Abstract
Protein disulfide isomerase (PDI), which consists of multiple domains arranged as abb′xa′c, is a key enzyme responsible for oxidative folding in the endoplasmic reticulum. In this work we focus on the conformational plasticity of this enzyme. Proteolysis of native human PDI (hPDI) by several proteases consistently targets sites in the C-terminal half of the molecule (x-linker and a′ domain) leaving large fragments in which the N terminus is intact. Fluorescence studies on the W111F/W390F mutant of full-length PDI show that its fluorescence is dominated by Trp-347 in the x-linker which acts as an intrinsic reporter and indicates that this linker can move between “capped” and “uncapped” conformations in which it either occupies or exposes the major ligand binding site on the b′ domain of hPDI. Studies with a range of constructs and mutants using intrinsic fluorescence, collision quenching, and extrinsic probe fluorescence (1-anilino-8-naphthalene sulfonate) show that the presence of the a′ domain in full-length hPDI moderates the ability of the x-linker to generate the capped conformation (compared with shorter fragments) but does not abolish it. Hence, unlike yeast PDI, the major conformational plasticity of full-length hPDI concerns the mobility of the a′ domain “arm” relative to the bb′ “trunk” mediated by the x-linker. The chaperone and enzymatic activities of these constructs and mutants are consistent with the interpretation that the reversible interaction of the x-linker with the ligand binding site mediates access of protein substrates to this site.
Keywords: Endoplasmic Reticulum (ER), Enzyme Mechanisms, Protein Conformation, Protein Domains, Protein Folding, Chaperone, Human Protein Disulfide Isomerase, Intrinsic Fluorescence, Mobility, Proteolysis Sites
Introduction
Disulfide bonds are vital for the stability of many secretory and cell-surface proteins and modulate protein activities in some cases (1). Disulfide bond formation, however, is an error-prone and rate-limiting step during protein folding (2). Protein disulfide isomerase (PDI)4, which is the archetypal and most abundant member of the endoplasmic reticulum (ER)-resident PDI family, plays a key role in catalyzing disulfide formation and rearrangement in eukaryotic cells (3, 4).
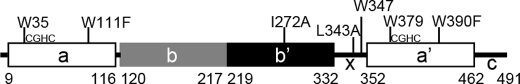
The overall domain construction of human PDI (hPDI) is abb′xa′c (Fig. 1), where domains a and a′ are two thioredoxin-like catalytic domains, b and b′ are two thioredoxin-like non-catalytic domains, x is a linker region, and c is a C-terminal acidic tail (5–7). The b′ domain provides the principal substrate binding site (7–10), and both the a and a′ domains are capable of catalyzing simple oxidoreductions and thiol-disulfide exchange reactions (11). However, when PDI catalyzes a complex disulfide-isomerization associated with protein folding, which involves both thiol/disulfide chemistry and substantive conformational change in the substrate, all four domains of PDI are required to function synergically, combining the chaperone activity with the catalytic activity (12–14). An enzyme performing such a role would be expected to show conformational plasticity, and hence, it is important to define the mobility of PDI in addition to its static structure(s).
FIGURE 1.
Schematic representation of the human PDI molecule. The figure shows the positions of the domains abb′xa′c and the position of tryptophan residues, active sites (CGHC sequences), and point mutations introduced in this work; residue numbering is for mature human PDI (after cleavage of the signal sequence).
No structure of full-length PDI from a multicellular organism has been reported so far, and this is assumed to arise from the interdomain mobility of the molecule and conformational heterogeneity (15, 16). The crystal structure at 4 °C of yeast PDI (yPDI), which has the same domain architecture as hPDI, shows that the four thioredoxin-like domains are arranged in the shape of a twisted “U” with domains a and a′ facing each other, representing two arms connected to a bb′ base, and that a continuous hydrophobic surface is formed at the inside of the U presumably for substrate protein binding (17). Small angle x-ray scattering data on hPDI (18) and data showing the close approach of the active sites in the a and a′ domains of bovine PDI (19) suggest that mammalian PDIs in solution adopt a similar overall conformation, whereas NMR data have confirmed the location of the ligand binding site in hPDI (9, 10). However, a distinct structure of yPDI was obtained at 22 °C that was no longer in the twisted U conformation but, instead, in a “boat” shape, with a ∼123° rotation of the a domain around the loop between the a and b domains. The a arm was further demonstrated to be more flexible than the a′ arm, and the flexibility of the arms, especially that of the a arm, was suggested to be essential for the enzymatic activity both in vitro and in vivo (20).
Regarding the flexibility of hPDI, it has recently been reported that the 19-residue x-linker, a region of defined structure (7), can adopt at least two conformations in the isolated b′x and bb′x fragments (the capped form or uncapped form) by interacting with the hydrophobic pocket in the b′ domain. Mutations were screened in b′x to make the x-linker favor one of these conformations, and the I272A and L343A mutants have been shown to shift the conformation of the x-linker toward the capped and uncapped conformation, respectively (15). Subsequent studies on b′x and bb′x demonstrated that the x-linker can compete with peptide ligands for binding to the b′ domain and has an effect on homodimerization that is also mainly contributed by the b′ domain (10, 16). The yPDI molecule also contains a loop of 17 residues between the b′ and a′ domains, mostly in extended conformation, similar to the 19-residue x-linker in hPDI; this loop is much longer than that of four residues between the a and b domains (17). The highlighting of the flexibility of the ab region rather than of the b′xa′ region in yPDI (20), thus, surprised us and prompted us to reconsider the overall plasticity of the full-length hPDI molecule.
In this work we attempted to define the conformational plasticity of full-length hPDI and to evaluate the role of conformational change in its catalytic and chaperone functions. We used susceptibility to proteases as a measure of flexibility and intrinsic protein fluorescence as a site-specific probe of conformation and worked with mutants and constructs lacking specific domains to provide more structural definition. Our data indicate that in contrast to the situation reported in yPDI (20), the C-terminal half (b′xa′ region) of hPDI is more flexible than the N-terminal half (ab region). The mobility of the x-linker region relative to the ligand binding site on the b′ domain, which was previously observed in b′x and bb′x constructs, is a significant feature in full-length PDI, and this mobility is moderated by the effect of the neighboring a′ domain.
EXPERIMENTAL PROCEDURES
Mutagenesis, Expression, and Purification
The genes encoding PDI, abb′x, bb′xa′ are cloned into pQE30 (Qiagen), and the resulting proteins contain N-terminal (MRGSH6GS) tags. The PDI gene was also cloned into pET24a (Novagen), and the resulting protein contained the C-terminal (His6) tag. Genes encoding PDI, W111F/W390F PDI, W111F abb′x, and b′x are cloned into pET23b (Novagen), and the resulting proteins contained (MH6M) as the N-terminal tags. The I272A and L343A mutants were constructed on the plasmids of their corresponding background proteins by overlap extension PCR (for pQE30) or site-directed mutagenesis (for pET23b), and the sequences of all the constructs were verified by DNA sequencing. Some of the above clones were kindly provided by Prof. L. W. Ruddock (University of Oulu). Proteins from pQE30 (18) or pET23b (16) were expressed and purified as described previously. Before further investigation, all preparations were assessed by native PAGE to confirm that they were monomeric (16). Protein concentrations were determined by the Bradford method with bovine serum albumin as a standard (21) or spectrophotometrically at 280 nm with the absorption coefficients 45.38 mm−1cm−1 for PDI and its I272A and L343A mutants, 34.76 mm−1cm−1 for W111F/390F PDI and its corresponding mutants, and 24.41 mm−1cm−1 for W111F abb′x and its mutants.
Limited Proteolysis Assay
PDI proteins were digested by proteases in 50 mm Tris-HCl buffer (pH 7.6) containing 0 mm (for trypsin), 5 mm (for proteinase K), or 10 mm CaCl2 (for chymotrypsin) and 10 mm dithiothreitol (for proteinase K and chymotrypsin) or 1 mm GSH (for trypsin). The reactions were terminated by phenylmethanesulfonyl fluoride and analyzed by reducing SDS-PAGE.
Mass Spectrometry
Protein samples were desalted into 100 mm ammonium acetate using a BioMax Ultrafree-0.5 PBCC centrifugal filter unit with a 5-kDa nominal cut-off. Six dilutions (10-fold) and concentration steps were performed. All data were acquired on a calibrated quadrupole-time of flight (Q-ToF I) instrument operated in positive ion mode with a precision of 3.5 ppm over the m/z range 500–3000. Each sample was manually infused using a nanoflow capillary after a 2-fold dilution in 30% acetonitrile, 1% formic acid.
Coomassie-stained gel pieces were processed and trypsin-digested using the manufacturer's recommended protocol on the MassPrep robotic protein handling system. The extracted peptides from each sample were analyzed by means of nanoliquid chromatography (LC)-electrospray ionization-MS/MS using a 45-min LC gradient. All MS and MS/MS data were corrected for mass drift using reference data collected from [Glu1]-Fibrinopeptide B (human, F3261; Sigma) sampled each minute of data collection. The data were used to interrogate the UniProt data base release 15.5 appended with the two PDI sequences supplied using ProteinLynx Global Server Version 2.3. Data base search parameters used included chymotrypsin and proteinase K as the primary reagent and trypsin as the secondary reagent. Each sample was analyzed twice; the first injection used 4.9 μl of the digest solution, and the second was ∼25 μl to maximize the numbers of identified peptides.
Intrinsic Fluorescence Measurements
Intrinsic fluorescence spectra of 5 μm proteins in 50 mm Tris-HCl buffer containing 150 mm NaCl (pH 7.6) were recorded at 310–400 nm at 25 °C with excitation at 290 nm.
Fluorescence Enhancement of 1-Anilino-8-naphthalene Sulfonate (ANS)
ANS was added to 50 μm in 50 mm Tris-HCl buffer containing 150 mm NaCl (pH 7.6) in the absence or presence of 5 μm PDI proteins and incubated for 20 min at 25 °C in the dark before the ANS fluorescence emission spectra (400–600 nm) were determined with excitation at 370 nm. The concentration of ANS was determined using an extinction coefficient at 350 nm of 5000 m−1cm−1 (22).
Fluorescence Quenching
Quenching experiments were performed by repeatedly adding 25 μl of 2 m potassium iodide (KI) to a 1-ml sample containing 5 μm purified protein in 20 mm phosphate buffer (pH 7.3) up to a final concentration of 0.33 m potassium iodide (8 additions). KI was also added to 1 ml of buffer as a blank. All fluorescence measurements were carried out in a cuvette with a magnetic stir bar to ensure thorough mixing after each addition, and determinations of intrinsic fluorescence intensity were corrected for dilution. Because the plot of F0/F (where F0 and F are, respectively, the fluorescence intensities at maximum emission wavelength of protein in the absence and presence of quencher) against [KI] was linear in every case, static quenching could be ignored, and the modified form of the Stern-Volmer equation (23, 24), F0/F = (1 + Ksv[KI])eV[KI], which includes the static quenching constant V, could be simplified as F0/F = 1+ Ksv[KI]. The Stern-Volmer constant (Ksv) for dynamic quenching was determined as the slope of the above plot.
Circular Dichroism Spectroscopy
CD spectra were measured at 25 °C in a 0.1-cm path length thermostatically controlled cuvette using a Pistar-180 instrument (Applied Photophysics). The spectra were acquired at 190–260 nm using a 1-nm step size and 2-nm slit widths. For heat-induced protein denaturation, CD signals at 222 nm were collected from 25 to 85 °C with 2 °C intervals controlled by a thermoelectric temperature controller. The real temperature in the cuvette was detected by an inner probe. All CD data were averages of five scans.
Chaperone Activity
Complete denaturation of rhodanese was carried out by incubation of 45 μm rhodanese in 6 m guanidine hydrochloride with 10 mm dithiothreitol for 1 h at room temperature. Refolding was initiated by 100-fold dilution of the denatured rhodanese into a 200 mm sodium phosphate buffer (pH 7.5) in the absence or presence of 2.25 μm PDI proteins at 20 °C. In another assay, 90 μm native rhodanese with 10 mm Na2S2SO3 was 60-fold diluted into the same phosphate buffer without or with 24 μm PDI proteins equilibrated in a water bath of 40 °C. Aggregation was monitored by recording the light scattering at 350 nm in both assays, and the suppression of aggregation during the refolding or heating was used to measure the chaperone activity of PDI proteins (25).
Isomerase Activity
Denatured and reduced RNase A, prepared as described (26), was reactivated at 8 μm in glutathione redox buffer (100 mm Tris acetate buffer containing 50 mm NaCl, 1 mm EDTA, 1 mm GSH, and 0.2 mm GSSG (pH 8.0)) in the presence of 3 μm PDI proteins at 25 °C, and RNase activity was determined by monitoring the absorbance increase at 296 nm due to hydrolysis of 4.5 mm cCMP. The slope of the initial linear increase in RNase activity after the lag time is taken as the measure of isomerase activity. The relative activity (%) was calculated as (A − A0)/(A1 − A0) × 100%, where A is the activity of PDI mutants or abb′x mutants, A1 is the activity of PDI or abb′x, and A0 is the blank in the absence of protein.
Reductase Activity
Insulin of 130 μm was added to 0.1 m potassium phosphate buffer containing 2.5 mm EDTA, 0.1 mm dithiothreitol (pH 7.5) in the absence or presence of 0.5 μm PDI/mutants or 2.5 μm abb′x/mutants to initiate the reaction, and the absorbance at 650 nm that represents light scattering from reduced and precipitated insulin chains was immediately recorded at 25 °C. The activity was calculated by the maximal slope of the curve relative to the lag time (27).
RESULTS
The b′xa′ Region of hPDI Is More Susceptible to Degradation Than the ab Region
We examined the intrinsic molecular flexibility in hPDI by limited proteolysis studies using proteinase K, chymotrypsin, and trypsin and working with full-length PDI and with species lacking specific domains. All constructs contained His6 tags, which facilitated analysis of the location of cleavages by blotting with anti-His6 antibodies.
As shown in Fig. 2A, digestion of full-length PDI (abb′xa′c) with proteinase K generated three new fragments, β, γ, and δ (lanes 6 and 9). The two smaller PDI fragments (γ, δ) could be the fragments abb′x and α (a digestion product of abb′x), respectively, in terms of their mobility in SDS-PAGE (compare lanes 6 to 4). No band corresponding to the size of bb′xa′ appeared in the digestion, implying that the a′ domain was much more sensitive than the a domain to be removed from PDI. Digestions of recombinant fragments abb′x and bb′xa′ were performed for comparison. The abb′x construct is quite resistant to proteinase K, giving rise over time to a slightly smaller product (α) (compare lanes 1, 4, and 7). When bb′xa′ was treated under the same conditions, no large product bands were detectable by Coomassie Blue with the exception of a faint band (ϵ) at around 25 kDa, although quantitative analysis of the bb′xa′ band intensity showed a decrease of 42% at 30 min (lane 8).
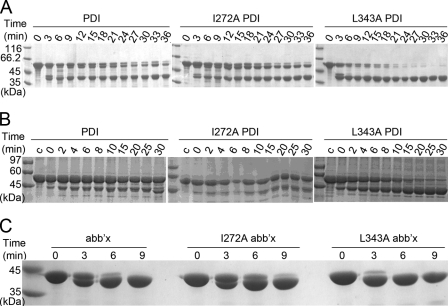
FIGURE 2.
SDS-PAGE analysis of proteolysis products of PDI and its fragments. PDI, abb′x, and bb′xa′ were digested by proteinase K (A and B). PDI was digested by chymotrypsin (C and D) and trypsin (E and F). Proteins with (MRGSH6GS) tags at the N termini (1 mg/ml) were incubated with proteinase K or chymotrypsin (2 μg/ml) at 25 °C for different times as indicated or trypsin at concentrations as indicated at 4 °C for 10 min. The reactions were terminated by adding phenylmethanesulfonyl fluoride to a final concentration of 0.5 mm and analyzed by reducing SDS-15% PAGE for staining by Coomassie Blue (A, C, and E) and for Western blot with anti-His6 monoclonal antibody (B, D, and F).
Because all of these PDI preparations contain a (MRGSH6GS) tag at the N terminus, blotting with anti-His6 monoclonal antibody was used to analyze the above profiles (Fig. 2B). All the bands detectable by Coomassie Blue in Fig. 2A were also visible in Fig. 2B, indicating that the N terminus is intact in every case and that proteinase K has removed varying amounts of sequence from the C terminus of these constructs. A further band (θ) was visible in the blot (lanes 5 and 8) as a digestion product of bb′xa′ slightly larger than ϵ but was not observed in the dye-stained gel. The data show clearly that exposure of PDI to proteinase K gives rise to a series of degradation products in which regions have been removed from the C terminus, but no large non-His6 products are observed, indicating that there is little degradation of the N-terminal region. The relative molecular masses of the various constructs and proteinase K-derived fragments were estimated by mobility on SDS-PAGE, and all the products can be rationalized on the basis of cleavages within the C-terminal tail region (β) or within the x-linker (supplemental Fig. S1).
As an independent check, a PDI construct with a different N-terminal tag, MH6M, was digested with proteinase K to generate an SDS-PAGE profile (supplemental Fig. S2A) in which a band of mobility comparable to band δ (Fig. 2A) was the most prominent product. The digest after 120 min was then analyzed by electrospray mass spectrometry, and the major product was found to have a mass of 38,035 Da, consistent with cleavages after two Met residues at residue −1 (i.e. at the junction between the N-terminal tag and the mature protein) and at residue Met-339 in the x-linker region (note that there was no Met at position −1 in the material analyzed in Fig. 2). This identification was confirmed by mass spectrometric analysis of tryptic digests of this product, which revealed peptides from the a, b, and b′ domains of PDI but none from the a′ domain (data not shown). It was further confirmed by the fact that proteinase K digestion of a full-length L343A mutant PDI (supplemental Fig. S2B) gave rise to a most prominent product of identical mass, indicating that the 38,035-Da product did not extend as far as residue 343.
To avoid conclusions based on the specificity of a single protease, we also carried out the digestion of hPDI by chymotrypsin. As shown in Fig. 2C, two major bands (2 and 3) and three weak bands (1, 4, and 5) appeared. The two major bands (2 and 3) around 45 kDa were both clearly recognized by anti-His6 monoclonal antibody (Fig. 2D), and bands 1 and 4 were also recognized by anti-His6 antibody after a longer time for exposure and development of the blot (data not shown), suggesting that all these large fragments (>35 kDa) still contain domain a at their N terminus and that the principal site(s) of cleavage by chymotrypsin is also in the C-terminal half of the molecule. Again as an independent check, the PDI construct with the MH6M N-terminal tag was digested with chymotrypsin to generate an SDS-PAGE profile in which a band around 45 kDa was the most prominent product (supplemental Fig. S2C). The digest after 120 min was then analyzed by mass spectrometry, and the major product was found to have a mass of 43,873 Da corresponding exactly to that of the product extending from the N-terminal tag through to Trp-379, adjacent to the active site in the a′ domain. Identification of the cleavage site as occurring at this position within the a′ domain was confirmed by two observations. First, mass spectrometric analysis of tryptic digests of this major product contained many tryptic peptides lying N-terminal to this site but none from locations C-terminal to this site (data not shown). Second, analysis of a chymotrypsin digestion of a corresponding full-length PDI containing the L343A mutation (supplemental Fig. S2D) gave rise to a major product of mass 43,833; the difference in mass of 40 between this product and that from wild-type PDI confirms that the site of cleavage is C-terminal to the position of this mutation.
In addition, we carried out digestion of hPDI by trypsin. As shown in Fig. 2E, with 500 μg/ml trypsin hPDI was hydrolyzed into 5 major components. Three bands above 34 kDa (a, b, and c) were recognized by anti-His6 monoclonal antibody (Fig. 2F) and must be fragments with domain a intact but a′ or part of a′ trimmed off. The two smaller fragments (d and e) were not recognized by anti-His6 antibody; band e (∼15 kDa) may be a single domain, and band d (∼24 kDa) may represent bb′. This result is consistent with that from using the other proteases and confirms that interdomain mobility in hPDI is more apparent in the b′xa′ region than in the ab region.
To check the possibility that the N-terminal (MRGSH6GS) tag protected the a domain from digestion, we removed this tag and made a new construct of PDI with a C-terminal (His6) tag. The major product of this protein digested by chymotrypsin has similar mobility to that of PDI with the MRGSH6GS tag at the N terminus (supplemental Fig. S3), and this product cannot be recognized by anti-His6 tag antibody (data not shown), indicating that the principal digestion sites are unaffected and not dependent on the location of the tag.
The x-Linker Can Adopt Alternative Conformations in Full-length PDI
Having established from protease studies that there is flexibility in the b′xa′ region of full-length PDI, we aimed to define the nature of the underlying conformational changes. Previous work on b′x and bb′x constructs has exploited the fluorescence and NMR properties of the unique tryptophan residue (Trp-347) in the x-linker as a conformation-sensitive probe and has established that the x-linker can adopt alternative conformations with respect to the b′ domain, either bound into the ligand binding pocket on b′ to give a capped conformation or displaced from this pocket, giving an uncapped conformation (15). We wished to determine if such alternative conformations occur in full-length PDI, building on the observation that the NMR properties of the indole NH of Trp-347 in full-length PDI indicate that it is mobile between alternative conformations (15).
In full-length PDI there are five tryptophan residues, Trp-35 and Trp-111 in the a domain, Trp-379 and Trp-390 in the a′ domain, and Trp-347 in the x region (see Fig. 1). The fluorescence of Trp-35 has been shown to be heavily quenched by the adjacent active site of the a domain in both dithiol and disulfide states (28), and we hypothesized that the fluorescence of Trp-379 in the a′ domain, which is situated just before the active site in an analogous position to Trp-35, would also be quenched. Hence, we constructed a double-mutant full-length PDI (W111F/W390F) in the hope that its fluorescence would be dominated by Trp-347 that could then act as an unambiguous probe of the conformation of x-linker in full-length PDI.
As shown in Fig. 3A, the intensity of the emission spectrum of W111F/W390F PDI is similar to that of b′x and is much lower than that of wild-type PDI. This is consistent with the suggestion that the fluorescence of W111F/W390F PDI is mainly contributed by Trp-347 in the x region. In isolated b′x, the I272A and L343A mutants have been shown to shift the conformation of the x-linker to the capped and uncapped conformation, respectively (15). Consequently, we constructed the I272A and L343A mutants of full-length PDI in the W111F/W390F mutant background and used their intrinsic fluorescence as models to investigate conformational change and the existence of alternative conformations (capped and uncapped) in full-length PDI.
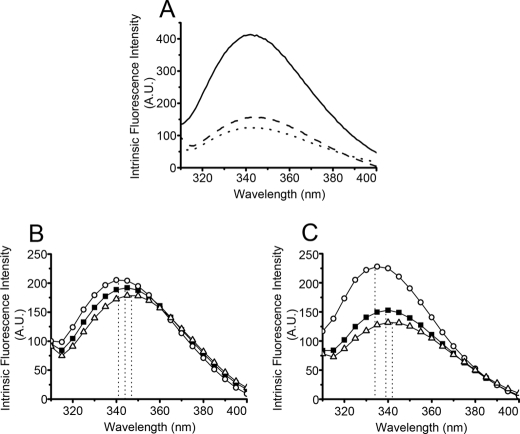
FIGURE 3.
Intrinsic fluorescence emission spectra of PDI, domain constructs, and mutants. Intrinsic fluorescence spectra of PDI (solid line), W111F/W390F PDI (dashed line), and b′x (dotted line) (A), I272A mutant (○) and L343A mutant (▵) in W111F/W390F PDI background (■) (B), and W111F abb′x background (■) (C). Dotted lines in B and C indicate the maximal emission wavelength. A.U., arbitrary units.
The intrinsic fluorescence spectra of W111F/W390F PDI and its I272A and L343A mutants were measured with excitation at 290 nm (Fig. 3B). The maximal emission wavelength of W111F/W390F PDI is 344 nm. The I272A mutant shows a blue shift with a emission maximum at 341 nm, and the L343A mutant shows a red shift with the emission maximum at 347 nm, suggesting that the shifts of conformation to a capped state by I272A and uncapped state by L343A observed in b′x also exist in full-length PDI. Because the maximum of fluorescence emission is difficult to determine precisely and does not necessarily define the properties of the full emission spectrum, we also used the full emission data from 310 to 400 nm to calculate the barycentric mean emission wavelength, the “center of gravity” of the spectrum, as an objective measure of spectral shift on mutants. This analysis confirmed that effects of the mutations in full-length W111F/W390F PDI were in the same directions as observed for b′x but smaller in magnitude (Table 1).
TABLE 1.
Intrinsic fluorescence properties of PDI mutants
| Protein | Barycentric mean wavelength (λm)a | Shifts in λmb | Ksvc |
|---|---|---|---|
| nm | nm | m−1 | |
| b′x | 343.8 | 0.92 ± 0.05 (n = 3) | |
| I272A b′x | 340.5 | −3.3 | 1.03 ± 0.02 (n = 2) |
| L343A b′x | 349.6 | +5.8 | 2.14 ± 0.06 (n = 2)d |
| W111F/W390F PDI | 348.7 | 0.84 ± 0.21 (n = 6) | |
| W111F/W390F/I272A PDI | 346.8 | −1.9 | 0.92 ± 0.22 (n = 5) |
| W111F/W390F/L343A PDI | 350.0 | +1.3 | 1.52 ± 0.17 (n = 4)d |
| W111F abb′x | 345.6 | 0e | |
| W111F/I272A abb′x | 342.9 | −2.7 | 0e |
| W111F/L343A abb′x | 347.0 | +1.4 | 1.26 ± 0.05 (n = 3)d |
a The barycentric mean wavelength between 310 and 400 nm was calculated as λm = ΣF(λ)×(λ)/ΣF(λ), where F(λ) is the fluorescence intensity at wavelength λ (29).
b Difference of λm of I272A or L343A mutants minus λm of the corresponding background proteins. −, blue shift; +, red shift.
c Data were expressed as the mean ± S.D.
d p < 0.001.
e No quenching detectable.
Fluorescence quenching experiments are an effective method for measuring the exposure to solvent of fluorophores within proteins, and hence, we used quenching of fluorescence by I− as a probe of tryptophan exposure in wild-type b′x in full-length W111F/W390F PDI and in their I272A and L343A mutants. Over the quencher range used, Stern-Volmer plots were linear (e.g. supplemental Fig. S4) and were analyzed to give simple dynamic quenching constants (Ksv). In the b′x background, the Ksv values of the wild-type and the I272A mutant are similar, 0.92 and 1.03 m−1 (Table 1), but a considerably larger value 2.14 m−1 was observed for the L343A mutant, consistent with Trp-347 being more exposed in the uncapped L343A mutant, as its fluorescence is more easily quenched. Similar results are also observed in the full-length protein. In W111F/W390F PDI background, the Ksv for the L343A mutant is again significantly higher than that for the background and for I272A mutant. These findings demonstrate that the L343A mutation in full-length PDI causes more exposure of Trp-347 compared with wild-type or I272A mutant, suggesting that this mutation promotes the uncapped conformation in full-length PDI.
We also used the less specific methods of ANS fluorescence and limited proteolysis to probe the conformational changes generated in full-length PDI by the I272A and L343A mutations. As shown in Fig. 4A and Table 2, wild-type PDI and L343A PDI show similar ANS fluorescence intensity, but I272A PDI shows lower ANS fluorescence intensity, indicating less hydrophobic area exposed, consistent with adopting the capped conformation in which the hydrophobic ligand binding site is obstructed. In the limited proteolysis assay, the full-length L343A PDI band almost disappeared after 21 min of digestion by proteinase K (Fig. 5A), whereas about half of I272A PDI and wild-type PDI is intact, indicating that L343A PDI is more sensitive to proteinase K than I272A PDI and wild-type PDI, consistent with a greater tendency to adopt an uncapped conformation. I272A PDI and PDI show similar protease sensitivity over this time scale; a band just below the full-length protein band becomes prominent (cf. band β in Fig. 2, A and B), possibly indicating removal of the C-terminal tail region. Digestion profiles of mutants of full-length PDI by chymotrypsin (Fig. 5B) gave similar results with the L343A mutant much more sensitive to this protease.
FIGURE 4.
ANS fluorescence spectra of I272A and L343A mutants in PDI and abb′x backgrounds. ANS (50 μm) was incubated without (□) or with 5 μm I272A mutant (○) and L343A mutant (▵) in PDI background (■) (A) and in abb′x background (■) (B) for 20 min at 25 °C in the dark before ANS emission spectra were determined with excitation at 370 nm. ● and ▴ are controls with no ANS and only buffer, respectively. A.U., arbitrary units.
TABLE 2.
ANS fluorescence properties of PDI, abb′x, and their mutants
| Protein | Enhancement factora |
|---|---|
| PDI | 8.3 ± 1 |
| I272A PDI | 5.5 ± 0.7 |
| L343A PDI | 7.3 ± 0.8 |
| abb′x | 6.7 ± 0.7 |
| I272A abb′x | 2.6 ± 0.3 |
| L343A abb′x | 6.6 ± 0.6 |
a Enhancement factor = (F(protein + buffer + ANS) − F(protein + buffer))/(F(ANS + buffer) − F(buffer)), where F is the fluorescence intensity at 480 nm. Data were expressed as the mean ± S.D. (n = 3).
FIGURE 5.
Limited proteolysis of I272A and L343A mutants in PDI and abb′x backgrounds. I272A and L343A mutants in PDI background (A) or in abb′x background (C) at 1 mg/ml were digested by 5 μg/ml proteinase K. Phenylmethanesulfonyl fluoride was added to 0.5 mm at different times as indicated to terminate the reaction for reducing SDS-15% PAGE gel. I272A and L343A mutants in PDI background at 2 mg/ml (B) were digested by 0.7 μg/ml chymotrypsin, and the reaction was terminated at various times with 5 mm phenylmethanesulfonyl fluoride.
The Effect of the a′ Domain on the Conformation of the x-Linker
We noticed that the shifts in fluorescence produced by I272A and L343A mutations are quantitatively smaller in full-length PDI (W111F/W390F) than in isolated b′x (Table 1 and Ref. 15), suggesting that the conformational change is moderated by the effects of the adjacent domains. We assumed that the presence of the a′ domain may affect the conformation of the x-linker. To address this question, we constructed the W111F abb′x truncation and its I272A and L343A mutants for fluorescence study of Trp-347. The maximal emission wavelength of W111F abb′x was 339 nm (Fig. 3C), which is a blue shift compared with that of W111F/W390F PDI (344 nm), indicating a more hydrophobic environment for Trp-347 in the truncated molecule with a′ domain removed. This conclusion is confirmed by the analysis of the barycentric mean emission wavelengths (Table 1). In this background the I272A mutation produces W111F/I272A abb′x, which shows emission maximum at 334 nm, a blue shift compared with 339 nm for W111F abb′x, whereas W111F/L343A abb′x shows a red shift with emission maximum at 342 nm; again, these results are confirmed by the analysis of barycentric mean emission wavelength (Table 1).
No fluorescence quenching by iodide was detected for W111F abb′x over the quencher concentration range used, indicating that the Trp-347 residue is buried and inaccessible in this truncated protein. However, the quenching of the Trp-347 in full-length PDI was detected to be 0.84 m−1. The above data indicate that the Trp-347 is more exposed to aqueous medium in the full-length protein, very likely due to the existence of the a′ domain, which makes the x-linker favor the uncapped conformer. The quenching experiments showed significant quenching of Trp-347 fluorescence in the W111F/L343A mutant of abb′x but not in W111F abb′x and the I272A mutant, also indicating that in abb′x the L343A mutation can induce formation of the uncapped state, with Trp-347 more exposed.
We also examined the hydrophobic exposure of abb′x and its I272A and L343A mutants by ANS fluorescence. As shown in Fig. 4B, I272A abb′x displays a much lower ANS fluorescence intensity than that of abb′x, indicating a capped conformation. L343A abb′x shows a similar ANS fluorescence intensity to abb′x. Comparison of the ANS enhancements for I272A abb′x and for I272A PDI (Table 2) suggests that the former is almost entirely in the capped conformation, which is prevented by the a′ domain in the full-length molecule.
From Fig. 2, A and B, it is known that abb′x is digested at its C terminus by proteinase K to give a slightly smaller product (α). In Fig. 5C, the effects of I272A and L343A mutations on proteinase K sensitivity of abb′x were explored. The upper band corresponding to abb′x disappeared by 9 min in abb′x but persisted in I272A abb′x, whereas L343A abb′x completely disappeared by 6 min of digestion. These results are consistent with the expected mutation-induced shifts between capped and uncapped conformations. But the fact that the difference observed in abb′x background is more marked than in full-length PDI background (Fig. 5, A and B) again demonstrates that the truncated protein shows clearer differentiation of capped and uncapped states than does full-length PDI.
To ensure our conclusions are not due to possible gross structural effects of PDI mutations, we measured the far-UV CD spectra of all the mutants. The mutants in either PDI (supplemental Fig. S5A) or abb′x (supplemental Fig. S5B) background showed very similar far-UV CD spectra, indicating that none of the mutations affect the secondary structures of PDI or abb′x. Thus, the Trp-to-Phe mutant proteins are good models for the background proteins while providing more site-specific intrinsic fluorescence probes of conformational change and environment of the x-linker. In addition, we tested the thermal stability of I272A and L343A mutants in both PDI (Fig. 6A) and abb′x backgrounds (Fig. 6B). The heat-induced unfolding curves of I272A and L343A mutants are also very similar to those of the background proteins, suggesting that the I272A and L343A mutations did not destabilize the structure of the background proteins. Hence, the differences we observed with the I272A and L343A mutations in the fluorescence and limited digestion assays arise from their effects on the balance between the capped and uncapped conformations. Furthermore, we found that full-length PDI (Fig. 6A) gradually loses a fraction of its secondary structure as the temperature rises from 25 to 50 °C, whereas the secondary structure of abb′x (Fig. 6B) is quite stable in the same temperature range and shows a single cooperative transition at around 60 °C. This indicates that the a′ domain is relatively unstable compared with abb′x, which is consistent with our finding that the a′ domain is more susceptible to digestion than the a domain.
FIGURE 6.
Heat-induced denaturation curves of I272A and L343A mutants in PDI and abb′x backgrounds. CD signals at 222 nm were collected during heating of 10 μm I272A (○) and L343A (▵) mutants in PDI (■) (A) and abb′x (■) (B) backgrounds in 50 mm Tris-HCl buffer containing 150 mm NaCl (pH 7.6).
The Capped Conformations Show Lower Chaperone Activity
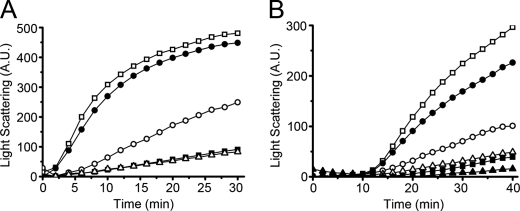
We next investigated the functional significance of these alternative conformations. As shown in Fig. 7A, I272A PDI showed significantly lower potency than PDI in suppressing aggregation of denatured rhodanese upon dilution, suggesting that less hydrophobic area was exposed and available for binding the rhodanese refolding intermediate in the I272A PDI molecule with the capped conformation. L343A PDI showed the same chaperone activity as that of PDI. abb′x and its mutants did not show measurable chaperone activity under these conditions (data not shown) but did show significant activity in an alternative assay, the ability to suppress heat-induced aggregation of rhodanese (Fig. 7B) with I272A abb′x showing lowest ability.
FIGURE 7.
Chaperone activity of I272A and L343A mutants in PDI and abb′x backgrounds. Denatured and reduced rhodanese (45 μm) was 100-fold diluted into 200 mm sodium phosphate (pH 7.5) in the absence (□) or presence of 2.25 μm PDI (■), I272A PDI (○), L343A PDI (▵), or BSA (●) at 20 °C (A). Native rhodanese of 90 μm with 10 mm Na2S2SO3 was 60-fold diluted into the same phosphate buffer without (□) or with 24 μm abb′x (■), I272A abb′x (○), L343A abb′x (▵), PDI (▴), or bovine serum albumin (●) equilibrated in a water bath of 40 °C (B). Aggregation was monitored by recording the light scattering at 350 nm. A.U., arbitrary units.
The Effects of the Alternative Conformations of the x-Linker on the Enzymatic Activity of PDI and abb′x
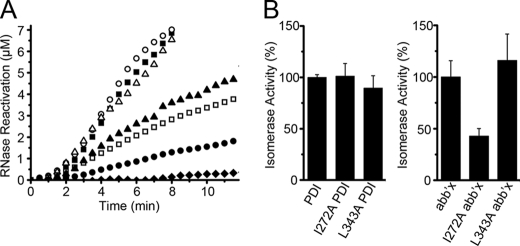
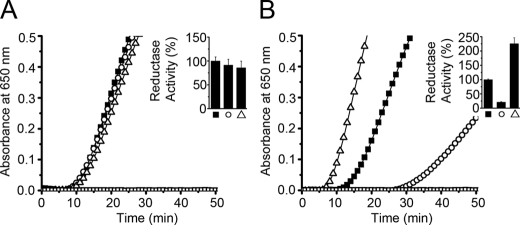
We also tested the protein disulfide isomerase (Fig. 8) and protein disulfide reductase (Fig. 9) activities of PDI and its mutants. The I272A and L343A mutants of full-length PDI are quite similar in both activities to wild-type PDI. However, marked differences were observed in the abb′x background. I272A abb′x showed significantly lower isomerase (43%) and reductase (21%) activity, while L343A abb′x showed higher isomerase (116%) and reductase (226%) activity.
FIGURE 8.
Isomerase activity of I272A and L343A mutants in PDI and abb′x backgrounds. Reactivation of denatured and reduced RNase A (8 μm) in glutathione redox buffer at 25 °C was determined by monitoring the absorbance increase at 296 nm due to hydrolysis of 4.5 mm cCMP in the absence (♦) or presence of 3 μm PDI (■), I272A PDI (○), L343A PDI (▵), abb′x (□), I272A abb′x (●), and L343A abb′x (▴) (A). The slope of the initial linear increase in RNase activity after the lag time (in A) was taken as the isomerase activity, and the enzyme activities of the background proteins (PDI and abb′x) were taken as 100%. Data are expressed as mean ± S.D. (n = 3) (B).
FIGURE 9.
Reductase activity of I272A and L343A mutants in PDI and abb′x backgrounds. Reduction of 130 μm insulin by 0.1 mm dithiothreitol in the absence (□) or presence of I272A mutants (○) and L343A mutants (▵) in PDI background (■) at 0.5 μm (A) and in abb′x background (■) at 2.5 μm (B) was monitored by recording the absorbance at 650 nm at 25 °C. The activity was expressed by using a ratio of the maximal slope of the absorbance curve over the lag time. The enzyme activities of the background proteins (PDI and abb′x) were taken as 100%. Data were expressed as mean ± S.D. (n = 3) (inset).
DISCUSSION
Protein disulfide isomerases are complex multidomain enzymes that catalyze oxidative folding or reductive unfolding of protein substrates depending on the overall redox conditions. In the oxidative folding reaction, reduced, partly oxidized, or mis-oxidized proteins with unfolded, partly folded, or mis-folded conformations undergo linked redox reactions and conformational changes to generate the correctly oxidized protein in its native conformation. Such reactions are intrinsically very slow at physiological pH, and in model systems they are significantly catalyzed by PDI, although the overall turnover numbers are of the order of seconds (26, 30). In recent years there has been increasing emphasis on the dynamic properties of enzymes in understanding their functions, and evidence has accumulated that the intrinsic protein dynamics of enzymes are linked to their catalytic properties (31). It is, therefore, of considerable interest to establish the intrinsic dynamic properties of PDI on time scales up to many seconds and to attempt to understand how these dynamics may be related to enzymic function.
As a step toward this goal we have attempted to define some features of the dynamics of human PDI in solution using limited proteolysis and intrinsic fluorescence as probes and using mutants and truncations of PDI to provide comparative data. Our proteolysis studies with a range of proteases and constructs indicate that the primary sites of cleavage all lie in C-terminal half of PDI, and we have identified specific cleavages occurring within the x-linker and a′ domain, implying greatest flexibility in this region.
To analyze the flexibility implied by the proteolysis data, we used the fluorescence of Trp-347 in the x-linker as an intrinsic probe and also exploited the effects of I272A and L343A mutations. These mutations were initially identified as generating significant blue and red shifts, respectively, in the fluorescence of Trp-347 within the human PDI b′x construct. Subsequent structural studies indicated that in I272A b′x, the x-linker was bound in the ligand binding site on the b′ domain, capping this site and placing Trp-347 in a buried hydrophobic environment (15), whereas wild-type b′x was a mixture of this conformer and a conformer in which Trp-347 was exposed to solvent, with the x-linker not bound to b′, giving an uncapped conformer with a tendency to dimerize (16).
To maximize the ability of Trp-347 to report on conformational changes in full-length PDI and to exploit fully the ability of the I272A and L343A to shift the environment of the x-linker, we mutated two tryptophan residues, Trp-111 and Trp-390, to phenylalanine in full-length PDI. We did not mutate the other two tryptophan residues, Trp-35 and Trp-379, as these are directly adjacent to the active sites in a and a′, respectively, and there was previous evidence that they were highly quenched (28). This was confirmed by our observation that the fluorescence emission spectrum of W111F/W390F PDI was almost identical to that of b′x and, hence, was dominated by emission from Trp-347.
We were, therefore, able to compare the fluorescence properties of nine distinct protein species, wild-type full-length PDI, the mutants I272A and L343A in this background, W111F/W390F full-length PDI and the corresponding mutations in this background, and the previously described b′x, I272A b′x, and L343A b′x. We exploited the position of the emission maximum of intrinsic fluorescence to analyze the hydrophobicity of the environment of Trp-347, the susceptibility to quenching by I− anions to probe the exposure of this residue to solvent, and the ability to enhance the fluorescence of the extrinsic probe ANS to assess the exposure of the hydrophobic ligand binding site on the b′ domain. All the L343A mutants showed the highest values for iodide quenching and ANS binding, consistent with their uncapped form with the ligand binding site unoccupied (available for ANS binding) and the x-linker exposed to solvent (susceptible to quenching). Conversely, all the I272A mutants showed low values for iodide quenching and ANS binding, consistent with their capped form with the ligand binding site occupied by the x-linker and the Trp-347 residues buried (as in the x-ray structure of I272A b′x). The background proteins showed intermediate behavior; their iodide quenching was like that of the I272A mutants (suggesting a capped conformation), whereas their ANS binding resembled that of the L343A mutants. These results suggest that the background proteins are predominantly in the capped conformation but that their x-linker can be readily displaced by binding of ANS to the hydrophobic ligand binding site, just as peptide ligands have been shown to displace x and drive the protein to the uncapped conformation (10). All the results from these studies provided evidence for motion of Trp-347 in full-length PDI between buried and exposed forms, implying that the x-linker can swing into and out of the binding site on the b′ domain to give capped and uncapped conformers of full-length PDI.
Comparative studies with a further three proteins, W111F abb′x and the I272A and L343A mutants in this background, show that without the a′ domain Trp-347 is in a more hydrophobic environment and is hardly quenched by I− anions. Thus, abb′x is quite similar to the monomeric forms of b′x and bb′x, which mainly adopt the capped conformation in solution (16). The presence of the a′ domain in full-length PDI prevents the x-linker from adopting a fully capped conformation; hence, the a′ domain moderates the flexibility of this region but does not abolish it. In this regard, ERp44, a member of the PDI family, is composed of three thioredoxin domains corresponding to the a, b, and b′ domains of PDI and a C-terminal tail but lacking an a′ domain. Interestingly, the hydrophobic pocket in b′ and the adjacent hydrophobic patch in a (proposed as the substrate binding site for ERp44) is partly covered by the C-tail, and removal of the C-tail did increase the activity of ERp44 (32). The C-tail of ERp44, thus, plays a role in regulating substrate binding and release.
Given the evidence that the I272A mutation favors a capped conformer, whereas the L343A mutation favors an uncapped conformer and the wild-type PDI interconverts between these conformers (15), we attempted to analyze the effects of these mutations on functional activities. The interpretation is not necessarily simple because a range of protein conformations can be expected to be involved in the functional catalytic cycle of PDI, and hence, the effect of perturbing the intrinsic balance between such conformations is not immediately predictable. The situation is simplest in relation to chaperone activity. Chaperone assays are performed with chaperone in excess over substrate protein, and the assays used here measure the suppression of substrate protein aggregation. Hence, chaperone activity determined by such assays is effectively a measure of the affinity of chaperone for unfolded or misfolded protein substrate. Hence, it is not surprising that we find chaperone activities correlating with the presence of uncapped conformers in both PDI and abb′x backgrounds and with other measures of the exposure of hydrophobic binding site such as the ability to enhance ANS fluorescence. We found that the fragment abb′x showed high chaperone activity at 40 °C but not at 20 °C, whereas full-length PDI showed high chaperone activity in both temperatures. According to our experimental results, it is suggested that at low temperature abb′x mainly adopts the capped conformer, so that the x-linker strongly competes with the substrate for binding on the b′ domain; high temperatures may enhance the mobility of the x-linker, resulting in a more uncapped state. In full-length PDI, the a′ domain promotes the move of the x-linker out of the capped conformer, thus facilitating substrate binding.
Assays of protein disulfide isomerase or reductase activities are performed as conventional enzyme assays with substrate in excess, but they only determine the appearance of the final product (active and oxidized ribonuclease or reduced insulin chains, respectively). For the reoxidation of ribonuclease, we know that the key slowest step involves disulfide isomerizations of ribonuclease species containing three disulfide bonds and that this is the case both in the absence and presence of PDI (33). But we do not know whether in the enzyme-catalyzed reactions substrate binding, oxidoreduction, substrate conformational change, enzyme conformational change, or substrate release steps are rate-determining, and hence, we cannot readily interpret differences in overall activity. We do know that ligands can displace the x-linker from the binding site so that ligand rather than x “caps” this site. The mutations in full-length PDI produce negligible changes in its overall protein disulfide isomerase and reductase activities, suggesting that the rate-determining step in these activities takes place after the formation of the complex of enzyme and substrate. In this complex the substrate protein will have displaced x, and the extent to which the binding site is capped in the free enzyme will be irrelevant. The construct abb′x has ∼20∼30% of the isomerase (34) and reductase activity (14) of full-length PDI and may operate by a more constrained mechanism, as it lacks the a′ domain, contains only one redox-active site and a less extensive hydrophobic binding site, and especially the x-linker has a greater tendency to adopt the capped conformer. In this significantly less active construct, the rate-determining step may be different, and affinity for the substrate protein may be the determining factor in overall activity.
The flexibility of hPDI described in this work clearly leads to a different picture from that arising from recent studies on yPDI where the junction between domains a and b was identified as a major site of flexibility. High plasticity of the C-terminal half of hPDI would also be expected in view of its functional promiscuity. It is known that hPDI is a multifunctional protein, and the b′xa′ region can fulfill the minimum requirement for function as a subunit of prolyl 4-hydroxylase (35) and for binding and functional redox interaction with Ero1-Lα (34), whereas the bb′xa′ region is necessary and sufficient to unfold and dissociate the cholera toxin A subunit 1 from the holotoxin (36).
However, much still remains to be discovered about the dynamic properties of PDI and about its interactions with substrates or other partner proteins. Future work on these issues should provide a more satisfying insight into the remarkable enzymic activities and other functions of PDI.
Supplementary Material
Acknowledgments
We thank the Waters Centre for Biological Mass Spectrometry and Proteomics in the Department of Biological Sciences, University of Warwick.
This work was supported by Chinese Ministry of Science and Technology Grants 2006CB806508 and 2006CB910903, Chinese Academy of Science Grants KSCX2-YW-R-119 and KSCX2-YW-R-256, and Biotechnology and Biological Sciences Funding Council Project Grant BB/D017807 and partnering award CPA1339 (to R. B. F.) and by Wellcome Trust VIP fellowships (to S. C. and A. K. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- PDI
- protein disulfide isomerase
- hPDI
- human PDI
- yPDI
- yeast PDI
- ER
- endoplasmic reticulum
- ANS
- 1-anilino-8-naphthalene sulfonate
- MS
- mass spectrometry.
REFERENCES
- 1.Hogg P. J. (2003) Trends Biochem. Sci. 28, 210–214 [DOI] [PubMed] [Google Scholar]
- 2.Mamathambika B. S., Bardwell J. C. (2008) Annu. Rev. Cell Dev. Biol. 24, 211–235 [DOI] [PubMed] [Google Scholar]
- 3.Ellgaard L., Ruddock L. W. (2005) EMBO Rep. 6, 28–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hatahet F., Ruddock L. W. (2009) Antioxid. Redox Signal. 11, 2807–2850 [DOI] [PubMed] [Google Scholar]
- 5.Freedman R. B., Gane P. J., Hawkins H. C., Hlodan R., McLaughlin S. H., Parry J. W. (1998) Biol. Chem. 379, 321–328 [DOI] [PubMed] [Google Scholar]
- 6.Alanen H. I., Salo K. E., Pekkala M., Siekkinen H. M., Pirneskoski A., Ruddock L. W. (2003) Antioxid. Redox Signal. 5, 367–374 [DOI] [PubMed] [Google Scholar]
- 7.Pirneskoski A., Klappa P., Lobell M., Williamson R. A., Byrne L., Alanen H. I., Salo K. E., Kivirikko K. I., Freedman R. B., Ruddock L. W. (2004) J. Biol. Chem. 279, 10374–10381 [DOI] [PubMed] [Google Scholar]
- 8.Klappa P., Ruddock L. W., Darby N. J., Freedman R. B. (1998) EMBO J. 17, 927–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denisov A. Y., Määttänen P., Dabrowski C., Kozlov G., Thomas D. Y., Gehring K. (2009) FEBS J. 276, 1440–1449 [DOI] [PubMed] [Google Scholar]
- 10.Byrne L. J., Sidhu A., Wallis A. K., Ruddock L. W., Freedman R. B., Howard M. J., Williamson R. A. (2009) Biochem. J. 423, 209–217 [DOI] [PubMed] [Google Scholar]
- 11.Darby N. J., Creighton T. E. (1995) Biochemistry 34, 11725–11735 [DOI] [PubMed] [Google Scholar]
- 12.Yao Y., Zhou Y., Wang C. (1997) EMBO J. 16, 651–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freedman R. B., Klappa P., Ruddock L. W. (2002) EMBO Rep. 3, 136–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darby N. J., Penka E., Vincentelli R. (1998) J. Mol. Biol. 276, 239–247 [DOI] [PubMed] [Google Scholar]
- 15.Nguyen V. D., Wallis K., Howard M. J., Haapalainen A. M., Salo K. E., Saaranen M. J., Sidhu A., Wierenga R. K., Freedman R. B., Ruddock L. W., Williamson R. A. (2008) J. Mol. Biol. 383, 1144–1155 [DOI] [PubMed] [Google Scholar]
- 16.Wallis A. K., Sidhu A., Byrne L. J., Howard M. J., Ruddock L. W., Williamson R. A., Freedman R. B. (2009) Protein Sci. 18, 2569–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian G., Xiang S., Noiva R., Lennarz W. J., Schindelin H. (2006) Cell 124, 61–73 [DOI] [PubMed] [Google Scholar]
- 18.Li S. J., Hong X. G., Shi Y. Y., Li H., Wang C. C. (2006) J. Biol. Chem. 281, 6581–6588 [DOI] [PubMed] [Google Scholar]
- 19.Hawkins H. C., Blackburn E. C., Freedman R. B. (1991) Biochem. J. 275, 349–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian G., Kober F. X., Lewandrowski U., Sickmann A., Lennarz W. J., Schindelin H. (2008) J. Biol. Chem. 283, 33630–33640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 22.Stryer L. (1965) J. Mol. Biol. 13, 482–495 [DOI] [PubMed] [Google Scholar]
- 23.Birks J. B. (1970) in Photophysics of Aromatic Molecules, pp. 443–447, Wiley-Intersciences, New York [Google Scholar]
- 24.Eftink M. R., Ghiron C. A. (1976) J. Phys. Chem. 80, 486–493 [Google Scholar]
- 25.Song J. L., Wang C. C. (1995) Eur. J. Biochem. 231, 312–316 [DOI] [PubMed] [Google Scholar]
- 26.Lyles M. M., Gilbert H. F. (1991) Biochemistry 30, 613–619 [DOI] [PubMed] [Google Scholar]
- 27.Martínez-Galisteo E., Padilla C. A., García-Alfonso C., López-Barea J., Bárcena J. A. (1993) Biochimie 75, 803–809 [DOI] [PubMed] [Google Scholar]
- 28.Lappi A. K., Lensink M. F., Alanen H. I., Salo K. E., Lobell M., Juffer A. H., Ruddock L. W. (2004) J. Mol. Biol. 335, 283–295 [DOI] [PubMed] [Google Scholar]
- 29.Chalton D. A., Lakey J. H. (2010) Anal. Chem. 82, 3073–3076 [DOI] [PubMed] [Google Scholar]
- 30.Weissman J. S., Kim P. S. (1993) Nature 365, 185–188 [DOI] [PubMed] [Google Scholar]
- 31.Henzler-Wildman K. A., Lei M., Thai V., Kerns S. J., Karplus M., Kern D. (2007) Nature 450, 913–916 [DOI] [PubMed] [Google Scholar]
- 32.Wang L., Wang L., Vavassori S., Li S., Ke H., Anelli T., Degano M., Ronzoni R., Sitia R., Sun F., Wang C. C. (2008) EMBO Rep. 9, 642–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shin H. C., Scheraga H. A. (2000) J. Mol. Biol. 300, 995–1003 [DOI] [PubMed] [Google Scholar]
- 34.Wang L., Li S. J., Sidhu A., Zhu L., Liang Y., Freedman R. B., Wang C. C. (2009) J. Biol. Chem. 284, 199–206 [DOI] [PubMed] [Google Scholar]
- 35.Pirneskoski A., Ruddock L. W., Klappa P., Freedman R. B., Kivirikko K. I., Koivunen P. (2001) J. Biol. Chem. 276, 11287–11293 [DOI] [PubMed] [Google Scholar]
- 36.Forster M. L., Mahn J. J., Tsai B. (2009) J. Biol. Chem. 284, 13045–13056 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.