Abstract
Sphingolipid metabolites regulate cell fate by acting on specific cellular targets. Although the influence of sphingolipids in cellular signaling has been well recognized, the exact molecular targets and how these targets influence cellular signaling mechanisms remain poorly understood. Toward this goal, we used affinity chromatography coupled with proteomics technology and identified acidic leucine-rich nuclear phosphoprotein-32A (ANP32A), an inhibitor of protein phosphatase 2A (PP2A) as a direct target of sphingosine, N,N′-dimethyl sphingosine (DMS) and phytosphingosine but not dihydrosphingosine or sphingosine 1-phosphate. Treatment of human umbilical vein endothelial cells (HUVEC) with DMS, which is not phosphorylated by sphingosine kinases, led to the activation of PP2A activity. Suppression of ANP32A with siRNA enhanced basal and DMS-activated PP2A activity suggesting that the sphingoid base binds to and relieves the inhibitory action of ANP32A on the PP2A complex. Indeed, DMS relieved the ANP32A-mediated inhibition of PP2A enzyme complex in vitro. Interestingly, DMS treatment induced the p38 stress-activated protein kinase (SAPK) and expression of cyclooxygenase (COX)-2 transcript and protein. Knockdown of ANP32A expression further induced p38 SAPK and COX-2. These data identify ANP32A as a novel molecular target of sphingoid bases that regulates cellular signaling events and inflammatory gene expression.
Keywords: Cyclooxygenase (COX) Pathway, Lipid, Phosphatase, PP2A, Sphingolipid
Introduction
Sphingomyelin, a major phospholipid in membranes, is metabolized into bioactive lipids such as ceramide, sphingosine, and sphingosine 1-phosphate (S1P)2 (1). S1P regulates cell migration, survival, and morphogenesis by binding and activating G protein-coupled S1P receptors (2, 3). In contrast, sphingosine and ceramide, which are produced intracellularly from the sphingomyelinase pathway, induce cell death and inhibit cell proliferation (4, 5). However, the mechanism of action of sphingolipids in the intracellular environment is poorly understood.
Sphingosine inhibits cell growth, regulates gene expression and induces apoptosis in a variety of mammalian cell lines (6, 7). In addition, it may inhibit tumor cell proliferation in vivo (8). Moreover, sphingoid bases has been shown to regulate gene expression; for example the inhibition of transcription of the CYP17 gene(9). In Saccharomyces cerevesiae, the sphingoid base (phytosphingosine) regulates actin organization, endocytosis, translation, and heat shock response (10, 11). Although treatment of cells with a sphingoid base regulates kinases and phosphatases (12), direct targets of sphingosine and how such molecules influence downstream cellular changes are poorly understood.
In this study, we searched for novel sphingosine-interacting proteins. We found that two proteins of the acidic, leucine-rich nuclear phosphoprotein-32 family (ANP32), ANP32A (a.k.a. pp32, PHAPI) and ANP32B (a.k.a APRIL), bind to sphingoid bases with high affinity and specificity. ANP32A binding by sphingoid base resulted in the activation of PP2A phosphatase activity, which up-regulated p38 SAPK and cyclooxygenase (COX)-2 expression in human endothelial cells. These data identify ANP32A as a novel intracellular target of sphingosine action in the regulation of cellular signaling and expression of inducible genes such as COX-2.
EXPERIMENTAL PROCEDURES
Materials
Sphingolipids and phospholipids were purchased from Avanti Polar Lipids or Matreya Inc. Aminopropylsilica was purchased from Silicycle (Quebec City, Canada). Antibodies were purchased from the indicated companies; GST, GAPDH, and β-actin antibodies (Sigma), histone 2B (Epitomics) phospho-p38 antibody (Cell Signaling), ANP32A, and p38 antibodies (Santa Cruz Biotechnologies), annexin I antibody (Transduction Labs). COX-2 antibody and PGE2 EIA kit was purchased from the Cayman Chemical Company and purified PP2A dimer was from Millipore.
Cell Culture and Extract Preparation
293T cells were cultured in DMEM supplemented with 10% FBS and antibiotics. HUVECs were cultured as described (13). Confluent cultures of cells were extracted with Buffer A (50 mm Tris, pH 8.0, 2 mm EDTA pH 9.0, 20 mm CHAPS, 10 mm NaF, 1.5 mm semicarbizide, 0.5 mm MgCl2, 1% Triton X-100) for 1 h. Supernatant (20,000 × g) was aliquoted and stored at −80 °C.
Identification of Sphingosine-binding Proteins
Total cell extracts (2 mg/ml) were added to streptavidin membrane spotted with the indicated amounts of biotinyl sphingosine (BSPO) or biotinyl sphingosine1-phosphate (BS1P) (Avanti Polar Lipids) and incubated for 1 h at 4 °C. Membranes were washed in Buffer A, and eluate was separated on a 10% SDS-PAGE gel, were stained with mass spectrometry compatible silver stain. The bands of interest were excised, and the gel pieces were washed and digested in situ with trypsin. Peptides thus generated were injected into a nanocapillary reverse-phase HPLC coupled to a nanoelectrospray ionization source of ThermoFinnigan LTQ quadrupole ion trap mass spectrometer (Finnigan LTQ, Thermo Finnigan, San Jose, CA). MS/MS spectra were searched against a local copy of the non-redundant human protein data base (56,709 entries, November 30, 2004 release version) from the NCI, National Institutes of Health, Advanced Biomedical Computing Center using the SEQUEST algorithm (SEQUEST-PVM version 27 (revision 0)) (14). SEQUEST parameters were as follows: all the filtering thresholds were off; mass tolerance of 1.5 Da for precursor ions and 0.5 Da for fragment ions; full tryptic constraint allowing one missed cleavage; and allowing oxidization (+16 Da) of methionine. The database search results were processed using the INTERACT program (15) and filtered with the following criteria: Xcorr cutoff values of 1.9, 2.2, and 3.7 for 1+, 2+, and 3+ peptides, respectively; ΔCn cutoff value of ≥ 0.1.
Protein Binding to Sphingosine-coupled Silica Beads
GST fusion proteins were purified by glutathione-Sepharose 4B (Amersham Biosciences) affinity chromatography using the Sarkosyl extraction method (16). Annexin I was purified as described (17).
The alkyl chain of the d-erythro-sphingosine backbone was tethered to aminopropyl silica gel by coupling a fully protected, activated w-carboxyl analog of sphingosine to the amino groups of 3-aminopropylsilica. The key transformations were the following sequence of reactions: addition of 1-(dec-9-ynyloxy)-4-methoxybenzene to (S)-Garner aldehyde, hydrolysis of the acetonide with Amberlyst ion-exchange resin, reduction of the triple bond with Red-Al, protection of the 1,3-diol as methoxymethyl (MOM) ethers, selective hydrolysis of the 4-methoxyphenyl ether, and oxidation of the terminal hydroxyl group to the corresponding w-carboxylic acid. After the protected sphingosine w-carboxyl derivative was converted to its p-nitrophenyl ester, reaction with 3-aminopropylsilica in dichloromethane provided the desired amido-tethered sphingosine on silica. The excess amino groups of aminopropylsilica were endcapped with acetic anhydride. The MOM and Boc-protecting groups on sphingosine were removed with bromotrimethylsilane. The silica bonded to the sphingosine ligand contained ∼0.37 mmol of ligand per g.
Sphingosine-coupled silica beads were incubated with 0.05 μg to 0.5 μg GST-ANP32B, GST-ANP32A, or annexin in 1 ml of buffer A. Beads were washed three times with 1 ml of Buffer A, eluted with SDS sample buffer, and separated on a 10% SDS-PAGE gel. Immunoblot analysis was performed with the GST antibody (1:10,000) or annexin I antibody (1:1,000).
Preparation of Liposomes and Protein Binding Assay
Phosphatidylcholine (950 μmol) and sphingolipids (50 μmol) were mixed together as solutions in choloroform, dried under argon, and resuspended in 1 ml of Buffer B (10 mm HEPES pH 7.2, 5 mm MgCl2, 100 mm KCl, and 10 mm NaCl) to a final concentration of 1 mm. Lipid mixtures were vortexed and subjected to 4 freeze (−80 °C) and thaw (25 °C) cycles, and were subsequently extruded (×13) through a 0.1 μm nucleopore filter at 55 °C. Liposomes were quantitated using a phosphorus analysis, which detects the content of PC (18), and analyzed by FACS. Liposomes were stored at room temperature for up to 2 weeks.
GST-ANP32A (0.5 μg) was incubated with 250 μm liposomes in 1 ml of Buffer B for 10 min and centrifuged at 100,000 × g. The liposome pellet was resuspended in SDS sample buffer, and analyzed on an SDS-PAGE gel. Immunoblot analysis was performed using a 1:10,000 dilution of a polyclonal ANP32A antibody.
PP2A Activity Assay
A PP2A assay kit (Upstate Biotechnology) was used to detect PP2A activity according to the manufacturer's instructions. In brief, HUVEC cell lysates were immunoprecipitated with monoclonal anti-PP2A antibody and placed in a PP2A assay (19). PP2A activity ranged from 15.7 to 36.3 units/mg/min and is represented as fold stimulation relative to control. In vitro PP2A assays were done using a radioactive enzyme assay as described below. Dephosphorylation reactions contained 25 mm Tris-HCI, pH 7.4, 1 mm DTT, 10 mm MgCl2. 0.005 units of purified PP2A (Millipore), 0–120 nm ANP32A purified protein, and 100,000 cpm [32P] histone substrate in a final volume of 0.05 ml. Assays utilizing [32P]phosphohistone as substrate also contained 0.2 fmol/ml histone. All lipid solutions were prepared fresh and dissolved in 10% ethanol to a concentration of 0–25 μm immediately prior to addition to assays. Five microliter aliquots were added to reactions giving 0–2.5 μm lipid and a final ethanol concentration of 1%. Reactions were run for 10 min at 30 °C. Reactions were terminated by addition of 0.1 ml of 20% TCA and 20 μg of BSA. Released [32P04] was quantitated by scintillation counting after removal of 0.1 μl of supernatant. PP2A activity ranged from 262 to 324 units/mg/min and is represented relative to control.
Inhibition of Gene Expression by siRNA
The siRNAs of human ANP32A (catalogue number sc-43856) and siRNA-A (catalogue number sc-37007) were purchased from Santa Cruz Biotechnology. Transfection of siRNA (80 nm was performed using Oligofectamine reagent as described by the manufacturer.
RNA Extraction and Real-time PCR
Total RNA was isolated, and qRT-PCR was performed as described (20).
PGE2 Assay
PGE2 concentrations in HUVEC cell media were measured by EIA following the manufacturer's protocol.
Statistical Analysis
Results are reported as means ± S.D. or ± S.E. Statistical significance for parametric data were determined using an unpaired t test for experiments comprising two groups. KD calculations were performed using GraphPad Prism. *, p ≤ 0.05; **, p ≤ 0.01 (two-tailed t test).
RESULTS
Identification of Sphingosine-binding Proteins
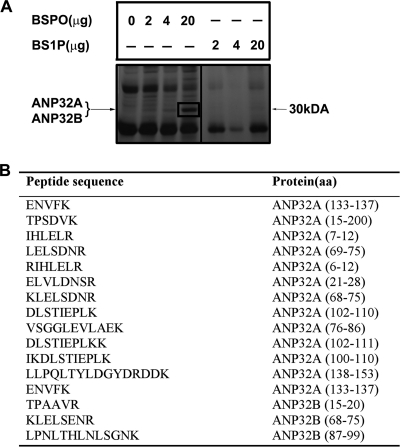
To identify novel sphingosine-binding proteins, human embryonic kidney (HEK)293T whole cell lysates were incubated with BSPO immobilized on a streptavidin membrane. BS1P immobilized on the streptavidin membrane was used as a specificity control. Proteins associated with sphingosine were eluted, separated by SDS-PAGE, and silver stained. A major band at ∼30 kDa bound specifically to BSPO and not to BS1P (Fig. 1A). The gel region corresponding to the ∼30-kDa band was excised, trypsizined, and analyzed by liquid chromatography tandem mass spectrometry (LC-MS/MS). The resulting MS/MS spectra were searched against the non-redundant human data base using SEQUEST algorithm. We identified two closely related proteins of the acidic leucine-rich nuclear phosphoprotein family ANP32A (also known as pp32 or PHAPI) and ANP32B (also known as PHAPII or APRIL)(Fig. 1B). These data suggest that ANP32A and APRIL are sphingosine-binding proteins.
FIGURE 1.
Identification of sphingosine-binding proteins. A, HEK293T whole cell extract (2 mg) was incubated for 1 h with BSPO or BS1P (2–20 μg) immobilized on a streptavidin membrane. After washing, BSPO, BS1P, or streptavidin membrane bound proteins were eluted with SDS sample buffer, separated by SDS-PAGE, and silver stained. The indicated band at 30 kDa representing proteins associated with BSPO was in-gel digested with trypsin and analyzed by LC-tandem mass spectrometry. B, 16 unique peptides that matched ANP32A and ANP32B are indicated.
Specificity of Binding of ANP32A and ANP32B to Sphingolipid Metabolites
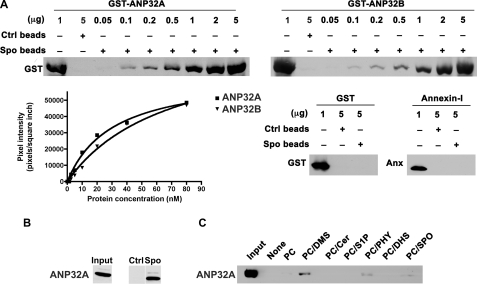
To assess the specificity of binding of ANP32A and ANP32B to sphingolipids, we used recombinant GST fusion proteins ANP32A and ANP32B (21). Upon increasing concentration of GST-ANP32A and GST-ANP32B, there is an increased specific and saturatable binding to sphingosine (Fig. 2A). Purified GST and annexin I, a phosphatidylserine-binding protein failed to bind to sphingosine. The apparent affinity (Kd) of GST-ANP32A and GST-ANP32B to sphingosine is 26.46 ± 5.44 nm and 23.66 ± 15.00 nm, respectively. We further performed a binding assay using a 293T whole cell lysate with sphingosine beads and control beads. Results indicate that the binding of endogenous ANP32A from 293T cells is specific to sphingosine beads and not to control beads (Fig. 2B).
FIGURE 2.
Interaction of ANP32A and ANP32B with sphingolipids. A, different amounts of purified GST-ANP32B, GST-ANP32A, GST, or annexin I were incubated with sphingosine or control beads (1 mg) for 10 min, eluted with SDS sample buffer, and separated by SDS-PAGE. Western blot analysis was performed using an α-GST antibody or α-annexin I antibody. Band intensities were quantitated using a densitometer. B, HEK293T cell extract (1 mg) was incubated with sphingosine beads or control beads (1 mg), washed, eluted with SDS sample buffer, and separated by SDS-PAGE. Western blot analysis was performed using an α-ANP32A antibody. Input represents 50 μg of whole cell lysate. C, GST-ANP32A bound to indicated liposomes (250 μm) was centrifuged, and ANP32A present in the pellet was detected by immunoblot analysis.
Next we tested if ANP32A:sphingosine interaction would occur when the lipid is incorporated into liposomes. We prepared liposomes composed of various sphingolipids, and tested them in an in vitro binding reaction with GST-ANP32A. The bound material was sedimented by ultracentrifugation and detected by immunoblot analysis. We found that ANP32A binds to liposomes containing PC/DMS, PC/sphingosine, and PC/phytosphingosine (Fig. 2C). In contrast, we did not detect binding of GST-ANP32A to liposomes containing PC, PC/ceramide, PC/S1P, or PC/dihydrosphingosine (Fig. 2C). These data suggest that ANP32A binds specifically to sphingosine and related sphingoid bases and that the presence of a double bond in eukaryotic sphingolipids (which is absent in dihydrosphingosine) is essential for this interaction.
Regulation of PP2A Activity by Sphingoid Base Interaction with ANP32A
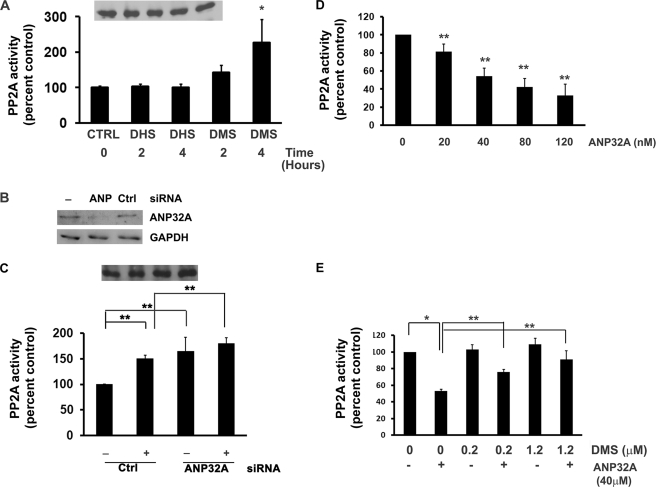
ANP32A is a multifunctional protein which inhibits PP2A activity (19). We investigated how sphingoid base binding to ANP32A affects its ability to inhibit PP2A. For cellular experiments, we used DMS, which binds to ANP32A strongly and cannot be phosphorylated to N,N′-dimethyl-S1P. We used human umbilical vein endothelial cells (HUVEC) which have been extensively characterized in studies of sphingolipid signaling (2, 22). When HUVEC were treated with DMS, cellular PP2A activity was increased significantly in a time-dependent manner. Maximal induction of PP2A activity occurred at 4 h with an increase of 2.26-fold (Fig. 3A). DHS, which did not bind to ANP32A, did not induce PP2A activity.
FIGURE 3.
DMS relieves the inhibitory action of ANP32A on PP2A. A, HUVEC were treated with 10 μm DMS or DHS for various times, and PP2A activity was determined as described under “Experimental Procedures.” PP2A was immunoprecipitated, and PP2A immunoblots are represented in the inset. Data represent mean ± S.D. of at least two independent experiments performed in duplicate. B, HUVEC were treated with ANP32A siRNA or control siRNA for 48 h. ANP32A and GAPDH protein levels were measured by immunoblot analysis. C, HUVEC were treated with ANP32A siRNA (80 nm) or control siRNA (80 nm) for 48 h, followed by treatment with DMS (10 μm) for 2 h or not and PP2A activity was determined. PP2A was immunoprecipitated, and PP2A immunoblots are represented in the inset. Data represent mean ± S.D. of at least two independent experiments performed in duplicate. D, PP2A dimer (containing A and C subunits) was incubated with increasing concentrations of ANP32A (0–120 nm), and its effects on PP2A activity was determined in vitro using the radioactive phosphatase assay, as described under “Experimental Procedures.” Data represent mean ± S.E. of at least three independent experiments performed in duplicate n = 6. E, purified ANP32A (40 nm) was incubated with various concentrations of DMS (0, 0.25, or 1.25 μm), and PP2A dimer activity were determined in vitro as described above. The data represent the mean ± S.E. of at least four independent experiments performed in duplicate n = 8.
To determine the effect of ANP32A on PP2A activity, we used ANP32A siRNA. As shown in Fig. 3B, ANP32A siRNA down regulated the expression of ANP32A protein in HUVEC whereas the scrambled siRNA did not. Down-regulation of ANP32A increased PP2A activity as compared with control siRNA (Fig. 3C). Treatment with DMS further increased PP2A activity. These data suggest that ANP32A is an inhibitor of PP2A and that binding to DMS may further relieve the inhibitory function of ANP32A on PP2A.
Incubation of purified ANP32A with PP2A in vitro resulted in dose-dependent inhibition of enzyme activity (Fig. 3D). However, co-incubation of DMS with ANP32A relieved this inhibition (Fig. 3E), suggesting that sphingoid bases bind to ANP32A and block its interaction with the PP2A enzyme complex.
DMS Induces p38 Stress-activated Protein Kinase and COX-2 Expression in an ANP32A-dependent Manner
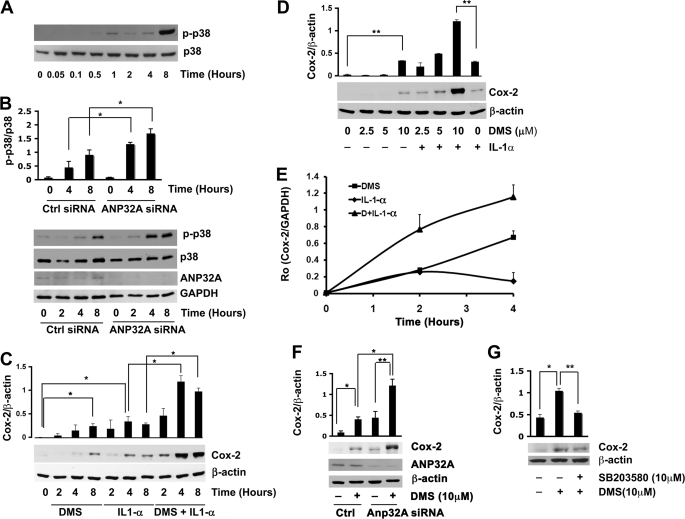
Because PP2A activity has been demonstrated to affect pro-inflammatory signaling pathways (23), we examined whether DMS can regulate the activity of the p38 stress-activated protein kinase (SAPK) and the expression of the inducible prostaglandin synthase COX-2 in HUVEC (24). Treatment with DMS led to a significant increase in phosphorylation of p38 SAPK from 1 to 8 h (Fig. 4A). ANP32A suppression with siRNA significantly amplified the effects of DMS on p38 SAPK activity at 4 and 8 h (Fig. 4B). These data suggest that DMS binding to ANP32A may be inducing p38 SAPK activity through the activation of PP2A.
FIGURE 4.
DMS induces p38 stress-activated protein kinase and COX-2 expression in an ANP32A-dependent manner. A, HUVEC were treated with DMS for the indicated time times. Phospho-p38 SAPK and p38 SAPK protein levels were measured by immunoblot analysis. Results are represented as mean ± S.D. of at least two experiments. B, HUVEC were treated with ANP32A siRNA or control siRNA for 48 h followed by treatment with DMS (10 μm) for the indicated time intervals. Phospho-p38 SAPK, p38 SAPK, ANP32A, and GAPDH protein levels were measured by immunoblot analysis. Phospho-p38 SAPK protein levels were normalized to p38 SAPK protein levels. Results are represented as mean ± S.D. of at least two experiments. C, HUVEC were incubated with IL-1α (10 ng/ml), DMS (10 μm), or DMS in combination with IL-1α for the indicated time intervals. COX-2 protein levels normalized to β-actin protein levels were measured by immunoblot analysis. Results are represented as mean ± S.D. of at least two experiments. D, HUVEC cells were incubated with IL-1α (10 ng/ml), DMS, or DMS in combination with IL-1α for the concentrations of DMS indicated or remained untreated for 8 h. COX-2 protein levels normalized to β-actin protein levels were measured by immunoblot analysis. E, HUVEC were incubated with 10 μm DMS, 10 ng/ml IL-1α, or DMS and IL-1α for the indicated time intervals. COX-2 mRNA levels normalized to GAPDH mRNA levels were measured using qRT- PCR as described under “Experimental Procedures.” Results are represented as mean ± S.D. (n = 3). The experiment was repeated twice with similar results. F, HUVEC were treated with ANP32A siRNA or control siRNA for 48 h followed by treatment with DMS (10 μm) for 8 h. COX-2 protein levels normalized to β-actin protein levels were measured by immunoblot analysis. Results are represented as mean ± S.D. of at least two experiments. G, HUVEC cells were pretreated with SB203580 (10 μm) for 30 min, incubated with DMS (10 μm) for 8 h. COX-2 protein levels were measured by immunoblot analysis and normalized to β-actin levels. Results are expressed as means ± S.D. of at least two experiments. Results are represented as mean ± S.D. of at least two experiments. *, p ≤ 0.05; **, p ≤ 0.01 (two-tailed t test).
We next tested the effects of DMS on COX-2 protein and mRNA expression by Western blot analysis and qRT-PCR, respectively (25). We used the pro-inflammatory cytokine IL-1α to induce COX-2 expression (26). COX-2 was induced by IL-1α in HUVEC as expected (Fig. 4, C and D). Interestingly, COX-2 expression was induced by DMS in a dose- and time-dependent manner. In addition, IL-1-α and DMS synergistically increased COX-2 polypeptide expression (Fig. 4, C and D). Moreover, DMS and IL-1Α alone increased COX-2 mRNA levels and showed synergy when added together (Fig. 4E).
We next tested the effect of ANP32A suppression on DMS induced COX-2 expression. ANP32A suppression significantly amplified the effects of DMS on COX-2 levels (Fig. 4F). Furthermore, treatment of HUVEC with DMS also induced PGE2 synthesis. DMS binding to ANP32A may be inducing COX-2 activity through the activation of PP2A and subsequent activation of p38 SAPK. Indeed, DMS induction of COX-2 was significantly attenuated by SB203580 (p38 inhibitor) (Fig. 4G).
DISCUSSION
In this report we have identified two novel but related sphingosine-binding proteins, ANP32A and ANP32B using affinity chromatography and proteomics technology. We have confirmed this interaction using a liposome assay and a sphingosine-coupled bead assay. We have also determined that ANP32A interacts specifically with phytosphingosine, dimethylsphingosine, and sphingosine. This indicates that ANP32A binds sphingosine-like molecules and not ceramides, which contain an acyl chain or S1P, which contains a phosphate group. Interestingly, dihydrosphingosine does not bind to ANP32A, suggesting that the presence of the unsaturated sphingoid base is essential. The results indicate that sphingoid bases in the sphingomyelinase pathway but not in the de novo biosynthetic pathway (e.g. dihydrosphingosine) interacts with ANP32A, suggesting a role in cellular signal transduction.
ANP32A is a multifunctional protein implicated in cellular signaling, apoptosis, tumor suppression, cerebellar neuronal function, and gene expression (27–29). It contains a leucine-rich repeat domain in the N terminus and an acidic C-terminal domain (30). Analysis of the mutants of ANP32A in a binding assay with sphingosine beads (supplemental Fig. S1) suggest that multiple lipid interaction sites may exist on this protein. However, ANP32A is also a direct inhibitor of PP2A activity (19). It is phosphorylated and shuttles between the nucleus and the cytoplasm to regulate RNA transport (21). However, regulation of ANP32A by upstream signaling pathways or cellular second messengers has not been described. Our finding that sphingosine binds to ANP32A and regulates its interaction with downstream signaling pathways is novel and suggests that it may mediate at least some of the biological functions of this sphingolipid.
We focused on the PP2A pathway as a potential downstream event regulated by sphingosine/ANP32A interaction. Even though a recent report showed that sphingosine activates the PP2A activity, the precise mechanism is not clear (31). Here we demonstrate that DMS is a strong inducer of PP2A activity in HUVEC. Mechanisms of how sphingoid base metabolites induce PP2A activity still remain unclear. Because suppression of ANP32A induces PP2A activity, DMS treatment further stimulates it and prevents the inhibitory effects of ANP32A on PP2A activity in vitro, we suggest that binding of DMS to ANP32A relieves its inhibitory actions on PP2A.
A recent report showed that activation of PP2A by sphingosine in Jurkat T cells led to activation of p38 SAPK (31). We show that DMS increased p38 SAPK activity in HUVECs. Indeed, suppression of ANP32A resulted in superinduction of p38 SAPK, suggesting that DMS/ANP32A activation of PP2A is involved. Thus, in endothelial cells, interaction of DMS with ANP32A led to strong induction of p38 SAPK, which is critically important and inflammatory and stress signaling.
Downstream of p38 SAPK, the inducible prostaglandin synthase COX-2 gene is strongly induced. Early studies showed that sphingosine induces PGE2 production (32). Our work in HUVEC also shows that DMS induces PGE2 synthesis. In addition, S1P was shown to induce COX-2 in epithelial cells (33). Our recent data suggest that S1P2 receptor activation in endothelial cells induces COX-2 expression by transcriptional activation (34). However, the novel findings in this report indicate that sphingoid base interaction with ANP32A induces COX-2 expression via a p38-dependent pathway. In addition to transcriptional activation, DMS may also stabilize the COX-2 mRNA from degradation and enhance translation by acting on post-transcriptional mechanisms.
Our finding that DMS binds to ANP32A and activates PP2A, p38 SAPK, and COX-2 gene expression suggests that ANP32A may also regulate other biological effects of sphingoid bases. Indeed, sphingosine and related sphingoid bases are implicated in apoptosis, gene expression and cell growth control in cell culture as well as in mouse models (6–9). Further, dietary sphingolipids are cancer chemopreventive (35, 36). Whether ANP32A is involved in such actions of sphingoid bases require further study. Similarly, ANP32B, which was implicated in the nuclear export of the RNA-binding protein HuR may also be involved in regulation of gene expression downstream of sphingoid bases. In conclusion, our results identify ANP32A as a novel intracellular target for sphingoid bases. Modulation of ANP32A by sphingoid bases may be involved in endothelial cell functions related to inflammation and angiogenesis.
Supplementary Material
Acknowledgment
The GST-ANP32A and GST-ANP32B plasmids were kindly provided by Dr. Henry Furneaux (UConn Health Center).
This work was supported, in whole or in part, by National Institutes of Health Grants HL67330 and CA77839 (to T. H.), CA113890 (to X. J.), and HL83187 (to R. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- S1P
- sphingosine 1-phosphate
- ANP32A/B
- acidic leucine-rich nuclear phosphoprotein A/B
- Anx-1
- annexin-I
- BSPO
- biotinyl-sphingosine
- Cer
- ceramide
- DHS
- dihydrosphingosine
- DMS
- N,N′-dimethyl sphingosine
- HUVEC
- human umbilical vein endothelial cells
- IL-1α
- interleukin-1α
- PC
- phosphatidylcholine
- PHAPI
- putative HLA class II-associated protein I
- Phy
- phytosphingosine
- PP2A
- protein phosphatase 2A
- SAPK
- stress-activated protein kinase
- siRNA
- small interfering RNA
- SPO
- sphingosine
- BS1P
- biotinyl sphingosine1-phosphate.
REFERENCES
- 1.Bartke N., Hannun Y. A. (2009) J. Lipid Res. 50, (suppl.) S91–S96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hla T., Lee M. J., Ancellin N., Paik J. H., Kluk M. J. (2001) Science 294, 1875–1878 [DOI] [PubMed] [Google Scholar]
- 3.Spiegel S., Milstien S. (2003) Nat. Rev. Mol. Cell Biol. 4, 397–407 [DOI] [PubMed] [Google Scholar]
- 4.Morales A., Lee H., Goñi F. M., Kolesnick R., Fernandez-Checa J. C. (2007) Apoptosis 12, 923–939 [DOI] [PubMed] [Google Scholar]
- 5.Cuvillier O. (2002) Biochim. Biophys. Acta 1585, 153–162 [DOI] [PubMed] [Google Scholar]
- 6.Ohta H., Sweeney E. A., Masamune A., Yatomi Y., Hakomori S., Igarashi Y. (1995) Cancer Res. 55, 691–697 [PubMed] [Google Scholar]
- 7.Nava V. E., Cuvillier O., Edsall L. C., Kimura K., Milstien S., Gelmann E. P., Spiegel S. (2000) Cancer Res. 60, 4468–4474 [PubMed] [Google Scholar]
- 8.Kohno M., Momoi M., Oo M. L., Paik J. H., Lee Y. M., Venkataraman K., Ai Y., Ristimaki A. P., Fyrst H., Sano H., Rosenberg D., Saba J. D., Proia R. L., Hla T. (2006) Mol. Cell. Biol. 26, 7211–7223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Urs A. N., Dammer E., Sewer M. B. (2006) Endocrinology 147, 5249–5258 [DOI] [PubMed] [Google Scholar]
- 10.Ferguson-Yankey S. R., Skrzypek M. S., Lester R. L., Dickson R. C. (2002) Yeast 19, 573–586 [DOI] [PubMed] [Google Scholar]
- 11.Zanolari B., Friant S., Funato K., Sütterlin C., Stevenson B. J., Riezman H. (2000) EMBO J. 19, 2824–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Megidish T., Cooper J., Zhang L., Fu H., Hakomori S. (1998) J. Biol. Chem. 273, 21834–21845 [DOI] [PubMed] [Google Scholar]
- 13.Hla T., Neilson K. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 7384–7388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sadygov R. G., Eng J., Durr E., Saraf A., McDonald H., MacCoss M. J., Yates J. R., 3rd. (2002) J. Proteome Res. 1, 211–215 [DOI] [PubMed] [Google Scholar]
- 15.Han D. K., Eng J., Zhou H., Aebersold R. (2001) Nat. Biotechnol. 19, 946–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frangioni J. V., Neel B. G. (1993) J. Cell Sci. 105, 481–488 [DOI] [PubMed] [Google Scholar]
- 17.Arur S., Uche U. E., Rezaul K., Fong M., Scranton V., Cowan A. E., Mohler W., Han D. K. (2003) Dev. Cell 4, 587–598 [DOI] [PubMed] [Google Scholar]
- 18.Ames B. N., Dubin D. T. (1960) J. Biol. Chem. 235, 769–775 [PubMed] [Google Scholar]
- 19.Li M., Makkinje A., Damuni Z. (1996) Biochemistry 35, 6998–7002 [DOI] [PubMed] [Google Scholar]
- 20.Ghosh M., Wang H., Ai Y., Romeo E., Luyendyk J. P., Peters J. M., Mackman N., Dey S. K., Hla T. (2007) J. Exp. Med. 204, 2053–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brennan C. M., Gallouzi I. E., Steitz J. A. (2000) J. Cell Biol. 151, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Argraves K. M., Obeid L. M., Hannun Y. A. (2002) Adv. Exp. Med. Biol. 507, 439–444 [DOI] [PubMed] [Google Scholar]
- 23.Avdi N. J., Malcolm K. C., Nick J. A., Worthen G. S. (2002) J. Biol. Chem. 277, 40687–40696 [DOI] [PubMed] [Google Scholar]
- 24.Dubois R. N., Abramson S. B., Crofford L., Gupta R. A., Simon L. S., Van De Putte L. B., Lipsky P. E. (1998) Faseb J. 12, 1063–1073 [PubMed] [Google Scholar]
- 25.Lasa M., Mahtani K. R., Finch A., Brewer G., Saklatvala J., Clark A. R. (2000) Mol. Cell. Biol. 20, 4265–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ristimäki A., Garfinkel S., Wessendorf J., Maciag T., Hla T. (1994) J. Biol. Chem. 269, 11769–11775 [PubMed] [Google Scholar]
- 27.Jiang X., Kim H. E., Shu H., Zhao Y., Zhang H., Kofron J., Donnelly J., Burns D., Ng S. C., Rosenberg S., Wang X. (2003) Science 299, 223–226 [DOI] [PubMed] [Google Scholar]
- 28.Pan W., da Graca L. S., Shao Y., Yin Q., Wu H., Jiang X. (2009) J. Biol. Chem. 284, 6946–6954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santa-Coloma T. A. (2003) Cerebellum 2, 310–320 [DOI] [PubMed] [Google Scholar]
- 30.Huyton T., Wolberger C. (2007) Protein Sci. 16, 1308–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoon C. H., Kim M. J., Park M. T., Byun J. Y., Choi Y. H., Yoo H. S., Lee Y. M., Hyun J. W., Lee S. J. (2009) Mol. Cancer Res. 7, 361–370 [DOI] [PubMed] [Google Scholar]
- 32.Ballou L. R., Chao C. P., Holness M. A., Barker S. C., Raghow R. (1992) J. Biol. Chem. 267, 20044–20050 [PubMed] [Google Scholar]
- 33.Pettus B. J., Bielawski J., Porcelli A. M., Reames D. L., Johnson K. R., Morrow J., Chalfant C. E., Obeid L. M., Hannun Y. A. (2003) Faseb J. 17, 1411–1421 [DOI] [PubMed] [Google Scholar]
- 34.Skoura A., Sanchez T., Claffey K., Mandala S. M., Proia R. L., Hla T. (2007) J. Clin. Invest. 117, 2506–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lemonnier L. A., Dillehay D. L., Vespremi M. J., Abrams J., Brody E., Schmelz E. M. (2003) Arch. Biochem. Biophys. 419, 129–138 [DOI] [PubMed] [Google Scholar]
- 36.Merrill A. H., Jr., Schmelz E. M., Wang E., Dillehay D. L., Rice L. G., Meredith F., Riley R. T. (1997) J. Nutr. 127, 830S–833S [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.