the toe-print signal for the combination GUG triplet-tRNAVal turned out to be much weaker (data not shown) suggesting that eIF2D was not able to equally stabilize all codon-anticodon pairs.
eIF2D Stabilizes the Complex of 40 S Ribosomal Subunits with Leaderless mRNAs
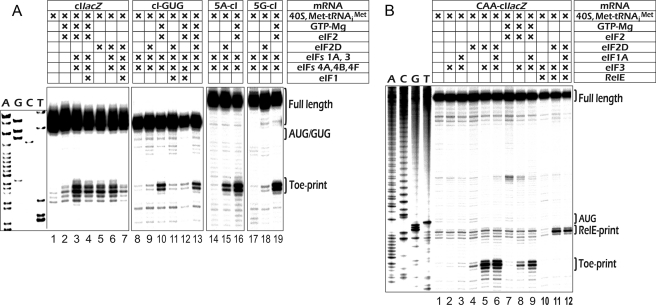
eIF2D was found to be inefficient in formation of 48 S preinitiation complexes with β-globin mRNA, a classical cap-dependent mRNA; the resulting toe print was rather weak (supplemental Fig. 3). This was again reminiscent of IF-M1, which had been shown to be inefficient in methionyl-puromycin synthesis, when total globin mRNA was used as a template instead of AUG triplet (19). From these results, we concluded that eIF2D could not substitute for eIF2 in scanning. However, we reasoned that eIF2D might work with a leaderless mRNA, another example of an mRNA that did not require scanning to initiate translation. As we have previously shown (13), a leaderless mRNA with unstructured 5′-end proximal sequence (cIlacZ) can form functional complexes (i.e. complexes competent for polypeptide elongation) with Met-tRNAiMet and 80 S ribosomes in the absence of initiation factors. However, similar complexes with ribosomal 40 S subunits (if any exist) were very unstable and did not give toe-print signals. Addition of eIF2 alone produced a weak toe print that was greatly enhanced by other canonical initiation factors in the absence of eIF1, whereas addition of the latter one to the resulting complex destabilized it (13, 30). These data were largely confirmed in this work (Fig. 4A, lanes 2–4). Contrary to eIF2, eIF2D alone was able to significantly stabilize the complex (Fig. 4A, lanes 5–7), but, remarkably, its strong stimulation effect was not affected by other factors, except eIF1. When the AUG triplet at the 5′ end of cIlacZ mRNA was replaced by GUG, eIF2D, unlike eIF2, was no longer able to promote the formation of 48 S complex (Fig. 4A, lanes 8–10), suggesting specific structural requirements for the nature of codon-anticodon interaction in the P-site.
FIGURE 4.
Effect of eIF2D on the formation of 48 S preinitiation complexes with various model constructs. A, assembly of the complexes with the leaderless mRNA construct cIlacZ and its derivatives. cIlacZ is an mRNA with the 5′-terminal gAUG sequence described by Andreev et al. (13). 5A-cI and 5G-cI are cIlacZ derivatives with GAAAA and GGGCC sequences added to its 5′ end, respectively, described in the same paper. cI-GUG is a leaderless cIlacZ construct where the 5′-terminal AUG was replaced by GUG. Other designations are as in Figs. 1 and 3. B, formation of the 48 S complex on the CAA-cIlacZ mRNA, which represents the sequence (CAA)19 added to the 5′ end of cIlacZ.
It is important to note that although eIF1 and eIF2D both contain a SUI1 domain, only eIF1 was able to destroy the 48 S complex formed on the cI-GUG mRNA in the presence of eIF2 (Fig. 4A, lanes 10–13). Similar conclusions can be made from results obtained with the β-globin mRNA, which forms aberrant initiation complexes at upstream near-cognate codons (UUGs and GUGs) in the absence of eIF1 (16, 18). eIF2D was unable to destroy such complexes that had been added instead of eIF1 (supplemental Fig. 3).
eIF2D Strongly Promotes Formation of Complexes of 40 S Ribosomal Subunits with RNAs Containing Single-stranded A-rich 5′-UTRs
The results described above prompted us to address the RNAs that contain 5′-leaders but presumably do not involve classical scanning with concomitant unfolding of the secondary structure within these 5′-UTRs. Fig. 4A, lanes 14–16, shows a successful formation of the 48 S complex with 5A-cI transcript, representing cIlacZ mRNA elongated at the 5′ end with GAAAA sequence. Remarkably, eIF2D failed to form the complex when using the RNA elongated with GGGCC sequence, presumably because of a high potential of G residues for base pairing. Next, we tested the cIlacZ mRNA elongated at the 5′ end with (CAA)19 sequence, known for its absolute single-stranded nature (31) and a relaxed dependence on the scanning machinery (30). Such a design resulted in the mRNA that had both a completely unstructured 5′-UTR and a single-stranded coding sequence adjacent to the initiation codon. As expected, this mRNA could form some amount of the eIF2-mediated 48 S initiation complex in the absence of scanning factors eIF1, eIF4F, eIF4A, and eIF4B (Fig. 4B, lanes 8 and 9). As follows from the data presented in Fig. 4B, lanes 4–6, this mRNA efficiently formed the 48 S complex with eIF2D instead of eIF2. Remarkably, eIF3 greatly stimulated formation of the complex, whereas eIF1A caused only very slight positive effect. We believe that the stimulation effect of eIF3 is most probably based on its known ability to recruit 40 S ribosomal subunits onto single-stranded RNAs even in the absence of additional initiation factors (see for instance Refs. 14, 32).
DISCUSSION
In this study, we report identification and initial characterization of a novel tRNA delivering factor possessing several unprecedented properties. The protein is encoded by sequence NM_006893. In 1989, a small part of this sequence containing a frameshift in the CDS has been previously characterized as a cDNA for human LGTN (33), a filamentous protein with covalently bound palmitic acid (25). It is a peripheral membrane receptor that binds and localizes phosphoglycoproteins at the cell surface (24, 34). However, several lines of evidence unambiguously indicate that the polypeptide encoded by NM_006893 is not actually related to LGTN. First, its molecular mass is 65 kDa (migrates in SDS gels as a 70-kDa protein), and the LGTN monomer it is about 10 kDa (25). Second, it does not contain any hydrophobic regions that might be attributed to a membrane-associated domain. Finally, eIF2D sequence analysis does not reveal any motifs relevant to membrane receptor function. Instead, two conserved domains, SUI1 and PUA, have been found in the protein, both of which most likely have intracellular functions.
Curiously, the confusion caused by the wrong identification of mRNA for LGTN resulted in several reports where researchers did not presumably realize that they studied expression and regulation of a gene not relevant to ligatin. In one of these reports (35), researchers investigated the regulation of the LGTN (actually eIF2D) gene in neurons and found out that the glutamate receptor activation and extracellular calcium entry into hippocampal neurons caused a long lasting down-regulation of LGTN mRNA and protein. The LGTN mRNA was shown to harbor determinants in their 3′-UTR for specific distribution within hippocampal neurons (36). Later, Wang et al. (37) identified LGTN (actually eIF2D) as one of the human hepatocellular carcinoma-associated antigens (HCA56), and showed that the eIF2D mRNA is ubiquitously expressed. This gene was also found deregulated in some other forms of human cancer (38, 39) and differentially expressed during development in invertebrates (40).
The eIF2D gene has orthologs in all eukaryotic genomes. It is well conserved in mammals (more than 80% identities) and among high eukaryotes (usually not less than 35% identity and 50% similarity). In yeast, the corresponding protein TMA64 (product of gene YDR117C) was found among proteins associated with ribosomes (41), whereas high throughput screening (42) showed its physical interaction with ribosomal protein S4 and RNA-helicase DED1 suggested to participate in translation. It is interesting that expression of the yeast protein increases under some stresses that result in inactivation of eIF2 (UPR-stress, anoxia, and oxidative stress) and is greatly enhanced during sporulation; at the same time, gene YDR117C is not essential for yeast; its deletion does not result in cell death, although sporulation is impaired (for references, see Saccharomyces Genome Data Base). Knock-out of the corresponding mammalian ortholog has not yet been performed.
The factor has two conserved domains, PUA and SUI1. Although the former one is implicated in many processes, the SUI1 domain is highly specific in occurrence and functions in both prokaryotes and eukaryotes. SUI1 is a well known functional domain present in translation initiation factor eIF1, where it plays an important role in accurate initiator codon recognition (30, 43, 44). During eukaryotic translation initiation, eIF1 binds near the P-site of the 40 S ribosome (45) and changes its conformation (46) in a way that the 40 S subunit becomes able to move along the mRNA in a process called scanning. It is thought that eIF1 interferes with the final accommodation of the initiator tRNA until the scanning ribosome reaches AUG codon in an appropriate nucleotide context (for review, see Ref. 10). At this point, eIF1 either dissociates or changes its location on the 40 S subunit, allowing Met-tRNAi to occupy the correct position in the P-site (47, 48). This leads to irreversible GTP hydrolysis in the Met-tRNAiMet·eIF2·GTP ternary complex and dissociation of eIF2 (49). The eIF1 ortholog in prokaryotes, YciH, has a similar “proofreading” activity (44).
The SUI1 fold is structurally similar to other RNA-binding domains (50) suggesting that this domain may directly interact with RNA in the initiation complex. Indeed, directed hydroxyl radical probing experiments (45) showed that particular regions within the SUI1 domain of human eIF1 do contact both 18 S rRNA and initiator tRNA, although in the latter case the signals were rather weak. Apart from eIF1 and eIF2D, there is only one more SUI1-containing protein encoded in human genomes. It is DRP/DENR, a protein whose expression was found to increase in cultured cells at high density (27). Interestingly, this protein has been shown to physically interact with MCT-1, a PUA domain-containing protein (28). The same report claims that the complex DENR·MCT-1 plays a role in translation initiation, being associated with the cap-binding complex. In yeast, their corresponding orthologs, TMA22 and TMA20, clearly interact with each other (see Saccharomyces Genome Data Base) and have been shown to be associated with the translational machinery (41). The complex of these two proteins may therefore represent a functional analog (or competitor) of the eIF2D in which PUA and SUI1 domains are not covalently linked. According to this, a recent large scale genetic interaction screen (51) showed a severe growth defect of the double tma64/tma20 mutant strain, suggesting at least partial redundancy in functions of the two proteins.
Although the functional role of eIF2D in eukaryotic cells remains to be established, data obtained in the course of this work allow us to draw some conclusions. The properties of this factor described here as well as literature data on its yeast ortholog strongly favor the conclusion that this protein is involved in some events related to translation. Our preliminary data show that eIF2D is localized in the cytoplasm rather in the nucleus and co-localizes with translation apparatus (at least with eIF3, see supplemental Fig. 4, and with PABPC1, data not shown). Sedimentation analysis of the cytoplasmic extract prepared from HEK293T cells with overexpressed eIF2D, combined with Western blot, showed that eIF2D is detected in the area of ribosomes (supplemental Fig. 5), thereby suggesting the interaction of the factor with the translational apparatus of mammalian cells. However, eIF2D is unlikely to be an obligatory component of the regular translation process. The same conclusion was drawn previously for IF-M1 (52). Its content in all laboratory cell lines we tested so far is very low and hardly revealed by Western blot. siRNA interference against its mRNA does not produce any visible effect and does not change significantly the growth properties of cultivated cells (data not shown). Thus, the most plausible idea is that eIF2D serves a specific mRNA(s) operating under unusual (abnormal) conditions.
At first sight, the ability of eIF2D to deliver not only the tRNAiMet but also some noninitiating tRNAs (tRNAPhe) to 40 S ribosomal subunits might suggest its implication in the elongation or termination rather than the initiation step of polypeptide synthesis. It should be stressed, however, that the novel factor directs tRNAs strictly to the P-site of the ribosome, the property that is characteristic of initiation factors only. Therefore, we think that the name “eukaryotic initiation factor 2D” for this factor is fully justified, at least tentatively.
eIF2D-promoted delivery of tRNAs to 40 S subunits does not involve GTP hydrolysis. The delivery to 40 S ribosomal subunits is not dependent on the presence or absence of the aminoacyl moiety, either. This is one more feature in which eIF2D is distinct from all known translation factors involved in tRNA delivery to ribosomes. This does not mean that the aminoacyl moiety is not required for later steps. This feature, however, may indicate that eIF2D does not specifically recognize the aminoacyl portion of aminoacyl-tRNAs. The presence of the SUI1 domain suggests that the protein may act by changing the conformation of the 40 S ribosomal subunit in a manner similar to eIF1, although in contrast to the latter factor eIF2D rather stabilizes tRNA binding within the P-site.
The absence of any clear effect of the combination of eIF1A, eIF3, eIF4F, eIF4A, and eIF4B on the eIF2D-promoted Met-tRNAiMet delivery to the 40 S ribosome programmed with the leaderless mRNA (Fig. 4A) is an argument against the interaction of eIF2D with these canonical factors. At the same time, eIF3 was necessary for eIF2D-dependent 48 S complex formation on the CAA-cIlacZ mRNA, although in the latter case eIF3 likely functions at the early stage to recruit the 40 S subunit onto mRNA (14, 32). Anyway, we do not exclude that on the ribosome eIF2D may have contact with some of the above initiation (or elongation) factors, and even with the eIF2·GTP·Met-tRNAiMet ternary complex if eIF2D is able to participate also in canonical initiation events.
Although the properties of eIF2D described in this study strongly favor its participation in the translation initiation on a specific mRNA(s), we cannot exclude that it is involved in the regulation of some general translational events, initiation, elongation, or termination, under specific conditions or in specific cells. Elucidation of the functional role of the protein is under way in our laboratory.
Supplementary Material
Acknowledgments
We thank M. Serebryakova for mass spectrometry analysis, V. Makhno and Y. Semenkov for the generous gift of E. coli tRNAfMet, and I. Boni and E. Alkalaeva for the generous gift of [35S]Met-tRNA and tRNAPhe, respectively. We are also very grateful to A. Hinnebusch for critical reading of the manuscript and valuable suggestions.
This work was supported by Grant 08-04-00399 from the Russian Foundation for Basic Research (to I. N. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- IRES
- internal ribosome entry site
- HCV
- hepatitis C virus
- RSW
- ribosomal salt wash
- RRL
- rabbit reticulocyte lysate
- TRITC
- tetramethylrhodamine isothiocyanate.
REFERENCES
- 1.Sonenberg N., Hinnebusch A. G. (2009) Cell 136, 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dever T. E., Roll-Mecak A., Choi S. K., Lee J. H., Cao C., Shin B. S., Burley S. K. (2001) Cold Spring Harbor Symp. Quant. Biol. 66, 417–424 [DOI] [PubMed] [Google Scholar]
- 3.Pestova T. V., Lomakin I. B., Lee J. H., Choi S. K., Dever T. E., Hellen C. U. (2000) Nature 403, 332–335 [DOI] [PubMed] [Google Scholar]
- 4.Ron D., Harding H. P. (2006) in Translational Control in Biology and Medicine (Mathews M. B., Sonenberg N., Hershey J. W., eds) pp. 345–368, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- 5.Terenin I. M., Dmitriev S. E., Andreev D. E., Shatsky I. N. (2008) Nat. Struct. Mol. Biol. 15, 836–841 [DOI] [PubMed] [Google Scholar]
- 6.Merrick W. C., Anderson W. F. (1975) J. Biol. Chem. 250, 1197–1206 [PubMed] [Google Scholar]
- 7.Zoll W. L., Horton L. E., Komar A. A., Hensold J. O., Merrick W. C. (2002) J. Biol. Chem. 277, 37079–37087 [DOI] [PubMed] [Google Scholar]
- 8.Komar A. A., Gross S. R., Barth-Baus D., Strachan R., Hensold J. O., Goss Kinzy T., Merrick W. C. (2005) J. Biol. Chem. 280, 15601–15611 [DOI] [PubMed] [Google Scholar]
- 9.Aravind L., Koonin E. V. (1999) J. Mol. Evol. 48, 291–302 [DOI] [PubMed] [Google Scholar]
- 10.Mitchell S. F., Lorsch J. R. (2008) J. Biol. Chem. 283, 27345–27349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dmitriev S. E., Pisarev A. V., Rubtsova M. P., Dunaevsky Y. E., Shatsky I. N. (2003) FEBS Lett. 533, 99–104 [DOI] [PubMed] [Google Scholar]
- 12.Pestova T. V., Shatsky I. N., Fletcher S. P., Jackson R. J., Hellen C. U. (1998) Genes Dev. 12, 67–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andreev D. E., Terenin I. M., Dunaevsky Y. E., Dmitriev S. E., Shatsky I. N. (2006) Mol. Cell. Biol. 26, 3164–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Terenin I. M., Dmitriev S. E., Andreev D. E., Royall E., Belsham G. J., Roberts L. O., Shatsky I. N. (2005) Mol. Cell. Biol. 25, 7879–7888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dmitriev S. E., Terenin I. M., Dunaevsky Y. E., Merrick W. C., Shatsky I. N. (2003) Mol. Cell. Biol. 23, 8925–8933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pestova T. V., Borukhov S. I., Hellen C. U. (1998) Nature 394, 854–859 [DOI] [PubMed] [Google Scholar]
- 17.Merrick W. C. (1979) Methods Enzymol. 60, 101–108 [DOI] [PubMed] [Google Scholar]
- 18.Andreev D., Hauryliuk V., Terenin I., Dmitriev S., Ehrenberg M., Shatsky I. (2008) RNA 14, 233–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams S. L., Safer B., Anderson W. F., Merrick W. C. (1975) J. Biol. Chem. 250, 9083–9089 [PubMed] [Google Scholar]
- 20.Dmitriev S. E., Andreev D. E., Terenin I. M., Olovnikov I. A., Prassolov V. S., Merrick W. C., Shatsky I. N. (2007) Mol. Cell. Biol. 27, 4685–4697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shanina N. A., Ivanov P. A., Chudinova E. M., Severin F. F., Nadezhdina E. S. (2001) Mol. Biol. (Mosk.) 35, 638–646 [PubMed] [Google Scholar]
- 22.Malygin A. A., Bochkaeva Z. V., Bondarenko E. I., Kosinova O. A., Loktev V. B., Shatskii I. N., Karpova G. G. (2009) Mol. Biol. (Mosk.) 43, 1070–1076 [PubMed] [Google Scholar]
- 23.Kelley L. A., Sternberg M. J. (2009) Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 24.Jakoi E. R., Zampighi G., Robertson J. D. (1976) J. Cell Biol. 70, 97–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jakoi E. R., Ross P. E., Ping Ting-Beall H., Kaufman B., Vanaman T. C. (1987) J. Biol. Chem. 262, 1300–1304 [PubMed] [Google Scholar]
- 26.Pérez-Arellano I., Gallego J., Cervera J. (2007) FEBS J. 274, 4972–4984 [DOI] [PubMed] [Google Scholar]
- 27.Deyo J. E., Chiao P. J., Tainsky M. A. (1998) DNA Cell Biol. 17, 437–447 [DOI] [PubMed] [Google Scholar]
- 28.Reinert L. S., Shi B., Nandi S., Mazan-Mamczarz K., Vitolo M., Bachman K. E., He H., Gartenhaus R. B. (2006) Cancer Res. 66, 8994–9001 [DOI] [PubMed] [Google Scholar]
- 29.Takaku H., Minagawa A., Takagi M., Nashimoto M. (2003) Nucleic Acids Res. 31, 2272–2278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pestova T. V., Kolupaeva V. G. (2002) Genes Dev. 16, 2906–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tzareva N. V., Makhno V. I., Boni I. V. (1994) FEBS Lett. 337, 189–194 [DOI] [PubMed] [Google Scholar]
- 32.Kolupaeva V. G., Unbehaun A., Lomakin I. B., Hellen C. U., Pestova T. V. (2005) RNA 11, 470–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jakoi E. R., Brown A. L., Ho Y. S., Snyderman R. (1989) J. Cell Sci. 93, 227–232 [DOI] [PubMed] [Google Scholar]
- 34.Marchase R. B., Koro L. A., Kelly C. M., McClay D. R. (1982) Cell 28, 813–820 [DOI] [PubMed] [Google Scholar]
- 35.Jakoi E. R., Panchision D. M., Gerwin C. M., DeLorenzo R. J. (1995) Brain Res. 693, 124–132 [DOI] [PubMed] [Google Scholar]
- 36.Severt W. L., Biber T. U., Wu X., Hecht N. B., DeLorenzo R. J., Jakoi E. R. (1999) J. Cell Sci. 112, 3691–3702 [DOI] [PubMed] [Google Scholar]
- 37.Wang Y., Han K. J., Pang X. W., Vaughan H. A., Qu W., Dong X. Y., Peng J. R., Zhao H. T., Rui J. A., Leng X. S., Cebon J., Burgess A. W., Chen W. F. (2002) J. Immunol. 169, 1102–1109 [DOI] [PubMed] [Google Scholar]
- 38.Dang C., Gottschling M., Manning K., O'Currain E., Schneider S., Sterry W., Stockfleth E., Nindl I. (2006) Oncol. Rep. 16, 513–519 [PubMed] [Google Scholar]
- 39.Lee M., Kistler C., Hartmann T. B., Li F., Dummer R., Dippel E., Booken N., Klemke C. D., Schadendorf D., Eichmüller S. B. (2007) Cancer Immunol. Immunother. 56, 783–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong Q. W., Mak W. Y., Chu K. H. (2008) Mar. Biotechnol. 10, 91–98 [DOI] [PubMed] [Google Scholar]
- 41.Fleischer T. C., Weaver C. M., McAfee K. J., Jennings J. L., Link A. J. (2006) Genes Dev. 20, 1294–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krogan N. J., Peng W. T., Cagney G., Robinson M. D., Haw R., Zhong G., Guo X., Zhang X., Canadien V., Richards D. P., Beattie B. K., Lalev A., Zhang W., Davierwala A. P., Mnaimneh S., Starostine A., Tikuisis A. P., Grigull J., Datta N., Bray J. E., Hughes T. R., Emili A., Greenblatt J. F. (2004) Mol. Cell 13, 225–239 [DOI] [PubMed] [Google Scholar]
- 43.Yoon H. J., Donahue T. F. (1992) Mol. Cell. Biol. 12, 248–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lomakin I. B., Shirokikh N. E., Yusupov M. M., Hellen C. U., Pestova T. V. (2006) EMBO J. 25, 196–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lomakin I. B., Kolupaeva V. G., Marintchev A., Wagner G., Pestova T. V. (2003) Genes Dev. 17, 2786–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Passmore L. A., Schmeing T. M., Maag D., Applefield D. J., Acker M. G., Algire M. A., Lorsch J. R., Ramakrishnan V. (2007) Mol. Cell 26, 41–50 [DOI] [PubMed] [Google Scholar]
- 47.Maag D., Fekete C. A., Gryczynski Z., Lorsch J. R. (2005) Mol. Cell 17, 265–275 [DOI] [PubMed] [Google Scholar]
- 48.Cheung Y. N., Maag D., Mitchell S. F., Fekete C. A., Algire M. A., Takacs J. E., Shirokikh N., Pestova T., Lorsch J. R., Hinnebusch A. G. (2007) Genes Dev. 21, 1217–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Algire M. A., Maag D., Lorsch J. R. (2005) Mol. Cell 20, 251–262 [DOI] [PubMed] [Google Scholar]
- 50.Fletcher C. M., Pestova T. V., Hellen C. U., Wagner G. (1999) EMBO J. 18, 2631–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., Sevier C. S., Ding H., Koh J. L., Toufighi K., Mostafavi S., Prinz J., St Onge R. P., VanderSluis B., Makhnevych T., Vizeacoumar F. J., Alizadeh S., Bahr S., Brost R. L., Chen Y., Cokol M., Deshpande R., Li Z., Lin Z. Y., Liang W., Marback M., Paw J., San Luis B. J., Shuteriqi E., Tong A. H., van Dyk N., Wallace I. M., Whitney J. A., Weirauch M. T., Zhong G., Zhu H., Houry W. A., Brudno M., Ragibizadeh S., Papp B., Pál C., Roth F. P., Giaever G., Nislow C., Troyanskaya O. G., Bussey H., Bader G. D., Gingras A. C., Morris Q. D., Kim P. M., Kaiser C. A., Myers C. L., Andrews B. J., Boone C. (2010) Science 327, 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Filipowicz W., Sierra J. M., Nombela C., Ochoa S., Merrick W. C., Anderson W. F. (1976) Proc. Natl. Acad. Sci. U.S.A. 73, 44–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.