Abstract
p190RhoGAP-A (p190) is a GTPase-activating protein known to regulate actin cytoskeleton dynamics by decreasing RhoGTP levels through activation of Rho intrinsic GTPase activity. We have previously shown that p190 protein levels are cell cycle-regulated, decreasing in mitosis, and that this decrease is mediated by the ubiquitin-proteasome pathway. In addition, overexpression of p190 results in decreased RhoGTP levels at the cleavage furrow during cytokinesis, p190 and the RhoGEF Ect2 play opposing roles in cytokinesis, and sustained levels of p190 in mitosis are associated with cytokinesis failure, all findings that suggest but do not directly demonstrate that completion of cytokinesis is dependent on reduced levels of p190. Here we report, using an RNAi reconstitution approach with a degradation-resistant mutant, that decreased p190 levels are required for successful cytokinesis. We also show that the multinucleation phenotype is dependent on p190 RhoGAP activity, determine that the N-terminal GBDS1 region is necessary and sufficient for p190 mitotic ubiquitination and degradation, and identify four N-terminal residues as necessary for the degradation of p190 in mitosis. Our data indicate that in addition to activation of RhoGEF(s), reduction of RhoGAP (p190) is a critical mechanism by which increased RhoGTP levels are achieved in late mitosis, thereby ensuring proper cell division.
Keywords: Cell Division, Microscopic Imaging, Mitosis, Rho, Ubiquitination, Cytokinesis, Degradation, Multinucleation, p190RhoGAP
Introduction
Mitosis is the final stage of the cell cycle where cell cytoplasm, organelles, and replicated DNA are equally separated to give rise to two daughter cells. Mitosis is divided into several, well defined, stages: prophase, prometaphase, metaphase, anaphase, and telophase. Cytokinesis, encompassing anaphase and telophase, begins shortly after sister chromatid separation with the formation of a cleavage furrow and proceeds until cell abscission is completed (1). Irreversible progression through the different mitotic stages depends largely on the proteasomal-dependent degradation of cell cycle regulatory proteins such as cyclin A, cyclin B, Securin, and Plk1 (2).
In addition to proteasomal degradation, an important driver of cytokinesis is the small GTPase RhoA (3). Rho family GTPases are regarded as molecular switches that cycle between the active (GTP-bound) and inactive (GDP-bound) states. Rho activation states are regulated by three classes of proteins: guanine nucleotide exchange factors (GEFs),2 GTPase-activating proteins (GAPs), and GDP dissociation inhibitors. Multiple lines of evidence show that RhoA regulates furrow formation and actomyosin ring contraction during cytokinesis (4). For example, inhibition of RhoA activity prevents cleavage furrow formation (5, 6), and RhoA and its activator, the GEF ECT2, localize to the cleavage furrow and midbody. These findings and others strongly support RhoA involvement in cytokinesis (7–9).
Opposing ECT2 activity is p190RhoGAP-A (p190), which provides a negative component of RhoA regulation (10). p190 has an N-terminal GTP binding domain (GBD), a middle domain (MD) that contains multiple protein interaction motifs, and a C-terminal GAP domain. This GAP domain is specific for the Rho family of GTPases, particularly RhoA (11, 12), and plays a vital role in the regulation of the actin cytoskeleton and its rearrangement in response to growth factor stimulation (13), integrin engagement (14, 15), and v-Src transformation (16).
Previous studies in our laboratory identified an additional function of p190 (17). These studies demonstrated that overexpression of p190 results in multinucleation, a phenotype indicative of cytokinesis failure. This event is dependent on the C-terminal region of p190, which contains the RhoGAP domain, a polyproline domain, and several kinase recognition sites (18). Immunofluorescence microscopy of mitotic cells revealed that endogenous p190 localizes to the cleavage furrow along with actin and upon overexpression interferes with the normal positioning and contraction of the furrow. Furthermore, p190 protein levels were observed to transiently decrease in late mitosis, a decrease that was mediated by the ubiquitin-proteasome pathway. These results raised the questions of whether the cell cycle-linked down-regulation of p190 is required for successful completion of cytokinesis and how mitotic p190 degradation is regulated.
Here, we demonstrate that the decrease in p190 levels observed in mitosis is required for successful completion of cytokinesis and that when it fails to occur, cell division is blocked, and multinucleation ensues. The minimal region on p190 required for significant ubiquitination was mapped to the GBDS1 domain. Moreover, four lysine residues near the extreme N terminus were identified as necessary for mitotic p190 degradation. Using point mutations, we further demonstrate that the multinucleated phenotype is dependent on p190 RhoGAP activity, and therefore, on aberrant Rho signaling. Together, these findings indicate that down-regulation of p190 is a necessary event for the completion of cytokinesis and avoidance of mitotic catastrophe. They also further define the mechanism by which cells achieve the well characterized increase in RhoGTP levels during cytokinesis.
EXPERIMENTAL PROCEDURES
Reagents
The following chemicals were used throughout this study: thymidine, nocodazole, MG132, DAPI (Sigma), lactacystin (Calbiochem), and doxycycline (Clontech). Monoclonal antibodies specific for HA tag (HA.11, Covance; 12CA5, a kind gift of M. Weber, University of Virginia), MAPK (B3B9, provided by M. Weber), p190 (BD Transduction Laboratories), β-actin (Clone AC-15, Sigma), and cyclin A and cyclin B1 (Santa Cruz Biotechnologies) antibodies were used in Western blotting.
Cell Culture and Synchronization
MDA-MB-468 Tet-on p190, ΔGBD, or vector-only inducible human breast cancer cells (17) and HeLa human cervical carcinoma cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen) and penicillin and streptomycin (Invitrogen). Growth media used for MDA-MB-468 cells also contained puromycin (130 ng/ml) and Geneticin (G418, 400 μg/ml) (Sigma). Cells were synchronized by treatment with 2 mm thymidine for 14–16 h, released for 8 h, and subsequently treated with 50 ng/ml nocodazole for 14–16 h. Cells were released from the nocodazole block and harvested by mitotic shake off at indicated times. Either lactacystin (10 μm) or MG132 (20 mm) was added during the time of release as indicated.
Plasmid Constructs
The following p190 constructs used in this study were previously described in Su et al. (17): ΔGBD, ΔGAP (Δ C terminus), GBD, Section 1 (S1), and GAP. The Y1105A, R1283A, 4KR, and GBDS1 4KR mutants were generated by using the QuikChange II site-directed mutagenesis kit (Stratagene) and expressed in the triple HA-tagged vector pKH3 (kind gift from I. Macara, University of Virginia). Other cDNA fragments of p190 used in this study were also cloned into the pKH3 vector at BamHI and EcoRI sites after generation by PCR using the following primer pairs: GBDIVS, 5′-GCCAGGATCCATGATGATGGCAAGAAAG-3′ and 5′-GCGCGAATTCTCACCACTTTAAGAACTCTGGC-3′; GBDS1, 5′-GCCAGGATCCATGATGATGGCAAGAAAG-3′ and 5′-AATTGAATTCCTGAAACGGCAACATTCCCC-3′; IVS/S1, 5′-GTCAGGATCCACAAGCAAAGGACAAG-3′ and 5′-AATTGAATTCCTGAAACGGCAACATTCCCC-3′; S2S3GAP, 5′-GTGAGGATCCCCTGTGAACTCTTTCCAGA-3′ and 5′-GGCCGAATTCCTGATAAGAAGACAAC-3′. All mutants obtained by PCR were confirmed by sequencing before use.
In Vitro Ubiquitination Assay
In vitro ubiquitination reactions were performed as described by Cockman et al. (19). Extracts were prepared from mitotic MDA-MB-468 cells, which provided all enzymes for the ubiquitination reaction. Briefly, cells were swollen in a hypotonic buffer (20 mm Tris, pH 7.5, 5 mm KCl, 1.5 mm MgCl2, and 1 mm DTT), homogenized, and centrifuged at 10,000 × g for 10 min at 4 °C. Aliquots of supernatants were stored at −70 °C. Ubiquitination reactions were performed at 30 °C for 4 h in a total volume of 40 μl that contained 2.5 μl of programmed reticulocyte lysate, 81 μg of hypotonic extract, 1× Energy-Regenerating System (Boston Biochem), 20 μg of ubiquitin (Sigma), 3 μm ubiquitin aldehyde (Boston Biochem), and 20 μm MG132. All constructs were in vitro transcribed and translated in the presence of 20 μCi of [35S]methionine (1175 Ci mmol−1; Amersham Biosciences) using TnT Quick Coupled Transcription/Translation System (Promega). Products of the reaction were analyzed by 7% SDS-PAGE and PhosphorImager scanning (Molecular Dynamics Storm Scanner). Densitometric analysis was performed using the ImageQuant software (Molecular Dynamics, Inc.). Quantitation of the extent of ubiquitination of each construct was determined by densitometric scanning of the area above the in vitro translated product (U lanes in Fig. 4B) in samples that contained the complete ubiquitination reaction and relating that value to the area above the luciferase control. These normalized values were then related to the picomoles of [35S]methionine-labeled TnT product present in the reaction mix. Picomoles of TnT product were calculated on the basis of the total moles of methionine present (hot and cold) in the translation reaction, the incorporation of [35S]methionine in the product, its molecular weight, and the number of methionines it contained. Values obtained from the Luc control were set at 1, and values of other samples were compared with it for generation of -fold ubiquitination/picomoles of protein.
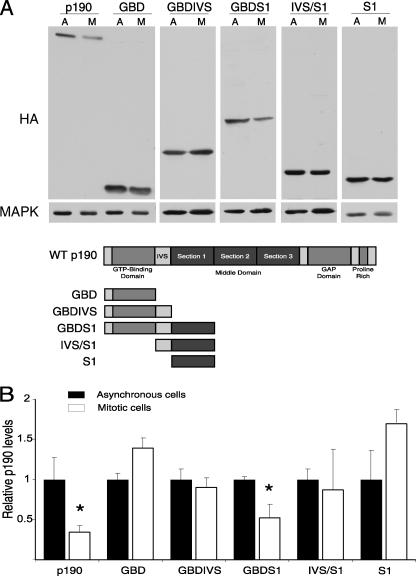
FIGURE 4.
The N-terminal GBDS1 region of p190RhoGAP is sufficient and necessary for its ubiquitination. A, shown are p190 plasmid constructs used in the in vitro ubiquitination assay. B, in vitro ubiquitination of p190 deletion mutants is shown. Plasmids were transcribed, translated, and 35S-labeled in vitro and subjected to in vitro ubiquitination as described under “Experimental Procedures.” A luciferase (Luc) construct was used as a negative control for the ubiquitination reactions. – lanes represent reactions performed in the absence of exogenous ubiquitin. U lanes represent reactions performed in the presence of exogenous ubiquitin. C, quantitation of ubiquitination assays is shown. Analysis of variance analysis and the Dunnett's test were employed to determine significant variances of -fold ubiquitination per pmol of p190 mutants and Luc control. Values are expressed as the mean ± S.E. (n = 3–5). # represents p < 0.02, comparing Luc to p190 mutant; * represents p < 0.02, comparing mutant to WT p190.
In Vivo Degradation Assay
HeLa cells were transfected with various p190 plasmids using Polyfect, as per the manufacturer's instructions. Indicated cDNA constructs were transfected into cells cultured in duplicate 100-mm dishes. Twenty-four hours post-transfection, one dish was synchronized as described, and cells were released from prometaphase arrest for 40 min. The control dish was maintained as an asynchronous cell population. Cells were lysed at 4 °C in radioimmune precipitation assay buffer (50 mm Tris, pH 7.2, 150 mm NaCl, 0.25% deoxycholate, 1% Nonidet P-40) supplemented with 0.5% aprotinin, 1 mm sodium orthovanadate, 12.5 μg/ml leupeptin, and 1 mm PMSF. Protein concentration was determined by the BCA protein assay (Pierce), and 100 μg cell extract was separated by SDS-PAGE, transferred to nitrocellulose, and subjected to Western blotting. HA-tagged p190 variants were detected by anti-HA monoclonal antibodies 12CA5 or HA.11 and HRP-conjugated anti-mouse secondary antibodies (Amersham Biosciences). MAPK or β-actin were used as loading controls. Densitometric analysis was performed using the software AlphaEase (Alpha Innotech Corp.).
RNAi Reconstitution Assay
The RNAi reconstitution assay was performed in MDA-MB-468 Tet-on ΔGBD p190 cells, in MDA-MB-468 Tet-on WT p190 cells, and in HeLa cells. The RNAi oligos were double-stranded and custom made by Dharmacon to silence the expression of human p190-A protein by targeting a unique sequence at the N terminus of the protein. The sequence of the double-stranded oligos was 5′-AAG AUG CAC AUU GUG GAG CAG-3′. MDA-MB-468 cells were treated with the p190 siRNA oligos (60 pmol) using the Oligofectamine reagent (Invitrogen) as per the manufacturer's instructions. After 24 h, cells were transfected with a second round of siRNA, and doxycycline was added (1 μg/ml) to induce the expression of stably transfected ΔGBD or WT p190. Twenty-four hours later, cells were harvested for Western blot analysis or fixed for multinucleation assays, described below. HeLa cells were treated with the siRNA using Oligofectamine reagent and, 24 h post-siRNA treatment, transfected with the pKH3-ΔGBD plasmid using the Polyfect reagent (Qiagen) as per the manufacturer's instructions. After an additional 24 h, cells were harvested for Western blot analysis or fixed for use in multinucleation assays.
Multinucleation Assay
MDA-MB-468 and HeLa cells were fixed by treatment with 4% paraformaldehyde for 20 min, washed 3 times with PBS, permeabilized with 0.2% Triton X-100 for 5 min, and blocked with 20% goat serum at room temperature for 1 h. After blocking, both cell lines were incubated with HA-11 antibody (1:1000 dilution) for 1 h and with Alexa 594-conjugated anti-mouse antibody (1:1000) for 30 min. Nuclear DNA was stained with 2 μg/ml DAPI stain in PBS for 3 min. Coverslips were mounted on microscope slides with VectaShield, and cells were viewed on a Leica microscope fitted for epifluorescence. The presence of multinucleated cells was determined by phase and DAPI staining.
Western Blotting
Cell lysates were prepared as described above and analyzed by SDS-PAGE. Proteins in the gel were wet-transferred to a nitrocellulose membrane and immunoblotted for p190, HA tag, MAPK, or β-actin as previously described (17). Immune complexes were detected with HRP-conjugated anti-mouse antibody, diluted to 1/5000.
RESULTS
A Decrease in p190 Levels during Mitosis Is Required for Successful Completion of Cytokinesis
We previously reported that p190 protein levels transiently decrease in late mitosis, an event mediated by ubiquitination and proteasomal degradation (17). Those results raised the question of whether the cell cycle-linked down-regulation of p190 is an event required for successful completion of cytokinesis.
To test this hypothesis, we used an RNAi reconstitution approach whereby endogenous p190 protein was silenced by transfection of a p190-specific siRNA and then rescued by the expression of an N-terminal deletion mutant (ΔGBD). We previously identified this mutant as resistant to mitotic degradation (17). Furthermore, the ΔGBD mutant also lacked the sequences targeted by the siRNA (Fig. 1A). We utilized MDA-MB-468 cells stably transfected with a tetracycline-inducible (tet-on) construct that, in response to regulated doxycycline treatment, expressed either ΔGBD p190 or WT p190 at levels similar to the endogenous p190 protein (Figs. 1, B and E). Western blotting analysis (Fig. 1B) showed that endogenous p190 protein levels in p190 siRNA-treated cells were reduced to ∼30% that of control siRNA-treated cells and that upon treatment with doxycycline, ΔGBD protein was expressed at levels similar to those of the endogenous p190 protein. Induction of the WT p190 protein in MDA-MB-468 cells after p190 siRNA treatment resulted in overall sustained levels of p190 that were similar to the controls. Fig. 1C shows that the ΔGBD mutant was expressed and protected from cellular degradation and RNAi in mitosis, when cyclin A and B1 levels, used as markers for mitotic progression, were reduced.
FIGURE 1.
A decrease in p190 levels during mitosis is required for successful completion of cytokinesis in doxycycline-inducible MDA-MB-468 cells. A, shown is the experimental design for the RNAi reconstitution. B, shown is a representative Western blot analysis of a reconstitution experiment. Doxycycline (Dox)-inducible expression of ΔGBD and WT p190 in MDA-MB-468 cells is shown after 48 h siRNA and 24 h of doxycycline treatments. 100 μg of whole cell lysate from each sample was analyzed by SDS-PAGE and then immunoblotted as indicated. -Fold expression of total p190 protein (full-length and ΔGBD) is indicated between the gel images. C, the ΔGBD mutant is expressed in mitosis. A reconstitution experiment was performed, and expression of cyclins A and B1 was monitored as a surrogate marker for mitotic progression. Noc, nocodazole; Asynch, asynchronously growing cell population. D, persistent expression of p190 in mitosis increases the frequency of cytokinesis failure and multinucleation. RNAi reconstitution followed by multinucleation assays was performed as described under “Experimental Procedures.” Quantification of multiple experiments is expressed as the ratio of % multinucleation versus control ± S.E. (n = 5). * represents p < 0.02 comparing ΔGBD reconstitution to control or WT p190 reconstitution. E, relative expression levels of WT or ΔGBD p190 are shown. Quantification is based on densitometric analysis of Western blots from numerous reconstitution experiments and is expressed as the -fold multinucleation versus control ± S.E. (n = 5), where control values were set at 1.
After siRNA reconstitution, cells were analyzed by immunofluorescence microscopy for extent of multinucleation. Fig. 1D demonstrates that cells reconstituted with the non-degradable ΔGBD mutant displayed a 2.5-fold increase in multinucleation over control cells (Luc siRNA, no doxycycline) and cells reconstituted with WT p190. When total p190 protein levels were compared among the various treatment groups, expression levels detected in both WT p190 and ΔGBD-reconstituted cells were similar to that of control cells and showed no statistically significant difference (Fig. 1E).
HeLa cells were also employed to test the requirement for a decrease in p190 levels for successful cytokinesis (Fig. 2). siRNA-mediated silencing of p190 was followed by transient introduction of the ΔGBD construct and assessment of multinucleation. Silencing approached 90% (panel A), and the amount of ΔGBD plasmid transfected was titrated to yield levels of ΔGBD expression equal to endogenous p190 (panels B and C). Fig. 2D shows that the number of multinucleated cells among ΔGBD transfectants was ∼11-fold higher than vector control, an increase from ∼3% to >33% in a 48-h period. Taken together, these data indicate that failure to down-regulate p190 in mitosis leads to an increase in cytokinesis defects, supporting the conclusion that degradation of p190 is required for successful cell division.
FIGURE 2.
A decrease in p190 levels during mitosis is required for successful completion of cytokinesis in transiently transfected HeLa cells. A, p190siRNA silences p190 expression in HeLa cells. Quantification of multiple experiments (n = 3) is represented in the graph. Inset, 100 μg of whole cell lysate was analyzed by Western blotting after HeLa cells were treated with 60 pmol of p190 siRNA for 48 h. B, results of titration of ΔGBD expression in HeLa cells are shown. HeLa cells were transfected with increasing amounts of the pKH3 ΔGBD plasmid as indicated for 48 h before harvesting. 100 μg of whole cell lysate was analyzed by SDS-PAGE and then immunoblotted with anti-p190 or anti-β-actin antibodies. C, results of RNAi reconstitution after transient transfection are shown. Representative experiment combining approaches described in A and B. D, persistent expression of the ΔGBD p190 mutant in HeLa cells resulted in increased multinucleation. Multinucleation assays were performed after cells were treated as described in A and B. Arrows indicate multinucleated cells. Quantification of multiple experiments is expressed as the -fold multinucleation versus control ±S.E. (n = 4), where control values were set at 1.
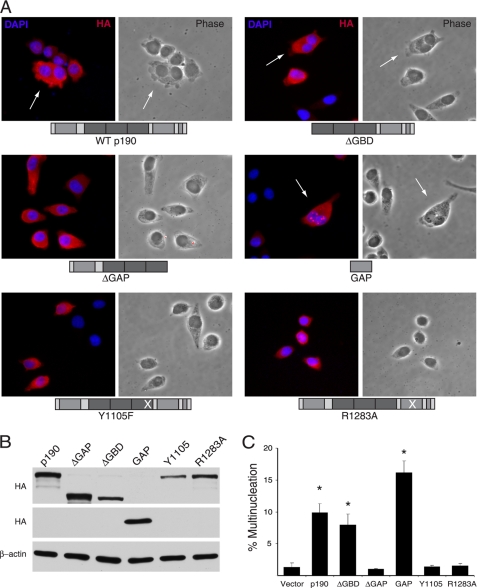
The GAP Activity of p190 Is Necessary and Sufficient to Induce the Multinucleation Phenotype
To better understand the mechanism by which improperly sustained p190 levels in mitosis resulted in multinucleation, we tested whether the GAP domain alone and its Rho-directed activity were necessary and sufficient for the effect. To this end, HeLa cells were transiently transfected with a panel of HA-tagged p190 mutants (depicted in Fig. 3A) and assessed for multinucleation (Fig. 3, A and C). Relative levels of expression were determined by Western blotting with α-HA antibodies (Fig. 3B). Fig. 3C shows that upon transfection of WT p190, ∼10% of expressing cells became multinucleated. Similar levels of mitotic failure were noted in cells expressing the ΔGBD deletion mutant, and control levels were observed in ΔGAP-expressing cells, as previously reported (17). The isolated GAP domain induced ∼16% multinucleation, a figure eight times greater than induced by vector alone, confirming that the GAP domain of p190 was sufficient for disruption of cytokinesis. To further examine the requirement for GAP activity in this event, two additional mutants were tested, Y1105F and R1283A. Both point mutants render p190 enzymatically inactive. Tyrosine 1105 is phosphorylated by Src, and this phosphorylation results in an increased association with p120RasGAP and activation of the GAP domain (20–22). Within the GAP domain, an arginine found at residue 1283, located in a putative G-protein binding helix pocket, is critical for GAP activity (23, 24). Panels A and C of Fig. 3 show that cells transfected with either the Y1105F or R1283A mutants exhibited multinucleation levels similar to those of vector control transfected cells (<2%). Together, these results indicate that the GAP activity of p190 is not only sufficient but also required for the multinucleation phenotype observed in p190-overexpressing cells and that c-Src-mediated phosphorylation of the protein is also required for this event.
FIGURE 3.
The GAP activity of p190 is necessary and sufficient to induce the multinucleation phenotype. A, immunofluorescence microscopy of HeLa cells transfected with the indicated constructs of p190 is shown. HeLa cells were transfected with 2 μg each of the different p190 constructs depicted. Cells were prepared for immunofluorescence microscopy analysis 48 h post-transfection. Arrows indicate multinucleated cells. Representative images are shown. B, Western blot analysis of the expression of p190 mutants is shown. 50 μg of whole cell lysate of transfected cells was analyzed by SDS-PAGE and immunoblotting using the anti-HA antibody, HA.11, or anti-β-actin antibody. C, shown is multinucleation analysis of transfected cells shown in panel A. Quantification of multiple experiments was performed by analysis of variance analysis, and statistical significance was determined by the Dunnett's test. Values are expressed as the mean % multinucleated cells ± S.E. (n = 4). * represents p < 0.02 comparing transfected mutants to vector.
The N-terminal GBDS1 Region of p190RhoGAP Is Sufficient for p190 Ubiquitination
To determine the domains of p190 required for its mitotic ubiquitination, a panel of deletion mutants was prepared and tested for its ability to be ubiquitinated in an in vitro ubiquitination assay (Fig. 4A). Fig. 4, B and C, show that full-length wt p190 protein was ubiquitinated 5-fold above control. Only two of the deletion mutants tested, ΔGAP and GBDS1, showed ubiquitination levels similar to full-length p190. The high levels of ubiquitination observed with the ΔGAP mutant suggested that the C terminus of p190 is dispensable for its ubiquitination. Similarly, high ubiquitination levels of the GBDS1 mutant suggest that most p190 ubiquitination occurs at the N terminus of the protein, indicating that sequences within the GBD domain and S1 of the middle domain are sufficient for p190 ubiquitination and may be important in mediating p190 mitotic degradation.
When the different subdomains found in the ΔGAP and GBDS1 mutants were separately tested for in vitro ubiquitination, a significant overall decrease in ubiquitination was observed. For example, testing of the isolated GBD domain yielded levels similar to those of control reactions, whereas the isolated S1 domain was modified more readily than control reactions but to a significantly lesser extent than the full-length protein. Other N-terminal deletion mutants, ΔGBD, IVS/S1, and S2S3GAP, exhibited levels of ubiquitination that were comparable with those of S1 alone. Together, these data indicate that the combined GBDS1 domains at the N terminus of p190 are sufficient and necessary for directing ubiquitination in vitro, although other regions can be poorly ubiquitinated in its absence.
The GBDS1 Region Is Necessary and Sufficient for Mitotic p190RhoGAP Degradation
Because the N terminus of p190 was necessary and sufficient for ubiquitination (Fig. 4), we next examined whether the GBD and S1 domains acted in a cooperative fashion to regulate the mitotic degradation of p190 and whether they were necessary and/or sufficient for degradation. Therefore, we tested smaller N-terminal deletion mutants to determine the minimal region of p190 that supported degradation. Fig. 5 shows that neither the GBD nor S1 alone was sufficient for degradation. However, the combined GBD and S1 domains (GBDS1) were degraded, indicating that this region is both sufficient and necessary for mitotic degradation of p190.
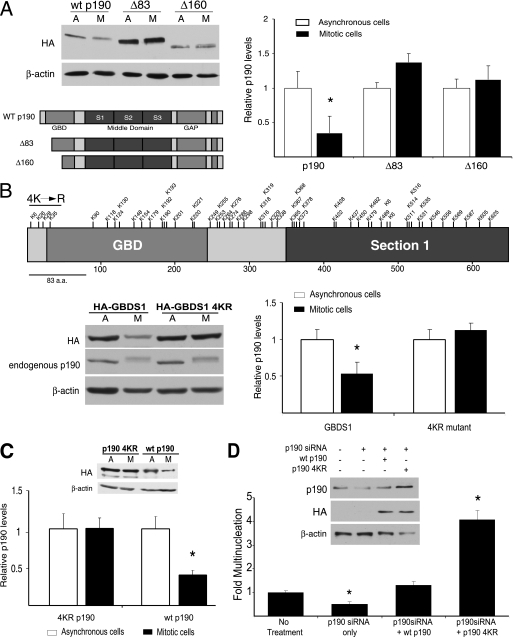
FIGURE 5.
The GBDS1 region is necessary and sufficient for mitotic degradation of p190. A, shown is degradation of N-terminal p190 fragments in HeLa cells. HeLa cells were transiently transfected with the pKH3 p190 expression plasmids indicated in the diagram. At 24 h post-transfection, cells were either maintained as an asynchronous population (A) or synchronized with nocodazole (M) and subsequently released for 40 min. 100 μg of whole cell lysate was subjected to SDS-PAGE followed by Western blotting with the anti-HA 12CA5 and anti-β-actin antibodies. B, quantification of p190 protein levels in asynchronous and mitotic cell populations is shown. Levels of HA-p190 were quantified by densitometric analysis of Western blots. The ratio of p190 to MAPK for the Asynchronous lanes of each mutant group was arbitrarily set to 1, and the corresponding Mitotic lanes (also corrected for amount of MAPK) were further normalized to that value. Values represent the normalized mean ± S.E. (n = 3–5). *. p < 0.02 relative to asynchronous cells expressing the corresponding p190 fragment.
N-terminal Lysine Residues Are Required for p190 Degradation
Because p190 degradation is a necessary mitotic event, we attempted to further define a small region within the molecule or a set of amino acid residues involved in regulating its mitotic stability. To do so, the N-terminal GBD domain was deleted by a nested set of one or two 83-amino acid lengths (leaving ⅔ or ⅓ of the GBD in the context of the WT p190), and these constructs, designated Δ83 and Δ160, were tested in degradation assays. Fig. 6A shows that both mutants were protected from degradation, as no significant changes in protein levels were observed, indicating that residues within the first 83 amino acids of p190 are critical for regulating its mitotic stability. Interestingly, multinucleation assays after introducing increasing amounts of either WT or Δ83 p190 by transient transfection revealed that at lower concentrations, the non-degradable form of p190 (Δ83) was more than twice as efficient in causing the multinucleation phenotype (supplemental Fig. S1). Yet, at greater concentrations, such differences disappear. Furthermore, ubiquitination assays comparing WT p190 to both Δ83 and Δ160 showed decreased ubiquitination levels for both mutants (supplemental Fig. S2), suggesting that regions or specific amino acids important to the ubiquitination and mitotic degradation of p190 were, in fact, to be found within the first 83 amino acids of the protein.
FIGURE 6.
N-terminal lysine residues direct the degradation of p190 in HeLa cells. A, N-terminal deletion mutants of p190 (Δ83 and Δ160) are insensitive to mitotic degradation. HeLa cells were transiently transfected with the indicated pkH3 p190 expression plasmids and subjected to degradation assays, SDS-PAGE analysis, and Western blotting, as described in Fig. 5A. HA-tagged p190 variants were immunoblotted with the HA.11 antibody. A representative gel image is shown. Levels of Δ83 and Δ160 were quantified by densitometric analysis of Western blots. The ratio of mutant to β-actin for the Asynchronous (A) lanes of each mutant group was arbitrarily set to 1, and the corresponding Mitotic lanes (M; also corrected for amount of β-actin) were further normalized to that value. Values represent the normalized mean ± S.E. (n = 3–5). *, p < 0.02 relative to asynchronous cells expressing the corresponding N-terminal mutant. B, the GBDS1 4KR mutant is insensitive to mitotic degradation. A diagram of the rationale for the generation of the GBDS1 4KR mutant is shown. HeLa cells were transiently transfected with the indicated pkH3 GBDS1 expression plasmids and subjected to a degradation assay and analysis as described in panel A. A representative gel image is shown. Quantification represents the normalized mean ± S.E. (n = 3–5). *, p < 0.02 relative to asynchronous cells expressing the corresponding N-terminal mutant. C, the full-length p190 4KR mutant is insensitive to mitotic degradation. HeLa cells were transiently transfected with the indicated pkH3 p190 expression plasmids and subjected to degradation assays and analysis as described in panel A. A representative gel image is shown. Quantification represents the normalized mean ± S.E. (n = 3–5). *, p < 0.02 relative to asynchronous cells expressing the corresponding N-terminal mutant. D, persistent expression of the p190 4KR mutant in HeLa cells resulted in increased multinucleation. Multinucleation assays were performed after RNAi reconstitution, where cells were treated with human-specific p190 siRNA for 24 h, transfected with either wtp190 or p190 4KR rat cDNA, and allowed to incubate for 24 more hours. Samples were then prepared for immunofluorescence microscopy. A representative gel image is shown as an inset. Quantification of multiple experiments is expressed as the -fold multinucleation versus Mock-treated ± S.E. (n = 3), where Mock treatment values were set at 1. * represents p < 0.02 when comparing to Mock or WT p190 reconstitution.
After examination of the amino acid sequence of the N-terminal GBDS1 region, the minimal fragment of p190 that supports both mitotic ubiquitination and degradation, we noticed that although nearly 60 lysines are found in this fragment, only 4 are located within the first 83 amino acids (Fig. 6B). We hypothesized that these four lysines, Lys-6, -26, -28, and -35, could be involved in the ubiquitin-proteasome pathway-dependent degradation of the GBDS1. We, therefore, tested whether point mutants, lysine to arginine substitutions, of these N-terminal residues would have an effect on mitotic GBDS1 stability. Single KR substitution mutants of the aforementioned lysines in the GBDS1 were individually tested in degradation assays. Results showed that all single KR mutants were degraded in a similar fashion as wild-type GBDS1 (supplemental Fig. S3). A mutant with substitutions at all four lysine sites, designated GBDS1 4KR, was then tested in degradation assays. Although the degradation of endogenous p190 protein, shown as a control, continues to be observed in the mitotic cell samples, the GBDS1 4KR mutant is spared from mitotic degradation (Fig. 6B).
To evaluate the relevance of these four N-terminal lysines in the context of the full-length molecule, a full-length p190 construct containing the four lysine to arginine substitutions (p190 4KR) was generated and transfected into HeLa cells, and its mitotic stability was compared against wild-type p190 protein in degradation assays. Fig. 6C shows that whereas wild-type p190 is degraded in mitosis, the p190 4KR mutant is resistant to mitotic degradation, confirming the relevance of these four N-terminal residues to the mitotic degradation of p190. Immunofluorescence microscopy was performed to compare the expression and localization of p190 4KR to that of the wild-type p190 (supplemental Fig. S4). Results demonstrate that after low level overexpression of these proteins, no differences in their localization patterns were observed, with nearly indistinguishable cytosolic localization of the degradation-insensitive 4KR mutant and wild-type p190.
Finally, to test whether the degradation-resistant p190 4KR mutant would yield an increase in the multinucleation phenotype similar to the one caused by the degradation-resistant ΔGBD mutant used in earlier assays (Figs. 1 and 2), an siRNA reconstitution using p190 4KR followed by a multinucleation assay was performed. The siRNA oligo used was specific to endogenous human p190, thus allowing the introduction and expression of rat p190 constructs (wild-type p190 and p190 4KR) in siRNA-treated HeLa cells. Results confirmed our previous findings showing that after silencing of the endogenous p190 pool, expression of the degradation-resistant p190 4KR mutant resulted in a 4-fold increase in observed multinucleation, whereas expression of the wild-type p190 caused no significant difference in multinucleation as compared with the luciferase siRNA control sample (Fig. 6D) further establishing the relevance of these four lysine residues in the regulation of mitotic p190 degradation.
DISCUSSION
Mitotic progression is inextricably dependent on the proteasome-dependent degradation of important regulatory molecules such as A- and B-type cyclins, Securin, and Plk1, among others (2). Furthermore, progression and completion of cytokinesis is also largely dependent on RhoA activity, which regulates the positioning as well as the establishment and contraction of the actomyosin ring at the cleavage furrow (3). Work in our laboratory provides a link between these two mitotic regulatory mechanisms. We previously reported that the expression levels of p190RhoGAP, a negative regulator of RhoA signaling, are cell cycle-regulated and strikingly decreased in late mitosis (17). In this report, we demonstrate that a decrease in p190 levels during mitosis is required for completion of cytokinesis, cell abscission, and avoidance of mitotic catastrophe. The timing of p190 degradation correlates with the detection of active Rho in the cleavage furrow (25, 26), suggesting that a key event that must occur in late mitosis is a decrease in RhoGAP activity. We confirm that the unregulated GAP activity of p190 is responsible for the observed multinucleation phenotype and report that four N-terminal lysine residues are required for p190 mitotic degradation. These findings suggest that the RhoGAP p190 is a critical regulator of cytokinesis.
Existing models ascertain the critical role that RhoA plays in regulating events in mitosis and cytokinesis. Several reports indicate that RhoA activity increases in late mitosis (25, 27), and it is widely accepted that this increased activation is necessary for successful completion of cytokinesis. How are increased RhoGTP levels attained in cytokinesis? Numerous studies have revealed the important role that the RhoGEF ECT2 plays in this activation event (28–31). However, a recent report (10) has shown that ECT2 and p190 have counteracting roles in regulating cytokinesis and demonstrates not only that they associate in mitosis but also colocalize at the cleavage furrow, where RhoA is found (32). The present study describes an additional molecular event that must occur to achieve the increased RhoGTP levels needed for cytokinesis. Here we show that a decrease in p190 levels during mitosis is required to avoid cytokinesis failure and multinucleation. We demonstrate that when cells are unable to lower mitotic levels of p190 protein, as is the case with the non-degradable ΔGBD, Δ83, and 4KR mutants (Figs. 1 and 2, and supplemental Fig. S1, and Fig. 6D, respectively), cytokinesis failure increases significantly. This is true not only in overexpression settings but also when the non-degradable protein is expressed at similar levels as the endogenous p190 protein, as is the case for the ΔGBD and 4KR mutants. Based on these results, we envision a model where the activities of p190 and ECT2 negatively and positively, respectively, regulate Rho activity levels until late in mitosis when increased RhoGTP is required for directing actomyosin ring positioning, establishment, and contraction. At that time, p190, the negative regulator of RhoA, is degraded, and its levels decreased sufficiently to allow full activation of RhoA to occur.
Current models of contractile ring formation posit that its organization is regulated by “RhoGTP activity zones” (33–35). These Rho zones are dynamic due to the cyclical nature of Rho activation yet spatially confined to a narrow region of the plasma membrane that serves as a signal for recruitment/organization of a contractile ring (36). In a recent study (26), our group showed that p190 participates in the establishment and stability of such Rho zones. Elevated levels of p190 during cytokinesis were shown to result in multiple cycles of abnormal contractile ring organization and site selection. When furrows were able to form, they were unstable, giving way to cytokinesis failure. Moreover, through FRET analysis, it was determined that the Rho activity zone in these cells is reduced significantly, giving rise to the aforementioned phenotypes. Altogether, one can conclude that a decrease in endogenous p190 levels during cytokinesis is required not only to increase RhoGTP levels but also to properly focus and stabilize the Rho activity zone, which will signal the organization and activation of the contractile ring.
A previous study by Su et al. (17) showed that the C terminus of p190 was required for mediating the multinucleation phenotype after p190 overexpression. Although that analysis defined a region containing the GAP domain as important for p190-induced multinucleation, it did not define p190 RhoGAP activity as critical to the event. In addition to the GAP domain, other important protein interaction domains are also located at the C terminus, such as the polyproline domain and numerous phosphorylation sites (18) that could potentially play a role in mediating the multinucleation phenotype. By using finer site-directed and deletion mutagenesis, we report that it is, in fact, the Rho GAP activity that is responsible for the multinucleation phenotype (Fig. 3). Mutation of Tyr-1105 and of Arg-1283 in p190 (the GAP activating, c-Src-dependent phosphorylation site and a GAP domain point mutant, respectively) prevents the multinucleation phenotype, whereas overexpression of the GAP domain alone elicits a strong phenotype stronger than that of the full-length protein (Fig. 3). This result further supports the idea that the down-regulation of p190 is necessary for increased RhoGTP levels and appropriate Rho zone focusing. It also suggests a role for c-Src tyrosine kinase through tyrosine 1105 phosphorylation in this process. c-Src involvement in regulating mitosis has been documented for more than two decades. The tyrosine kinase activity of c-Src increases in M phase (37, 38) specifically during metaphase progression and cytokinesis, and this increase can be blocked by treatment with Src family kinase inhibitors (39–41). Thus, regulation of p190RhoGAP in mitosis may be a new mechanism of mitotic regulation by c-Src and Src family kinases.
The increased levels of DNA content observed in multinucleated cells could be achieved by two potential mechanisms, namely, endoreplication or cytokinesis failure. Endoreplication is an alternate cell cycle where a cell genome is replicated without undergoing mitosis or cytokinesis (42). Because endoreplication lacks all vestiges of mitosis such as chromosome condensation/segregation and mitotic spindle formation, we believe that the observed multinucleation phenotype after p190 overexpression is mediated by cytokinesis failure and not endoduplication. In unpublished results obtained by means of confocal microscopy, we clearly and consistently observe chromosome condensation, congression, and segregation as well as mitotic spindle formation in p190-overexpressing cells. Additionally, a recent report (26) shows by means of time-lapse microscopy that the multinucleation phenotype arises from the inability of daughter cells to complete furrow contraction and separate from one another rather than from a previous endoduplication event.
Structure-function analysis of regions of the molecule regulating p190 degradation directed our interest to the GBDS1 N-terminal fragment of the protein, as it was heavily ubiquitinated in vitro and required for mitotic degradation (Figs. 4 and 5). Results indicated that only the combined GBDS1 and not either of the individual domains (GBD or S1) was sufficient for ubiquitination and degradation. Furthermore, deletion of the first 83 amino acids of p190 resulted in protection of the fragment from mitotic degradation (Fig. 6A) as well as a decrease in ubiquitination (supplemental Fig. S2). Together, these results suggested that a degradation motif or sequence might exist in the first 83 amino acids, yet none was found. However, in contrast to the rest of the GBDS1, which is laden with lysine residues, only four isolated lysines are found within the first 83 residues of the p190 sequence. Alanine substitution of all these lysines at once, in the context of both the GBDS1 fragment and the full-length protein, stabilized p190 during mitosis (Fig. 6, B and C), whereas the mutation of single lysines did not affect the degradation of p190. This finding was not at all unexpected given that in numerous other instances, mutations of single lysines have been shown to bear little to no effect on the stabilization or modification of proteins that are ubiquitin targets (43–46). Thus, we define these four residues as the minimal region of p190 regulating its degradation. From these data, we envision a mechanism where there are sequences spread between the GBD and S1 regions that are critical for mediating p190 degradation. For example, a sequence in S1 could function as a recognition site for the ubiquitination machinery, whereas lysines in the GBD domain, potentially Lys-6, -26, -28, and -35, could act as ubiquitin-accepting residues. It is also possible that in the native conformation of the protein, intramolecular interactions between these two regions are necessary to expose ubiquitin accepting or recognition sites elsewhere in the protein. Moreover, it is also possible that sequences along the GBD or S1 may be further modified or involved in protein-protein interactions that, in combination, are required for degradation so that individual domains are found to be stable.
Our results propose that the degradation of p190 in mitosis is a carefully regulated and important event. In a process in which energy efficiency and conservation are paramount, the systematic degradation of p190 in late mitosis is indicative of its critical role as a negative regulator of RhoA activation and, therefore, of cytokinesis.
Supplementary Material
Acknowledgment
We thank P. Todd Stukenberg for constructive and insightful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant R01CA039438 (to S. J. P.), supplemental Grant CA039438–20S1 (to S. A. S. M.), and Cancer Training Grant T32 CA009109 (to the University of Virginia Cancer Center), all from the NCI.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- GEF
- guanine nucleotide exchange factors
- GAP
- GTPase-activating proteins
- GBD
- GTP binding domain
- MD
- middle domain
- S1
- section 1
- IVS
- intervening sequence.
REFERENCES
- 1.Glotzer M. (2001) Annu. Rev. Cell Dev. Biol. 17, 351–386 [DOI] [PubMed] [Google Scholar]
- 2.Peters J. M. (2002) Mol. Cell 9, 931–943 [DOI] [PubMed] [Google Scholar]
- 3.Balasubramanian M. K., Bi E., Glotzer M. (2004) Curr. Biol. 14, R806–R818 [DOI] [PubMed] [Google Scholar]
- 4.Barr F. A., Gruneberg U. (2007) Cell 131, 847–860 [DOI] [PubMed] [Google Scholar]
- 5.Kishi K., Sasaki T., Kuroda S., Itoh T., Takai Y. (1993) J. Cell Biol. 120, 1187–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mabuchi I., Hamaguchi Y., Fujimoto H., Morii N., Mishima M., Narumiya S. (1993) Zygote 1, 325–331 [DOI] [PubMed] [Google Scholar]
- 7.Takaishi K., Sasaki T., Kameyama T., Tsukita S., Tsukita S., Takai Y. (1995) Oncogene 11, 39–48 [PubMed] [Google Scholar]
- 8.Prokopenko S. N., Brumby A., O'Keefe L., Prior L., He Y., Saint R., Bellen H. J. (1999) Genes Dev. 13, 2301–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tatsumoto T., Xie X., Blumenthal R., Okamoto I., Miki T. (1999) J. Cell Biol. 147, 921–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mikawa M., Su L., Parsons S. J. (2008) Cell Cycle 7, 2003–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Settleman J., Narasimhan V., Foster L. C., Weinberg R. A. (1992) Cell 69, 539–549 [DOI] [PubMed] [Google Scholar]
- 12.Ridley A. J., Self A. J., Kasmi F., Paterson H. F., Hall A., Marshall C. J., Ellis C. (1993) EMBO J. 12, 5151–5160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang J. H., Gill S., Settleman J., Parsons S. J. (1995) J. Cell Biol. 130, 355–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakahara H., Mueller S. C., Nomizu M., Yamada Y., Yeh Y., Chen W. T. (1998) J. Biol. Chem. 273, 9–12 [DOI] [PubMed] [Google Scholar]
- 15.Arthur W. T., Burridge K. (2001) Mol. Biol. Cell 12, 2711–2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fincham V. J., Chudleigh A., Frame M. C. (1999) J. Cell Sci. 112, 947–956 [DOI] [PubMed] [Google Scholar]
- 17.Su L., Agati J. M., Parsons S. J. (2003) J. Cell Biol. 163, 571–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang W., Betson M., Mulloy R., Foster R., Lévay M., Ligeti E., Settleman J. (2008) J. Biol. Chem. 283, 20978–20988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cockman M. E., Masson N., Mole D. R., Jaakkola P., Chang G. W., Clifford S. C., Maher E. R., Pugh C. W., Ratcliffe P. J., Maxwell P. H. (2000) J. Biol. Chem. 275, 25733–25741 [DOI] [PubMed] [Google Scholar]
- 20.Roof R. W., Haskell M. D., Dukes B. D., Sherman N., Kinter M., Parsons S. J. (1998) Mol. Cell. Biol. 18, 7052–7063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haskell M. D., Nickles A. L., Agati J. M., Su L., Dukes B. D., Parsons S. J. (2001) J. Cell Sci. 114, 1699–1708 [DOI] [PubMed] [Google Scholar]
- 22.Hu K. Q., Settleman J. (1997) EMBO J. 16, 473–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li R., Zhang B., Zheng Y. (1997) J. Biol. Chem. 272, 32830–32835 [DOI] [PubMed] [Google Scholar]
- 24.Tatsis N., Lannigan D. A., Macara I. G. (1998) J. Biol. Chem. 273, 34631–34638 [DOI] [PubMed] [Google Scholar]
- 25.Yoshizaki H., Ohba Y., Parrini M. C., Dulyaninova N. G., Bresnick A. R., Mochizuki N., Matsuda M. (2004) J. Biol. Chem. 279, 44756–44762 [DOI] [PubMed] [Google Scholar]
- 26.Su L., Pertz O., Mikawa M., Hahn K., Parsons S. J. (2009) Exp. Cell Res. 315, 1347–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maddox A. S., Burridge K. (2003) J. Cell Biol. 160, 255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimura K., Tsuji T., Takada Y., Miki T., Narumiya S. (2000) J. Biol. Chem. 275, 17233–17236 [DOI] [PubMed] [Google Scholar]
- 29.Yüce O., Piekny A., Glotzer M. (2005) J. Cell Biol. 170, 571–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishimura Y., Yonemura S. (2006) J. Cell Sci. 119, 104–114 [DOI] [PubMed] [Google Scholar]
- 31.Chalamalasetty R. B., Hümmer S., Nigg E. A., Silljé H. H. (2006) J. Cell Sci. 119, 3008–3019 [DOI] [PubMed] [Google Scholar]
- 32.Nishimura Y., Nakano K., Mabuchi I. (1998) FEBS Lett. 441, 121–126 [DOI] [PubMed] [Google Scholar]
- 33.Piekny A., Werner M., Glotzer M. (2005) Trends Cell Biol. 15, 651–658 [DOI] [PubMed] [Google Scholar]
- 34.Bement W. M., Miller A. L., von Dassow G. (2006) BioEssays 28, 983–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bement W. M., Benink H. A., von Dassow G. (2005) J. Cell Biol. 170, 91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller A. L., von Dassow G., Bement W. M. (2008) Biochem. Soc. Trans. 36, 378–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chackalaparampil I., Shalloway D. (1988) Cell 52, 801–810 [DOI] [PubMed] [Google Scholar]
- 38.Kuga T., Nakayama Y., Hoshino M., Higashiyama Y., Obata Y., Matsuda D., Kasahara K., Fukumoto Y., Yamaguchi N. (2007) Arch. Biochem. Biophys. 466, 116–124 [DOI] [PubMed] [Google Scholar]
- 39.Moasser M. M., Srethapakdi M., Sachar K. S., Kraker A. J., Rosen N. (1999) Cancer Res. 59, 6145–6152 [PubMed] [Google Scholar]
- 40.Ng M. M., Chang F., Burgess D. R. (2005) Dev. Cell 9, 781–790 [DOI] [PubMed] [Google Scholar]
- 41.Kasahara K., Nakayama Y., Nakazato Y., Ikeda K., Kuga T., Yamaguchi N. (2007) J. Biol. Chem. 282, 5327–5339 [DOI] [PubMed] [Google Scholar]
- 42.Edgar B. A., Orr-Weaver T. L. (2001) Cell 105, 297–306 [DOI] [PubMed] [Google Scholar]
- 43.Zelcer N., Hong C., Boyadjian R., Tontonoz P. (2009) Science 325, 100–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Welbourn S., Jirasko V., Breton V., Reiss S., Penin F., Bartenschlager R., Pause A. (2009) J Gen. Virol. 90, 1071–1080 [DOI] [PubMed] [Google Scholar]
- 45.Kim J. H., Sohn S. Y., Benedict Yen T. S., Ahn B. Y. (2008) Biochem. Biophys. Res. Commun. 366, 1036–1042 [DOI] [PubMed] [Google Scholar]
- 46.Batonnet S., Leibovitch M. P., Tintignac L., Leibovitch S. A. (2004) J. Biol. Chem. 279, 5413–5420 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.