Abstract
Drug resistance-associated mutations in HIV-1 reverse transcriptase (RT) can affect the balance between polymerase and ribonuclease H (RNase H) activities of the enzyme. We have recently demonstrated that the N348I mutation in the connection domain causes selective dissociation from RNase H-competent complexes, whereas the functional integrity of the polymerase-competent complex remains largely unaffected. N348I has been associated with resistance to the non-nucleoside RT inhibitor (NNRTI), nevirapine; however, a possible mechanism that links changes in RNase H activity to changes in NNRTI susceptibility remains to be established. To address this problem, we consider recent findings suggesting that NNRTIs may affect the orientation of RT on its nucleic acid substrate and increase RNase H activity. Here we demonstrate that RNase H-mediated primer removal is indeed more efficient in the presence of NNRTIs; however, the N348I mutant enzyme is able to counteract this effect. Efavirenz, a tight binding inhibitor, restricts the influence of the mutation. These findings provide strong evidence to suggest that N348I can thwart the inhibitory effects of nevirapine during initiation of (+)-strand DNA synthesis, which provides a novel mechanism for resistance. The data are in agreement with clinical data, which demonstrate a stronger effect of N348I on susceptibility to nevirapine as compared with efavirenz.
Keywords: AIDS, Drug Action, Drug Resistance, Reverse Transcription, Viral Polymerase
Introduction
HIV-12 reverse transcriptase (RT) is a major target for antiretroviral drugs that are currently used in the clinic. The enzyme consists of a heterodimer containing a 66-kDa (p66) and a 51-kDa (p51) subunit. The larger subunit is composed of two domains: the polymerase domain, containing the fingers, palm, thumb, and connection subdomains, as well as the ribonuclease H (RNase H) domain (1). HIV-1 RT is a multifunctional enzyme that converts the (+)-strand RNA genome into double-stranded DNA (2). As RT synthesizes the first DNA strand, namely (−)-strand DNA, by extending the 3′-end of a tRNA primer, the RT-associated RNase H activity degrades the transcribed RNA of the newly synthesized DNA·RNA hybrid. During this process, a short polypurine tract (PPT) near the 3′-end of the viral RNA genome is resistant to RNase H degradation and later serves to prime synthesis of the second DNA strand, the (+)-strand DNA. Specific RNase H cleavage is also required to remove the tRNA and PPT primers (3). Although both polymerase and RNase H activities are essential for viral replication, drugs that inhibit RNase H activity have yet to be developed. The approved nucleoside analogue RT inhibitors (NRTIs) and non-nucleoside analogue RT inhibitors (NNRTIs) target the polymerase active site. However, recent studies have indicated that changes in efficiency and specificity of the RT-associated RNase H activity may also affect susceptibility to some members of these two classes of drugs (4–14).
NRTIs act through chain termination, although the incorporation of these inhibitors into the growing DNA chain is reversible. In this instance, a pyrophosphate donor, such as ATP, can excise the nucleotide analogue, providing an important mechanism for resistance. Mutations in HIV-1 RT that confer resistance to the NRTI, 3′-azidothymidine, increase rates of excision (15–21). More recently, it has been shown that mutations in the connection and RNase H domains of HIV-1 RT confer resistance to NRTIs and NNRTIs (9, 10, 22–26). These mutations appear to exert their effects on susceptibility to 3′-azidothymidine more indirectly. In particular, the connection domain mutation N348I has been demonstrated to reduce RNase H activity (9, 27). Once the chain-terminating nucleotide is incorporated, the enzyme is still capable of moving further downstream. This action supports polymerase-independent RNase H cleavages that result in shorter fragments. The N348I mutant is deficient in generating these shorter fragments; however, binding in the polymerase-dependent mode is not significantly affected (27). The selective dissociation from RNase H-competent complexes and, in turn, reductions in RNase H activity provide more time for the excision reaction, ensuring the rescue of DNA synthesis. This mechanism may in part explain why some connection and RNase H domain mutations contribute to further decreases in susceptibility to certain NRTIs (27, 28); however, the mechanism by which these mutations decrease susceptibility to NNRTIs remains elusive.
NNRTIs bind to an allosteric site that is 10 Å from the polymerase active site (1, 29–31) and 60 Å from the RNase H active site (1). A number of possible mechanisms that help to explain the inhibitory effects of NNRTIs during DNA synthesis have been proposed (32, 33). For example, enzyme kinetic studies have shown that these inhibitors interfere with the chemical step of nucleotide incorporation (8, 34, 35). It has also been suggested that the initiation of (+)-strand DNA synthesis is particularly sensitive to NNRTI inhibition (36). During the initiation of (+)-strand DNA synthesis, RT can bind its nucleic acid substrate in one of two orientations (37, 38). The polymerase-competent mode is characterized through interaction between the polymerase active site and the 3′-end of the primer. Conversely, in the RNase H-competent mode, the RT enzyme binds with the polymerase active site in the vicinity of the DNA template so that the RNase H active site is positioned over the chimeric RNA·DNA strand that is cleaved at the junction. Thus, the efficient NNRTI-mediated inhibition during initiation of (+)-strand DNA synthesis may be explained by changes in the ratio of polymerase- to RNase H-competent complexes. NNRTIs appear to promote the latter orientation, which inhibits DNA synthesis and facilitates the primer removal. Fluorescence resonance energy transfer-based single-molecule studies provide strong support for this notion (39, 40). Here, we asked whether NNRTI resistance-conferring mutations have the potential to reverse these effects, which could provide a novel mechanism of resistance to this class of inhibitors.
The results of this study show that the two NNRTIs nevirapine (NVP) and efavirenz (EFV), increase the primer removal reaction. N348I is able to counteract the effect of NVP and, to a lesser degree, EFV. Based on these findings, we conclude that N348I can exert NVP resistance-conferring effects during (+)-strand initiation.
EXPERIMENTAL PROCEDURES
Enzymes and Nucleic Acids
Heterodimeric HIV-1 RT p66/p51 of the HXB-2 strain, termed “wild type” (WT) RT was expressed and purified as described previously (41). Mutant enzymes were prepared with the use of the Stratagene QuikChangeTM kit according to the manufacturer's protocol. These include the RNase H active site mutant E478Q, the connection domain mutant N348I, the double mutant E478Q/N348I, and classic NNRTI resistance mutations K103N and Y181C. DNA and chimeric oligonucleotides were obtained from IDT, and the RNA oligonucleotide was from TriLink (where uppercase lettering denotes DNA, and lowercase denotes RNA). Oligonucleotides were either 5′-end-labeled with [γ-32P]ATP (PerkinElmer Life Sciences) and T4 polynucleotide kinase (Fermentas) or 3′-end-labeled with [α-32P]dTTP (PerkinElmer Life Sciences) and the RNase H-deficient E478Q mutant in order to prevent degradation of the RNA. Reactions persisted for 90 min at 37 °C, and labeled products were purified on a 15% polyacrylamide gel (7 m urea, 50 mm Tris borate, 1 mm EDTA, pH 8.0) and eluted overnight (500 mm NH4-acetate).
(−)-Strand DNA Synthesis
A 2.5-fold excess of the 5′-end-labeled primer-binding site (PBS)-derived DNA primer 22dpol (5′-AGGTCCCTGTTCGGGCGCCACT-3′) or RNA template 52r (5′-ggaaaucucuagcaguggcgcccgaacagggaccugaaagcgaaagggaaac-3′) was annealed to the complementary strand for 5 min at 90 °C in buffer (50 mm Tris-HCl, pH 7.8, 50 mm NaCl), followed by a gradual decrease in temperature to 25 °C. These DNA·RNA hybrids will be denoted as *22dpol·52r and 22dpol·*52r, respectively.
To compare RNase H activity between WT and N348I, the 22dpol·*52r substrate was preincubated with a 5-fold excess of either WT or mutant RT (500 nm), 50 mm Tris-HCl, pH 7.8, 50 mm NaCl, and 200 nm EDTA (reaction mixture) at 37 °C. To assess how NNRTIs affect RNase H activity, the reaction mixture was preincubated with increasing concentrations of NVP or EFV. To ensure single-turnover conditions, reactions were initiated with a mixture of 6 mm Mg2+ and 4 mg/ml heparin (BioShop) and stopped after 5 min with 100% formaldehyde containing trace amounts of xylene cyanol and bromphenol blue. Products were separated on a 15% polyacrylamide gel and analyzed with a PhosphorImager (Amersham Biosciences).
To measure inhibition of full-length DNA synthesis with WT RT and mutant enzymes, the *22dpol·52r substrate was preincubated with increasing concentrations of NNRTIs, and 2 μm dNTPs. Each reaction was carried out under multiple-turnover conditions, initiated with 6 mm Mg2+, and stopped after 7 min as described.
Initiation of (+)-Strand DNA Synthesis
To mimic the initiation of (+)-strand DNA synthesis, a 2.5-fold excess of the 5′-labeled PPT-derived chimeric primer 17r8d (5′-uuaaaagaaaaggggggACTGGAAG-3′) was annealed to the complementary DNA template 57d (5′-CGTTGGGAGTGAATTAGCCCTTCCAGTCCCCCCTTTTCTTTTAAAAAGTGGCTAAGA-3′) and will be denoted *17r8d·57d. This substrate was used to monitor polymerization and RNase activity in the absence and presence of nucleotide and NNRTIs under single- and multiple-turnover conditions. To examine full-length DNA synthesis and the resulting RNase H cleavage of both WT and N348I, a 3-fold excess of the 3′-labeled PPT-derived chimeric primer 17r3d (5′-uuaaaagaaaaggggggACT-3′) was annealed to the complementary DNA template 57d (5′CGTTGGGAGTGAATTAGCCCTTCCAGTCCCCCCTTTTCTTTTAAAAAGTGGCTAAGA-3′) and will be denoted 17r3d*·57d. The reaction mixture was preincubated with 100 nm RT, 25 μm dNTP mix and was initiated with 6 mm Mg2+ in a time course in the absence or presence of either 500 nm NVP or 10 nm EFV.
Filter-based Assay for Determination of NNRTI Inhibition
To obtain IC50 values for both NVP and EFV on WT and mutant enzymes, DNA polymerase activity was tested with 100 μg/ml activated calf thymus DNA (Amersham Biosciences). The substrate was incubated with 45 nm WT RT, 50 mm Tris-HCl, pH 7.8, 50 mm NaCl, 5 μm each dATP, dCTP, and dGTP, 1 μm [3H]dTTP, and varying concentrations of NVP or EFV at 37 °C. The reactions were started with 6 mm Mg2+ and stopped after 20 min with 600 μl of cold 10% trichloroacetic acid, 1% NaPPi, followed by nucleic acid precipitation on ice for 30 min. The samples were filtered and washed with 10% trichloroacetic acid, 1% NaPPi to measure the labeled DNA by scintillation counting.
RESULTS
Experimental Design
(−)-Strand DNA synthesis occurs in conjunction with cleavage of the genomic RNA template. Per definition, polymerase-dependent cleavage occurs when the polymerase active site is in contact with the 3′-end of the growing DNA chain (42). Polymerase-independent cleavage occurs at any point when the 3′-end of the DNA primer is not in contact with the polymerase active site (i.e. when the enzyme continues to move in the 5′-direction of the template in the absence of DNA synthesis) (Fig. 1A). To assess the effect of N348I during (−)-strand DNA synthesis, we have previously used a PBS-derived system consisting of a DNA primer annealed to a 5′-end-labeled RNA template (27). The data revealed decreases in polymerase-independent cleavage associated with this mutation. During the initiation of (+)-strand DNA synthesis, HIV-1 RT can also display polymerase-independent RNase H cleavage. However, the conformation is distinct from complexes during (-)-strand DNA synthesis. During (+)-strand initiation, RT can bind its substrate in two different orientations (Fig. 1B) that permit DNA synthesis or RNase H activity. Here we devised a model system that mimics (+)-strand initiation and, for comparison, a system that mimics (−)-strand DNA synthesis to test the hypothesis that N348I confers resistance to NNRTIs by affecting the primer removal reaction.
FIGURE 1.
HIV-1 RT forms distinct complexes during (−)- and (+)-strand DNA synthesis. A, during (−)-strand DNA synthesis, the polymerase-dependent conformation occurs when the polymerase active site (white cylinder) is in contact with the 3′-end of the DNA primer. The RNase H active site (black arrow) is situated 18 base pairs upstream and can simultaneously cleave the RNA template. Once the enzyme has progressed along the template, DNA synthesis is not possible, and polymerase-independent RNase H activity occurs. B, synthesis of the (+)-strand DNA occurs when RT sits in a polymerase-dependent orientation. This conformation is in equilibrium with the polymerase-independent orientation at which point the RNase H active site is near the 3′-end of the primer and cleaves at the RNA·DNA junction to remove the PPT.
Effects of N348I on DNA·RNA and RNA·DNA Primer·Templates
We employed the PBS-based DNA·RNA system (22dpol·*52r) and compared the RNase H cleavage pattern of WT HIV-1 RT with the N348I mutant. In the absence of the next complementary nucleotide substrate, we observed diminished formation of polymerase-independent RNase H products (−12) with the mutant enzyme (Fig. 2, A and B), which is in agreement with our previous data (27). Increasing concentrations of the next complementary nucleotide further reduce formation of shorter reaction products. The formation of intermediate reaction products (−14, −15), in favor of the short −12 product, is evident for WT RT. These results are expected given that the incoming nucleotide traps the polymerase-competent complex, which delays progressive RNase H cleavage.
FIGURE 2.
The effect of N348I on RNase H degradation during (−)-strand DNA synthesis. A, polymerase-dependent RNase H cleavage (−18, −19), intermediate products (−14, −15), and polymerase-independent RNase H cleavage (−12) were monitored on the PBS sequence 22dpol·*52r, in the presence of increasing concentrations of the next nucleotide. The corresponding sequence indicates the location of these cuts. B, quantification of WT compared with N348I RNase H cleavage is achieved by comparing all cleavage products with respect to total RNase H activity (the sum of all cuts).
To determine the effects of N348I on RNA·DNA primer·templates that are generated during the initiation of (+)-strand DNA synthesis, we used a chimeric primer (*17r8d) that allowed us to simultaneously monitor both nucleotide incorporation and the removal of the primer following initiation (37). For WT HIV-1 RT and the N348I mutant, we observed similar efficiencies for single nucleotide incorporation events as the concentration of substrate increased (Fig. 3A). Most importantly, the primer removal reaction is severely compromised with the N348I mutant, even in the absence of nucleotide substrate. For WT HIV-1 RT, we observed a gradual decrease in RNase H activity under these conditions (Fig. 3B). The fact that the diminution in RNase H activity does not directly translate into increases in DNA synthesis points to the existence of an unproductive population of complexes or unbound RT (43). Overall, these findings show that N348I diminishes polymerase-independent RNase H cleavage during both (−)-strand DNA synthesis and initiation of (+)-strand DNA synthesis.
FIGURE 3.
The effect of N348I on polymerization and RNase H degradation during (+)-strand DNA synthesis. A, removal of the PPT primer (indicated as the 17r cleavage product) was examined in the presence of increasing concentrations of nucleotide. The 17r cleavage occurs at the RNA·DNA junction indicated on the sequence below. B, RNase H activity was quantified as a percentage of initial substrate.
Combined Effects of NNRTIs and N348I on DNA·RNA and RNA·DNA Primer·Template Substrates
We next studied the effects of NNRTIs on RNase H cleavage in the context of the N348I mutant. Initially, we used the PBS system to assess the reaction on regular DNA·RNA substrates used during (−)-strand DNA synthesis (Fig. 4A). In the absence of inhibitor, WT RT showed approximately 2 times the amount of polymerase-independent cleavage when compared with N348I under the same conditions (Fig. 4B). As concentrations of NVP or EFV increased, the amount of polymerase-dependent and polymerase-independent cleavage for N348I-containing complexes approach WT levels, with no significant differences between the two inhibitors. These findings demonstrate that NNRTIs and N348I are antagonistic with respect to RNase H cleavage during (−)-strand synthesis.
FIGURE 4.
RNase H degradation of the PBS substrate is enhanced by NVP and EFV. A, in the presence of increasing concentrations of NVP and EFV, polymerase-dependent (−18, −19) versus -independent cleavage (−12) of the 52r template was monitored for WT and N348I-contaning RT. B, graphical representation of subsequent RNase H cleavage seen with respect to the sum of polymerase-dependent and -independent cuts.
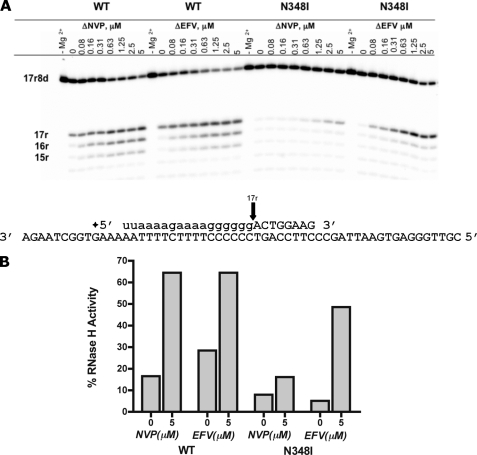
We then investigated the efficiency of RNase H cleavage on the *17r8d·57d substrate that is generated during the initiation of (+)-strand DNA synthesis to assess whether NNRTIs restore RNase H activity in this system as well (Fig. 5A). In the absence of inhibitor, the base-line RNase H activity of WT RT was approximately 3.5 times higher than that of N348I (Fig. 5B). However, as the concentration of EFV increased, the formerly diminished RNase H activity was restored to nearly wild type levels. Conversely, NVP was unable to reverse the diminished RNase H activity to the same extent. These findings differ from the results obtained with the DNA·RNA primer·template system that showed similar effects for both inhibitors.
FIGURE 5.
N348I counteracts the effect of NVP on the primer removal reaction. A, RNase H activity of WT and N348I was monitored in the presence of increasing concentrations of NVP and EFV. B, graphical representation of RNase H cleavage products 17r, 16r, and 15r seen in A, presented as a percentage of total RNase H product compared to the initial amount of substrate.
Effects of Known NNRTI Resistance Mutations on (+)-Strand DNA Synthesis
Classic mutations associated with NNRTI resistance have also been shown to affect RNase H activity in various systems (7, 44–46). However, it remains to be seen whether these mutations alter RNase H cleavage during (+)-strand DNA synthesis. We studied two of the most common mutations in this regard. K103N confers resistance to NVP and EFV, whereas Y181C confers resistance to NVP only. We compared these enzymes with WT RT and N348I in an assay that monitored simultaneous single nucleotide incorporation events and primer removal using the same model system as described in the legend to Fig. 3. We found that all mutations can decrease RNase H activity to a certain degree (Fig. 6, A and B); however, N348I shows the strongest effect. The efficiency of the primer removal reaction followed the order WT > Y181C > K103N ≫ N348I. This pattern was amplified in the absence of an enzyme trap, which allowed more time for the accumulation of subsequent RNase H cleavage products (supplemental Fig. 1).
FIGURE 6.
The effects of NNRTIs on the initiation of (+)-strand DNA synthesis. Incorporation of a single nucleotide (17r9d) and RNase H activity (17r, 16r, 15r) were compared between N348I and classical NNRTI resistance mutations K103N and Y181C. This equilibrium was monitored in the presence of increasing concentrations of NVP (A) and EFV (B).
In this assay, we also examined the dose-dependent response of NVP and EFV. Both NNRTIs cause inhibition of polymerization while increasing the primer removal reaction. The mutations severely reduced the ability of NVP to enhance the primer removal reaction (Fig. 6A). Conversely, N348I can no longer exert this effect in the presence of EFV (Fig. 6B). As a result, RNase H activity approaches nearly WT levels with each mutant enzyme tested. We measured the ratio of RNase H cleavage at high concentrations of inhibitor over base-line activity for each enzyme. These data show that EFV causes the greatest increase in RNase H activity with N348I (Table 1).
TABLE 1.
Effects of NNRTIs on RNase H activity during (+)-strand DNA synthesis
| Enzyme | -Fold increases in RNase H activitya |
|
|---|---|---|
| NVP | EFV | |
| -fold | ||
| WT | 3.0 | 4.2 |
| N348I | 2.8 | 6.8 |
| K103N | 1.7 | 5.4 |
| Y181C | 2.1 | 3.3 |
a -Fold increases in RNase H activity are represented as a ratio of RNase H activity of each enzyme in the presence of either 5 μm NVP or 0.31 μm EFV compared with base-line RNase H activity of each enzyme.
RNase H-dependent Contribution to NVP Resistance
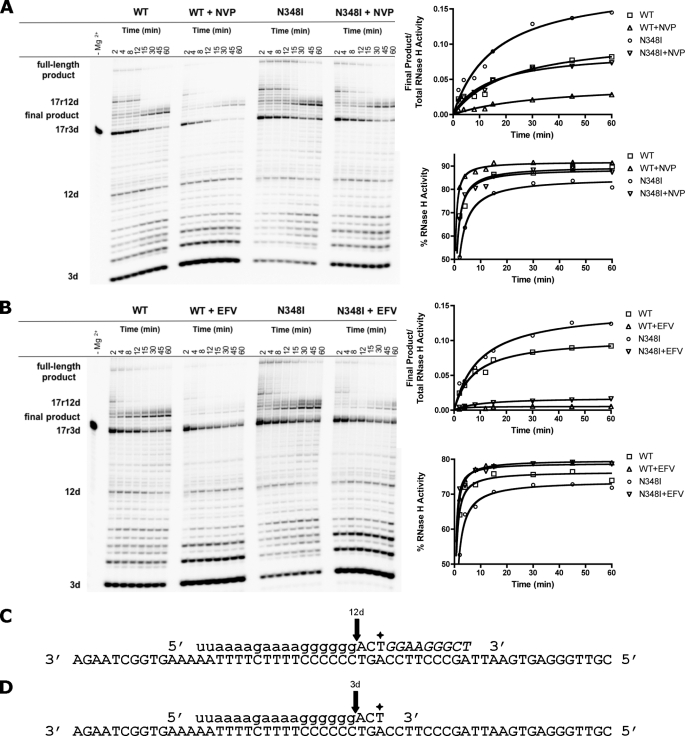
To assess whether the diminished RNase H activity associated with N348I translates into increased production of full-length DNA, we monitored the reaction under multiple-turnover conditions in the presence of all four nucleotides. We used a 3′-end-labeled chimeric primer that allowed us to simultaneously monitor DNA synthesis and RNase H cleavage. The 17r3d* primer mimics the reaction product after incorporation of three nucleotides. We have previously shown with this system that RT pauses after the incorporation of 12 nucleotides, allowing RT to dissociate, change orientations, and cleave the PPT primer (37). Prior to the reaction, the enzyme can bind the substrate in the polymerase- or RNase H-competent orientation. The latter binding mode produces the short 3d fragment that is not further extended (Fig. 7). In contrast, the pausing site, referred to as 17r12d, disappears with time, and the corresponding RNase H cleavage product (12d) appears. The reaction with WT RT shows that the 12d product then disappears, whereas the final extension product emerges. Although the overall pattern remains unchanged, the presence of NVP and EFV shows diminished DNA synthesis and concomitant increases in RNase H activity. N348I increases formation of the final DNA product, in particular in the presence of NVP (Fig. 7A) but not EFV (Fig. 7B). The gain in extension product correlates with diminished RNase H activity. Reductions in RNase H activity are most evident when comparing the amount of the short 3d product. These findings show that binding of the mutant enzyme in the RNase H-competent orientation is diminished. Most importantly, N348I is dominant over NVP, and the primer removal reaction remains compromised. The diminished RNase H activity correlates with increases in levels of DNA synthesis that are comparable with WT RT in the absence of inhibitor. A significant effect of N348I on the inhibitory potential of EFV is not evident.
FIGURE 7.
The effect of NNRTIs and N348I on (+)-strand DNA synthesis under multiple-turnover conditions. The PPT-based sequence 17r3d*·57d was used to monitor full-length DNA synthesis and the corresponding cleavage of each product. After the incorporation of 12 nucleotides, RT dissociates, changes orientations, and cleaves at the RNA·DNA junction (12d product). As time increases, the final product accumulates at 60 min in conjunction with the disappearance of the 12d cleavage. Two alternate possibilities also exist. RT can process though the 17r12d pausing site, forming the full-length DNA product, or can cleave the initial substrate 17r3d, resulting in the 3d product. This process was monitored with both WT and N348I-containing RT in the presence of 500 nm NVP (A) and 10 nm EFV (B). To demonstrate that resistance is RNase H-dependent, formation of the final product was quantified with respect to initial substrate and total RNase H cleavage. The graphical representation of RNase H activity in the absence and presence of inhibitor is expressed as previously described. C and D, the oligonucleotide sequence for the chimeric hybrid duplex. The arrows indicate cleavage products 12d, after the incorporation of nine additional nucleotides (italics), which results in 17r12d product (C) and 3d, the cleavage of the initial primer (D). Both cuts are the result of the RNase H cut at the RNA·DNA junction.
RNase H-independent Contribution to NVP Resistance
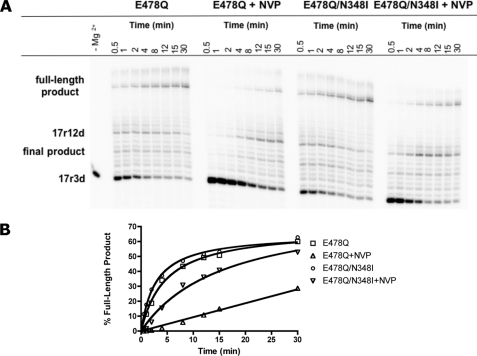
In order to study whether the effect on RNase H activity is the dominant mechanism that helps to explain N348I resistance, we investigated (+)-strand DNA synthesis in the absence of RNase H activity. Here we used the RNase H-deficient mutant enzyme E478Q, in comparison with the double mutant E478Q/N348I (Fig. 8A). Time course experiments revealed the 17r12d product at the pausing site and the full-length product that still contains the RNA primer. NVP diminishes formation of the full-length product, as expected. We observe similar levels of full-length product when the E478Q mutant is compared with the E478Q/N348I double mutant. In addition, the double mutant remains partially sensitive to NVP (Fig. 8B). It is therefore conceivable that in addition to an RNase H-dependent mechanism, an RNase H-independent mechanism also contributes to NVP resistance.
FIGURE 8.
RNase H-independent full-length DNA synthesis. A, the RNase H active site mutant enzyme, E478Q, was used with the 3′-end-labeled PPT chimeric primer to monitor full-length synthesis in the absence of RNase H activity. The formation of each synthesis product was monitored over 30 min in the absence and presence of 500 nm NVP. B, formation of full-length products are quantified in the corresponding graph.
To further address this issue, we measured IC50 values for N348I and classical NNRTI-resistant mutants on both DNA·RNA and DNA·DNA substrates. Using the PBS-derived DNA·RNA (*22dPol·52r) substrate, we observed resistance profiles for K103N and Y181C consistent with phenotypic susceptibility measurements (Table 2). However, N348I causes only a subtle 2.7-fold increase in IC50 to NVP, and no significant change is seen with EFV. We determined very similar values and trends with DNA·DNA substrates (Table 3). Together, these findings indicate that an RNase H-independent mechanism may in part contribute to NVP resistance on each of the various substrates tested in this study. However, 3-fold increases in IC50 values are smaller than the 4–27-fold increases measured in cell culture. We therefore conclude that N348I confers significant levels of resistance to NVP through diminished RNase H cleavage during (+)-strand initiation.
TABLE 2.
IC50 values for NVP and EFV measured on DNA·RNA primer·templates
| Enzyme | NVP |
EFV |
||
|---|---|---|---|---|
| IC50a | -Fold increaseb | IC50a | -Fold increaseb | |
| μm | -fold | μm | -fold | |
| WT | 16.0 ± 2.5 | 0.88 ± 0.10 | ||
| N348I | 43.0 ± 2.0 | 2.7 | 1.10 ± 0.05 | 1.3 |
| K103N | >300 | NAc | >5 | NAc |
| Y181C | >300 | NAc | 0.63 ± 0.07 | 0.7 |
a IC50 is the inhibitory concentration of either NVP or EFV that reduced full-length DNA synthesis by 50% using a DNA·RNA hybrid. Values were calculated by fitting 8 data points to a sigmoidal dose-response equation using GraphPad Prism (version 4.0b). S.D. values were determined on the basis of two independent experiments.
b -Fold increase is calculated as a ratio of the IC50 value of the mutant enzyme as compared with the wild-type enzyme.
c NA, not applicable; unable to calculate.
TABLE 3.
IC50 values for NVP and EFV measured on DNA·DNA primer·templates
| Enzyme | NVP |
EFV |
||
|---|---|---|---|---|
| IC50a | -Fold increaseb | IC50a | -Fold increaseb | |
| μm | -fold | nm | -fold | |
| WT | 2.03 ± 0.1 | 2.73 ± 0.25 | ||
| N348I | 6.13 ± 1.0 | 3.0 | 3.80 ± 0.95 | 1.4 |
| K103N | 56.77 ± 5.1 | 28.0 | 21.83 ± 4.25 | 8.0 |
| Y181C | 78.37 ± 8.1 | 38.6 | 2.47 ± 0.06 | 0.9 |
a IC50 is the inhibitory concentration of either NVP or EFV that reduced the nucleotide incorporation activity of enzyme by 50% using activated calf thymus DNA. Values were calculated by fitting 10 data points to a sigmoidal dose-response equation using GraphPad Prism (version 4.0b). S.D. values were determined on the basis of three independent experiments.
b -Fold increase is calculated as a ratio of the IC50 value of the mutant enzyme as compared with the wild-type enzyme.
DISCUSSION
In light of the many different biochemical mechanisms that have been associated with the inhibitory effects of NNRTIs, it is difficult to assess the specific contribution of possible reductions in (+)-strand initiation to the overall antiviral effect. We reasoned that a specific antiviral effect (if any) is likely to be counteracted by a specific mechanism of resistance that targets the same reaction with the opposite outcome. To address this question, we asked whether NNRTI resistance-conferring mutations in HIV-1 RT could neutralize the inhibitor specifically during initiation of (+)-strand DNA synthesis. We focused on the N348I mutation in the connection domain of HIV-1 RT because our previous data revealed deficiencies of this mutation in polymerase-independent RNase H cleavage, albeit during regular (−)-strand DNA synthesis (27).
In this study, we directly demonstrate that both NNRTIs tested (NVP and EFV) increase the primer removal reaction during the initiation of (+)-strand DNA synthesis in the context of WT HIV-1 RT, which is consistent with previous assumptions. Under these conditions, the enzyme prefers to utilize the RNA strand of the RNA·DNA hybrid as a template and not as a primer. Conversely, the N348I mutation diminishes the primer removal reaction, whereas DNA synthesis remains largely unaffected. Most importantly, we demonstrate that the N348I mutation counteracts the inhibitory effect mediated by NVP, which provides a novel mechanism for resistance to NVP.
Classic NNRTI resistance conferring mutations, such as K103N and Y181C, are clustered around the binding site for these compounds. Structural and biochemical data have shown that Y181C, among many other NNRTI-associated mutations, diminishes the affinity to the inhibitor (39, 47, 48). K103N appears to interfere with the access of the inhibitor to its designated binding site (49). More recently, it has been shown that mutations in the connection and RNase H domain can reduce RNase H cleavage on DNA·RNA hybrids (50). The authors concluded that such reductions translate into resistance, because DNA synthesis may be reinitiated before the primer·template complex dissociates irreversibly. Our experimental data are inconsistent with this model. The effects of NNRTIs on RNase H cleavage are not considered in this model, and we show that increasing concentrations of NNRTIs can reverse N348I-mediated deficits in RNase H cleavage on regular DNA·RNA hybrids. Moreover, we demonstrate that N348I causes only minor reductions (3-fold) in sensitivity to NVP on both DNA·DNA and DNA·RNA substrates. Although these data suggest that an RNase H-independent mechanism may in part contribute to the resistant phenotype, the effect is subtle. In line with this suggestion, recent modeling studies have indicated that allosteric changes may affect inhibitor binding (9, 24). Our experiments with an enzyme containing N348I against a background of the RNase H active site mutation E478Q revealed also minor RNase H-independent contribution to resistance during initiation of (+)-strand. These findings could be explained by allosteric effects that can influence inhibitor and/or nucleotide binding, which is consistent with our previous data pointing to increases in processive DNA synthesis with the N348I mutant (27, 51).
The ability of N348I to counteract the effects of NNRTIs on the primer removal reaction is specific to NVP. EFV increases RNase H activity to a similar extent as seen with NVP; however, the reversal in the presence of N348I is far less pronounced. The properties of EFV as a tight binding inhibitor may help to explain these findings. The RT enzyme appears to form a stable complex with the RNA·DNA substrate, and the inhibitor ultimately increases RNase H activity. In contrast, NVP dissociates more frequently from the complex, which, in turn, can facilitate reorientation of the mutant RT on its substrate and continuation of DNA synthesis. These data are in agreement with previous susceptibility measurements in cell culture that have shown 4–27-fold increases in IC50 values for NVP and insignificant increases with EFV (9, 24–26).
We do not observe significant differences between NVP and EFV when RNase H cleavage is monitored on regular DNA·RNA substrates that mimic reverse transcription during (−)-strand DNA synthesis. These findings raise the question as to why the nature of the inhibitor is more relevant during initiation of (+)-strand DNA synthesis. N348I decreases polymerase-independent RNase H cleavage during (−)-strand DNA synthesis as a result of a decrease in the affinity for shorter double-stranded fragments (27). A possible structural explanation for this is that the strong interaction between the polymerase domain and the double-stranded segment of the substrate are lost, and the effect of connection domain mutations becomes more relevant when the overall affinity decreases. On the other hand, it is conceivable that the influence of the bound NNRTI on RNase H activity is limited when the polymerase domain is only in contact with the single-stranded template overhang. The structure of the RNase H-competent complex that is formed during (+)-strand initiation is different in this context (Fig. 9). Here the polymerase domain is still in contact with the double-stranded, RNase H-resistant PPT hybrid; therefore, the bound NNRTI is still able to increase RNase H activity. As a result, the binding properties of NNRTIs (i.e. tight binding versus frequent dissociation) probably become more important in this conformation.
FIGURE 9.
Conformational differences between RT complexes that permit polymerase-independent RNase H cleavage during (−)-strand DNA synthesis and (+)-strand initiation. The RT enzyme is schematically shown (white oval) with its polymerase active site (white cylinder) within the polymerase domain (red oval) and its RNase H active site (black arrow). The NNRTI-BP (blue diamond) is occupied by an NNRTI (yellow diamond) in the polymerase domain and is depicted to be in close proximity to the polymerase active site. During (−)-strand DNA synthesis, as subsequent cleavage occurs, RNase H cleavage becomes polymerase-independent, at which point the polymerase domain gradually loses contact with the double-stranded DNA·RNA. We suggest that at this stage, the effect of N348I within the connection domain (green oval) is dominant and that NNRTIs lose their ability to enhance RNase H cleavage. The effects of N348I are much more pronounced during the initiation of (+)-strand synthesis. When RT is in the polymerase-independent orientation, the polymerase domain is still in contact with the double-stranded RNA·DNA, and both NVP and EFV increase the removal of the PPT primer. This effect is counteracted by N348I solely in the presence of NVP because its tendency to dissociate allows the N348I mutation to dominate.
Taken together, the diminished RNase H activity associated with N348I in HIV-1 RT is a factor that can be linked to resistance to both NRTIs and NNRTIs. However, although resistance to NRTIs can theoretically occur at any stage during reverse transcription, our findings provide strong evidence that the effect of N348I on susceptibility to NVP occurs specifically at the level of (+)-strand initiation. Given that N348I is a naturally occurring mutation, these results in turn suggest that the antiviral effect of NVP is at least in part mediated during this stage. Overall, the data show that the mechanisms by which a specific connection domain mutation decreases susceptibility to NRTIs and NNRTIs can be diverse. This study argues against a unifying mechanism.
Supplementary Material
Acknowledgment
We thank Dr. Egor Tchesnokov for inspiring discussion.
This work was supported by the Canadian Institutes for Health Research (CIHR).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- HIV-1
- human immunodeficiency virus, type 1
- RT
- reverse transcriptase
- RNase H
- ribonuclease H
- NNRTI
- non-nucleoside reverse transcriptase inhibitor
- PPT
- polypurine tract
- NRTI
- nucleoside reverse transcriptase inhibitor
- NVP
- nevirapine
- EFV
- efavirenz
- WT
- wild type
- PBS
- primer-binding site.
REFERENCES
- 1.Kohlstaedt L. A., Wang J., Friedman J. M., Rice P. A., Steitz T. A. (1992) Science 256, 1783–1790 [DOI] [PubMed] [Google Scholar]
- 2.Coffin J. M., Hughes S. H., Varmus H. E. (1997) Retroviruses, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 3.Götte M., Li X., Wainberg M. A. (1999) Arch Biochem. Biophys 365, 199–210 [DOI] [PubMed] [Google Scholar]
- 4.Nikolenko G. N., Svarovskaia E. S., Delviks K. A., Pathak V. K. (2004) J. Virol. 78, 8761–8770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nikolenko G. N., Palmer S., Maldarelli F., Mellors J. W., Coffin J. M., Pathak V. K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2093–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaw-Reid C. A., Feuston B., Munshi V., Getty K., Krueger J., Hazuda D. J., Parniak M. A., Miller M. D., Lewis D. (2005) Biochemistry 44, 1595–1606 [DOI] [PubMed] [Google Scholar]
- 7.Wang J., Dykes C., Domaoal R. A., Koval C. E., Bambara R. A., Demeter L. M. (2006) Virology 348, 462–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia Q., Radzio J., Anderson K. S., Sluis-Cremer N. (2007) Protein Sci. 16, 1728–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yap S. H., Sheen C. W., Fahey J., Zanin M., Tyssen D., Lima V. D., Wynhoven B., Kuiper M., Sluis-Cremer N., Harrigan P. R., Tachedjian G. (2007) PLoS Med. 4, e335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nikolenko G. N., Delviks-Frankenberry K. A., Palmer S., Maldarelli F., Fivash M. J., Jr., Coffin J. M., Pathak V. K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 317–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hang J. Q., Li Y., Yang Y., Cammack N., Mirzadegan T., Klumpp K. (2007) Biochem. Biophys. Res. Commun. 352, 341–350 [DOI] [PubMed] [Google Scholar]
- 12.Delviks-Frankenberry K. A., Nikolenko G. N., Boyer P. L., Hughes S. H., Coffin J. M., Jere A., Pathak V. K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10943–10948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Radzio J., Sluis-Cremer N. (2008) Mol. Pharmacol. 73, 601–606 [DOI] [PubMed] [Google Scholar]
- 14.Zelina S., Sheen C. W., Radzio J., Mellors J. W., Sluis-Cremer N. (2008) Antimicrob. Agents Chemother. 52, 157–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arion D., Kaushik N., McCormick S., Borkow G., Parniak M. A. (1998) Biochemistry 37, 15908–15917 [DOI] [PubMed] [Google Scholar]
- 16.Meyer P. R., Matsuura S. E., Mian A. M., So A. G., Scott W. A. (1999) Mol. Cell 4, 35–43 [DOI] [PubMed] [Google Scholar]
- 17.Götte M., Arion D., Parniak M. A., Wainberg M. A. (2000) J. Virol. 74, 3579–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arion D., Sluis-Cremer N., Parniak M. A. (2000) J. Biol. Chem. 275, 9251–9255 [DOI] [PubMed] [Google Scholar]
- 19.Meyer P. R., Matsuura S. E., Schinazi R. F., So A. G., Scott W. A. (2000) Antimicrob. Agents Chemother. 44, 3465–3472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldschmidt V., Marquet R. (2004) Int. J. Biochem. Cell Biol. 36, 1687–1705 [DOI] [PubMed] [Google Scholar]
- 21.Menéndez-Arias L. (2010) Antiviral Res. 85, 210–231 [DOI] [PubMed] [Google Scholar]
- 22.Brehm J. H., Koontz D., Meteer J. D., Pathak V., Sluis-Cremer N., Mellors J. W. (2007) J. Virol. 81, 7852–7859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delviks-Frankenberry K. A., Nikolenko G. N., Barr R., Pathak V. K. (2007) J. Virol. 81, 6837–6845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hachiya A., Kodama E. N., Sarafianos S. G., Schuckmann M. M., Sakagami Y., Matsuoka M., Takiguchi M., Gatanaga H., Oka S. (2008) J. Virol. 82, 3261–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hachiya A., Shimane K., Sarafianos S. G., Kodama E. N., Sakagami Y., Negishi F., Koizumi H., Gatanaga H., Matsuoka M., Takiguchi M., Oka S. (2009) Antiviral Res. 82, 115–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta S., Fransen S., Paxinos E. E., Stawiski E., Huang W., Petropoulos C. J. (2010) Antimicrob. Agents Chemother. 54, 1973–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ehteshami M., Beilhartz G. L., Scarth B. J., Tchesnokov E. P., McCormick S., Wynhoven B., Harrigan P. R., Götte M. (2008) J. Biol. Chem. 283, 22222–22232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brehm J. H., Mellors J. W., Sluis-Cremer N. (2008) Biochemistry 47, 14020–14027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smerdon S. J., Jäger J., Wang J., Kohlstaedt L. A., Chirino A. J., Friedman J. M., Rice P. A., Steitz T. A. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 3911–3915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren J., Esnouf R., Garman E., Somers D., Ross C., Kirby I., Keeling J., Darby G., Jones Y., Stuart D., et al. (1995) Nat. Struct. Biol. 2, 293–302 [DOI] [PubMed] [Google Scholar]
- 31.Ding J., Das K., Moereels H., Koymans L., Andries K., Janssen P. A., Hughes S. H., Arnold E. (1995) Nat. Struct. Biol. 2, 407–415 [DOI] [PubMed] [Google Scholar]
- 32.Sluis-Cremer N., Tachedjian G. (2008) Virus Res. 134, 147–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren J., Stammers D. K. (2008) Virus Res. 134, 157–170 [DOI] [PubMed] [Google Scholar]
- 34.Spence R. A., Kati W. M., Anderson K. S., Johnson K. A. (1995) Science 267, 988–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L. Z., Kenyon G. L., Johnson K. A. (2004) J. Biol. Chem. 279, 38424–38432 [DOI] [PubMed] [Google Scholar]
- 36.Grobler J. A., Dornadula G., Rice M. R., Simcoe A. L., Hazuda D. J., Miller M. D. (2007) J. Biol. Chem. 282, 8005–8010 [DOI] [PubMed] [Google Scholar]
- 37.Götte M., Maier G., Onori A. M., Cellai L., Wainberg M. A., Heumann H. (1999) J. Biol. Chem. 274, 11159–11169 [DOI] [PubMed] [Google Scholar]
- 38.Fuentes G. M., Rodríguez-Rodríguez L., Fay P. J., Bambara R. A. (1995) J. Biol. Chem. 270, 28169–28176 [DOI] [PubMed] [Google Scholar]
- 39.Abbondanzieri E. A., Bokinsky G., Rausch J. W., Zhang J. X., Le Grice S. F., Zhuang X. (2008) Nature 453, 184–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu S., Abbondanzieri E. A., Rausch J. W., Le Grice S. F., Zhuang X. (2008) Science 322, 1092–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Le Grice S. F., Grüninger-Leitch F. (1990) Eur. J. Biochem. 187, 307–314 [DOI] [PubMed] [Google Scholar]
- 42.Furfine E. S., Reardon J. E. (1991) J. Biol. Chem. 266, 406–412 [PubMed] [Google Scholar]
- 43.Wöhrl B. M., Krebs R., Goody R. S., Restle T. (1999) J. Mol. Biol. 292, 333–344 [DOI] [PubMed] [Google Scholar]
- 44.Gerondelis P., Archer R. H., Palaniappan C., Reichman R. C., Fay P. J., Bambara R. A., Demeter L. M. (1999) J. Virol. 73, 5803–5813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Archer R. H., Dykes C., Gerondelis P., Lloyd A., Fay P., Reichman R. C., Bambara R. A., Demeter L. M. (2000) J. Virol. 74, 8390–8401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Archer R. H., Wisniewski M., Bambara R. A., Demeter L. M. (2001) Biochemistry 40, 4087–4095 [DOI] [PubMed] [Google Scholar]
- 47.Das K., Ding J., Hsiou Y., Clark A. D., Jr., Moereels H., Koymans L., Andries K., Pauwels R., Janssen P. A., Boyer P. L., Clark P., Smith R. H., Jr., Kroeger Smith M. B., Michejda C. J., Hughes S. H., Arnold E. (1996) J. Mol. Biol. 264, 1085–1100 [DOI] [PubMed] [Google Scholar]
- 48.Ren J., Nichols C., Bird L., Chamberlain P., Weaver K., Short S., Stuart D. I., Stammers D. K. (2001) J. Mol. Biol. 312, 795–805 [DOI] [PubMed] [Google Scholar]
- 49.Hsiou Y., Ding J., Das K., Clark A. D., Jr., Boyer P. L., Lewi P., Janssen P. A., Kleim J. P., Rösner M., Hughes S. H., Arnold E. (2001) J. Mol. Biol. 309, 437–445 [DOI] [PubMed] [Google Scholar]
- 50.Nikolenko G. N., Delviks-Frankenberry K. A., Pathak V. K. (2010) J. Virol. 84, 5238–5249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.von Wyl V., Ehteshami M., Symons J., Bürgisser P., Nijhuis M., Demeter L. M., Yerly S., Böni J., Klimkait T., Schuurman R., Ledergerber B., Götte M., Günthard H. F. (2010) J. Infect. Dis. 201, 1054–1062 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.