Abstract
The slc4a10 gene encodes an electroneutral Na+-dependent HCO3− importer for which the precise mode of action remains unsettled. To resolve this issue, intracellular pH (pHi) recordings were performed upon acidification in the presence of CO2/HCO3− by 2′,7′-bis(carboxyethyl)-5,6-carboxyfluorescein (BCECF) fluorometry of stably slc4a10-transfected NIH-3T3 fibroblasts. slc4a10 expression induced a significant Na+-dependent pHi recovery, which was accompanied by an increase in the intracellular Na+ concentration evaluated by use of the Na+-sensitive fluorophore CoroNa Green. The estimated Na+:HCO3− stoichiometry was 1:2. Cl− is most likely the counterion maintaining electroneutrality because (i) Na+-dependent pHi recovery was eliminated in Cl−-depleted cells; (ii) acute extracellular Cl− removal led to a larger alkalization in slc4a10-transfected cells than in control cells; and (iii) the 4,4′-diisothiocyanato-stilbene-2,2′-disulfonic acid (DIDS)-sensitive and Na+- and HCO3−-dependent 36Cl−-efflux during pHi recovery was significantly greater in acidified slc4a10-transfected cells than in control cells. Charged amino acids specific to slc4a gene family members that transport Na+ and are expected to move more HCO3− molecules/turnover were targeted by site-directed mutagenesis. Na+-dependent pHi recovery was reduced in each of the single amino acid mutated cell lines (E890A, E892A, H976L, and H980G) compared with wild type slc4a10-transfected cells and completely eliminated in quadruple mutant cells. In conclusion, the data suggest that slc4a10 expressed in mammalian cells encodes a Na+-dependent Cl−/HCO3− exchanger in which four specific charged amino acids seem necessary for ion transport.
Keywords: Cell pH, Chloride Transport, Membrane Proteins, Site-directed Mutagenesis, Sodium Transport, Bicarbonate Transport
Introduction
Tight control of intracellular pH (pHi)2 is crucial for maintaining a variety of basic cellular functions (1). Regulation of pHi largely depends on acute intracellular buffering and longer term extrusion or import of acid/base equivalents. Integral plasma membrane proteins such as the Na+-dependent HCO3− transporters from the solute carrier gene family 4 (slc4a) mediate these functions. The slc4a10-gene product is primarily expressed in the central nervous system and is important for normal brain function (2–4). In the choroid plexus, the slc4a10 gene product is a main basolateral Na+ loader and is most likely necessary for secretion of cerebrospinal fluid because disruption of the slc4a10 in mice leads to decreased brain ventricle sizes (4). In humans, a mutation in the SLC4A10 promoter region resulted in severe complex partial epilepsy and mental retardation (5).
Within the gene family, slc4a10 shares the highest amino acid homology with the electroneutral Na+:HCO3− cotransporter Nbcn1 (slc4a7) and the Na+-dependent Cl−/HCO3− exchanger, Ndcbe (slc4a8). The transport mode of the slc4a10 gene product has been investigated by more research groups defining the protein as an electroneutral Na+ and HCO3−-dependent transporter which is inhibited by 4,4′-diisothiocyanatostilbene-2,2′-disulfonate (DIDS) (2, 6, 7). The studies, however, differ in opinion regarding the involvement of Cl− in the transport process. Thus, the human SLC4A10 gene product, first designated with the abbreviation Ncbe for Na+-dependent Cl−/HCO3− exchanger (2), was recently suggested renamed as Nbcn2 for electroneutral Na+:HCO3− cotransporter 2, displaying Cl−/Cl− self-exchange instead of Cl−/HCO3− exchange activity (7). The knowledge on the primary structure of the slc4a10-derived protein is not sufficient to suggest which parts of the transmembrane region are involved in ion translocation and can, thus, not help predict the mode of action for this Na+-dependent HCO3− transporter. More structure-function data are available on the anion exchanger Ae1 (slc4a1), which seems to present two hinge-like loops within the transmembrane region with access to both the extra- and intracellular environment (8). These loops were proposed to be involved in attraction and perhaps even translocation of ions by Ae1.
In this investigation, we aimed to define the transport mode of the slc4a10 gene product by analyzing the targeted ionic and structural requirements for transport by the protein. The study was conducted on mammalian NIH-3T3 fibroblasts devoid of endogenous Na+-dependent HCO3− transport stably transfected with single copies of the coding region of rodent slc4a10. We found three lines of evidence for the functional involvement of Cl− in transport by the slc4a10-derived protein: (i) the Na+:HCO3− stoichiometry was estimated to 1:2; (ii) Na+-dependent pHi regulation was dependent on the presence and the gradient of Cl−; and (iii) slc4a10 transfection induced a DIDS-sensitive and HCO3−-dependent increase in Cl− efflux during Na+-dependent pHi regulation. Although there is a high degree of homology among the gene family members in the hinge-like regions, we aimed to identify amino acid residues of importance for transporter function based on bioinformatics and the literature on Ae1 mutagenesis. The analysis supports the importance of each of the four charged amino acids for Na+:HCO3− import by the slc4a10-derived polypeptide. We recommend preserving the original name of the slc4a10 gene product, Ncbe.
EXPERIMENTAL PROCEDURES
cDNA Constructs
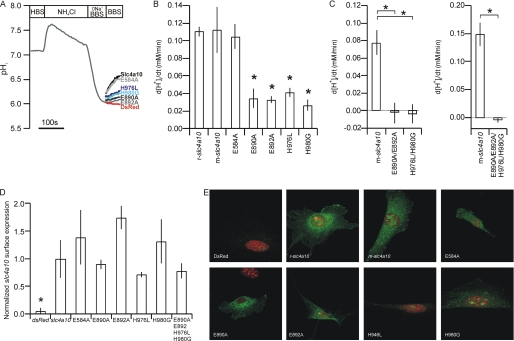
The coding sequences of rat slc4a10 (rb2Ncbe, NBCn2-D, AY579374) and mouse slc4a10 (corresponding to rb1Ncbe, NBCn2-B, AB033759) were obtained by reverse transcription-PCR as described previously (9) and inserted into pcDNA5/FRT vectors (flipase recognition target; Invitrogen). The rat slc4a7 (Nbcn1d) gene was purchased from Geneart (Regensburg, GE) and inserted into the same vector. Mutation sites in the slc4a10 were chosen by comparative analysis of the predicted amino acid sequence in all members of the slc4a family and across the species: mouse, rat, dog, and human. The hinge-like loops of Ae1 have been suggested as sites for ion translocation. There are only four charged candidate amino acids in these domains and the flanking transmembrane domains of slc4a-derived polypeptides that vary systematically among the members of the gene family. Putative Na+-attracting motifs in the hinge-like loop regions were identified as the negatively charged amino acids found in Na+-dependent but not in Na+-independent slc4a family members i.e. Ae1–4. Alanine substitutions were introduced at positions Glu890 and Glu892 (Fig. 1A) by site-directed mutagenesis (Stratagene). As negative control, a similar mutation was made in Glu584 just after the first transmembrane domain. Two putative anion-attracting motifs were selected based on the appearance of positively charged amino acids in the hinge-like loops. One amino acid is specific to slc4a family members believed to transport more than two HCO3− molecules/Na+ (Nbce1, Nbce2, and Ndcbe). The other amino acid is charged in the anion exchangers, polar in Nbcn1 and has the pH-sensitive histidine in all other Nbcs. Mutations were introduced at positions His976 and His980 by replacing the histidines with leucine and cysteine, respectively. Resulting cDNA was sequenced and cloned into the pcDNA5/FRT vector. A construct encoding a nucleus-targeted red fluorescent protein, pDsRed2-Nuc (Clontech) was also cloned into that vector and used as a functionally negative control for slc4a10 throughout this study.
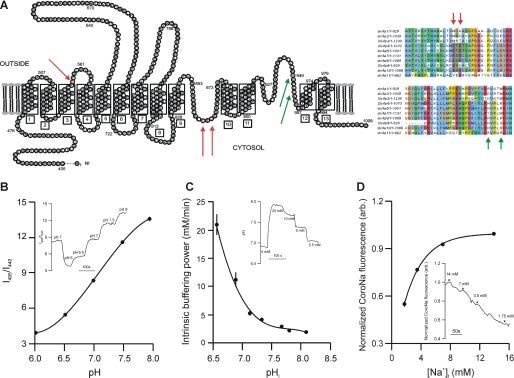
FIGURE 1.
A, proposed topological model for the mouse slc4a10-derived polypeptide based on the similar model from slc4a1, Ae1. The mutated amino acids are highlighted in red and by arrows. Right panels show the amino acid alignment of the hinge-like loops of slc4a-derived polypeptides. Mutated amino acids are marked with arrows. (Modified with permission from J. Casey.) B, calibration of excitation fluorescence ratio (I495/I440) into pHi values. Inset shows fluorescence ratio trace and extracellular pH values from one experiment. C, determination of pHi buffering capacity, βint. Inset shows pHi trace and extracellular [NH4+ + NH3] values from one experiment. D, calibration of intracellular CoroNa fluorescence to [Na+]i values. Inset shows fluorescence recording and extracellular [Na+] values from one experiment. Error bars = S.E.
Cell Culture
FRTs containing NIH-3T3 fibroblasts (Invitrogen) were grown in Dulbecco's modified Eagle's medium glutaMAXTM supplemented with 10% donor bovine serum. Cells were stably transfected with a single copy of cDNA constructs into a predefined locus using the Flp-InTM system (Invitrogen). Transfection and protein expression were validated by PCR and sequencing, surface biotinylation, Western blotting, and immunocytochemistry.
Fluorophores
For pHi measurements, cells were loaded in 2 μm BCECF-AM in a HEPES-buffered salt solution (HBS; Table 1) for 15 min. For intracellular [Na+] measurements, cells were loaded in 10 μm CoroNa Green sodium indicator for 30 min in HBS. All incubations were performed in a dark chamber heated to 37 °C. Fluorophores were purchased from Invitrogen.
TABLE 1.
Experimental solutions
| Solution | HBS | 0Na+ |
NH4+ |
0Cl− |
HiK+ |
BBS | 0Na+ |
NH4+ |
0Cl− |
0NaCl |
|---|---|---|---|---|---|---|---|---|---|---|
| HBS | HBS | HBS | HBS | BBS | BBS | BBS | BBS | |||
| Na+ | 145.0 | 125.0 | 145.0 | 10.0 | 145.0 | 125.0 | 145.0 | |||
| K+ | 3.6 | 3.6 | 3.6 | 3.6 | 138.6 | 3.6 | 3.6 | 3.6 | 3.6 | 3.6 |
| Ca2+ | 1.8 | 1.8 | 1.8 | 1.8 | 1.8 | 1.8 | 1.8 | 1.8 | 1.8 | 1.8 |
| Mg2+ | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 |
| NH4+ | 20.0 | 20.0 | ||||||||
| Cl− | 138.6 | 138.6 | 138.6 | 138.6 | 114.6 | 114.6 | 114.6 | |||
| SO42− | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 |
| HCO3− | 24.0 | 24.0 | 24.0 | 24.0 | 24.0 | |||||
| Glucose | 5.5 | 5.5 | 5.5 | 5.5 | 5.5 | 5.5 | 5.5 | 5.5 | 5.5 | 5.5 |
| HEPES | 10.0 | 10.0 | 10.0 | 10.0 | 10.0 | 10.0 | 10.0 | 10.0 | 10.0 | 10.0 |
| NMDG | 145.0 | 121.0 | 121.0 | |||||||
| Gluconate | 138.6 | 114.6 | 114.6 | |||||||
| Choline | 24.0 | 24.0 | ||||||||
| PO42− | 2.0 | 2.0 | 2.0 | 2.0 | 2.0 | 2.0 | 2.0 | 2.0 | 2.0 | 2.0 |
| pH | 7.4 | 7.4 | 7.4 | 7.4 | 7.4 | 7.4 | 7.4 | 7.4 | 7.4 | 7.4 |
Intracellular pH and [Na+] Measurements
Cells were grown to approximately 50% confluence on glass coverslips. Fluorophore-loaded cells were mounted in a closed perfusion chamber (358-μl RC-21BR or 36-μl RC-20; Warner Instruments) and perfused with a linear flow rate of 0.8 mm/s at 37 °C. For pHi measurements, the dye excitation periods of 20 ms were alternating between 495-nm and 440-nm light from a monochromator (Till Photonics). The light emission at 510–535 nm was recorded by a 12-bit cooled monochrome CCD camera (QImaging, Retiga EXi). QED InVivo imaging software (Media Cybernetics) was used to control wavelength, light exposure time (20 ms), frequency (one image pair each 4 s), and 4 × 4 binning (to 348 × 260 pixel images), as well as the data collection from regions of interest (one/individual cell). The mean values for cells from one coverslip represent n = 1. Where indicated, fluorophore-loaded cells were mounted in a coverslip holder and placed in a cuvette containing experimental solution in a heated cuvette house as alternative to fluorescence microscopy. For pHi measurements, dye excitation alternated between 495-nm and 440-nm monochromator light at 1 Hz, and the light emission at 510–535 nm was recorded by a photomultiplier tube of a Quantum Master UV-visual QM-4 spectrofluorometer (Photon Technologies International). The ratio derived from the BCECF measurements was calibrated to intracellular pHi by clamping intracellular pH to stepwise changing extracellular pH by 10 μm nigericin in a high K+ buffer (10) (Fig. 1B). The Na+-dependent d[H+]i/dt was calculated as the product of dpHi/dt and the total buffering power, βtot. The βtot was calculated as the sum of the intrinsic buffering power (βint) and the contribution of the CO2/HCO3− buffering system as previously described (10, 11) (Fig. 1C). For the single wavelength [Na+]i dye CoroNa Green, the excitation/emission wavelengths were 492/516 nm. CoroNa fluorescence was calibrated to [Na+]i values by varying extracellular [Na+] in the presence of the ionophore monensin, 10 μm (Fig. 1D). Each time course recording was followed by a one-point calibration of [Na+]i to 14 mm with monensin. The changes were fully reversible, and neither photobleaching nor probe leak was observed during the experiments. The composition of experimental solutions is listed in Table 1.
36Cl− Flux Measurements
Cells were grown to 80% confluence in 12-well plates (Life Sciences) and loaded with 2 μCi/ml H36Cl (added equimolar amount of NaOH) or Na36Cl in HBS for 2 h in a heating chamber at 37 °C (GE Healthcare or Risø National Laboratory, Denmark, respectively). Cells were acidified for 3 min by adding NH4Cl to the solution (final concentration 20 mm) and washed four times in a Na+-free CO2/HCO3−-buffered salt solution. The last wash was added 125 μm of the Cl− channel blocker 5-nitro-2-(3-phenylpropylamino)-benzoate, NPPB, and was collected for scintillation counting. After the final wash, the cells were incubated with Na+-containing BBS with the same Cl− channel blocker. The solution was collected for scintillation counting after 2 min. In parallel wells, 200 μm DIDS was added to block slc4a10 gene product activity in the continued presence of the Cl− channel blockers. To compare values between wells of varying cell density, we normalized the counts for the background Cl− in the final wash in Na+-free BBS. For assessment of intracellular Cl− depletion, cells were loaded with 36Cl− as described above. After two washes in HBS, cells were incubated in Cl−-free HBS for 0, 1, 5 or 30 min before removal of the supernatant. After 2 washes in Cl−-free HBS and addition of 0.2 m NaOH, the cell lysates were collected for scintillation counting. Cl− influx was measured by exposing cells to 8 μCi/ml Na36Cl for 2 min corresponding to the initial period of Na+-dependent pHi recovery after NH4Cl prepulsing in the presence of 1 mm furosemide. Cells were washed three times in cold HBS before lysis and counting. The influx values were corrected for variations in cell number (3H2O space). All samples were run in duplicate and measured in a liquid scintillation counter (RackBeta 1211; LKB Wallac). The Cl− efflux rate constants were determined as fractional loss of isotope/min.
Immunocytochemistry
Cell cultures were fixed and immune-stained as described previously (12). In brief, 50–75% confluent cells were fixed in 4% paraformaldehyde in phosphate-buffered solution (PBS: 167 mm Na+, 2.8 mm H2PO4−, 7.2 mm H2PO42−, pH 7.4), permeabilized in 0.2% saponin, and blocked in 10% FCS, 0.1% BSA, and 0.05% saponin in PBS. After additional blocking with 1% BSA, 0.2% fish gelatin, 0.05% saponin, and 0.05 m glycine in PBS, cells were stained with anti-slc4a10 antibody (3, 9) in PBS with 0.5% BSA and 0.05% saponin and then incubated with Alexa Fluor 488 goat anti-rabbit IgG (Invitrogen). Cells were counterstained using Topro3 nuclear stain (Invitrogen).
Surface Biotinylation and Immunoblotting
Stably transfected cells were grown to 80% confluence in 25-cm2 surface area flasks. Cell surface biotinylation was achieved by adding biotin to washed cells at 4 °C as detailed previously (13). After cell lysis and sonication, the biotinylated proteins from the cleared homogenate were isolated using a neutravidin column. Eluates from the columns were added 1.5% (w/vol) SDS, 40.0 mm 1,4-dithiothreitol, 6% (v/v) glycerol, 10 mm Tris(hydroxymethyl)-aminomethane, pH 6.8, and bromphenol blue. Samples were heated for 15 min at 65 °C and stored at 4 °C until use. Proteins were separated by SDS-PAGE (12). Proteins were then electrotransferred onto nitrocellulose membranes and blocked by 5% nonfat dry milk in a Tween-containing PBS. The membranes were incubated with the same primary antibody as above and subsequently with horseradish peroxidase-conjugated goat anti-rabbit IgG secondary antibody (Dako, Glostrup, Denmark). The signal was detected by an ECL kit (Amersham Biosciences).
Statistical Analysis
In general, data were analyzed by two-tailed t test using GraphPad Instat software. An analysis of variance test with Dunnett post hoc test was performed for multiple comparisons of unpaired pHi recoveries. A paired, nonparametrical analysis of variance test (Friedman) was performed for surface biotinylation data, where Gaussian distribution could not be expected. Values of p < 0.05 were considered an appropriate level of statistical significance.
RESULTS
slc4a10-transfected Cells Display Na+-dependent pHi Recovery and Na+ Import following Acidification
The Na+:HCO3− transport capacity of rat slc4a10-transfected cells was investigated by pHi measurements. The slc4a10-transfected cells exhibited significant Na+-dependent pHi recovery following NH4Cl-induced acidification compared with the dsRed-transfected negative controls (Fig. 2A; pHi acidification level slc4a10, 6.29 ± 0.07; dsRed, 6.22 ± 0.05, n = 6, not significant). As expected for a Na+:HCO3− transporter, the activity was fully dependent on the presence of CO2/HCO3− and was blocked by 200 μm DIDS (Fig. 2B). An unexpected Na+/H+ exchange activity was induced by transfection with both slc4a10 and dsRed. This transport was inhibited by 5-(N-ethyl-N-isopropyl) amiloride during pHi recoveries in all experiments. In parallel to the pHi recovery, there was a significant increase in [Na+]i in slc4a10-transfected cells compared with the control cells (Fig. 2C).
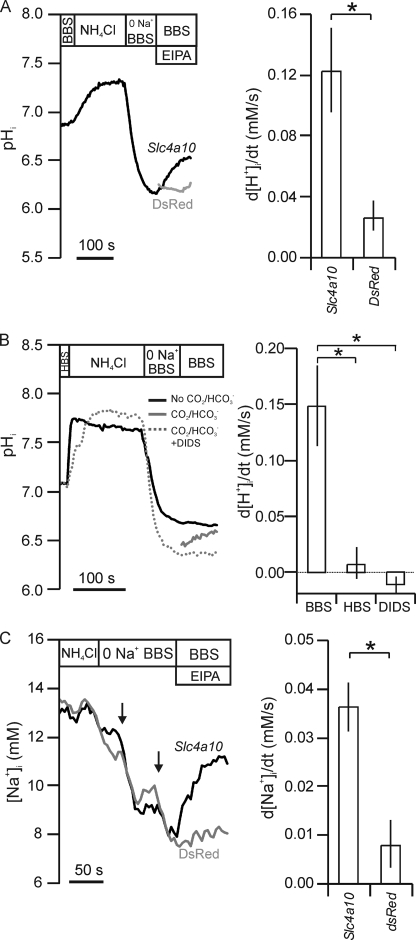
FIGURE 2.
Na+ and HCO3− import in stably rat slc4a10-transfected NIH-3T3 cells. A, example of pHi recording from BCECF-loaded slc4a10-transfected (black) and negative control dsRed-transfected cells (gray) by fluorescence microscopy. Cells were acidified by a 20 mm NH4Cl prepulse followed by a wash in Na+-free solution in larger volume perfusion chambers. The pHi recovery rates (d[H+]i/dt) were determined from the curve slope after readdition of Na+. Bar graph shows mean d[H+]i/dt values ± S.E. (error bars) of six such experiments. B, recording of pHi changes during NH4Cl prepulsing and recovery after acidification in the presence and absence of CO2/HCO3− and with the inhibitor DIDS in BBS. Bar graph shows mean values ± S.E. of similar experiments on slc4a10-transfected cells in the presence and absence of CO2/HCO3− (BBS and HBS, respectively) or with the inhibitor DIDS in BBS, 200 μm (n = 5). C, [Na+]i recordings in CoroNa Green-loaded slc4a10-transfected (black) and negative control cells (gray). The rate of change in intracellular Na+ concentration (d[Na+]i/dt) was determined after readdition of Na+. Arrows indicate focus adjustment. Bar graph represents mean d[Na+]i/dt values ± S.E. of six experiments. Error bars = S.E.
slc4a10-transfected Cells Transport Na+ and HCO3− with an Apparent Stoichiometry of 1Na+:2HCO3−
To estimate the stoichiometry of the Na+:HCO3− transport, pHi and [Na+]i were measured after NH4Cl prepulsing in smaller perfusion chambers to increase the fluid exchange rate (Fig. 3A, left and center panels, respectively). The net pHi recovery rate and [Na+]i change were determined the first 20 s after returning acidified cells to Na+-containing CO2/HCO3−-buffered solution and subtracted the corresponding values obtained in dsRed-transfected cells. The experiments indicate that 1.93 acid/base equivalents are transported for each Na+ molecule (Fig. 3A, right panel, p = 0.002, n = 6). To establish the accuracy of this estimate, the measurements were repeated by cuvette-based fluorometry in which a larger number of cells are measured simultaneously and with virtually instantaneous fluid exchange (Fig. 3B). The pHi recovery rate and [Na+]i change were determined the first 5 s after returning acidified cells to Na+-containing CO2/HCO3−-buffered solution and subtracting the corresponding values obtained in dsRed-transfected cells. The experiments suggested a Na+:HCO3− stoichiometry of 1:2.19 (Fig. 3C, left panel, p = 0.001, n = 6). The same cell culture line transfected with the slc4a7 gene (Nbcn1) also displayed Na+-dependent pHi recovery and corresponding [Na+]i changes. With the same method and data analysis, the apparent Na+:HCO3− stoichiometry for Nbcn1 was 1:1.17 (Fig. 3C, right panel, n = 6).
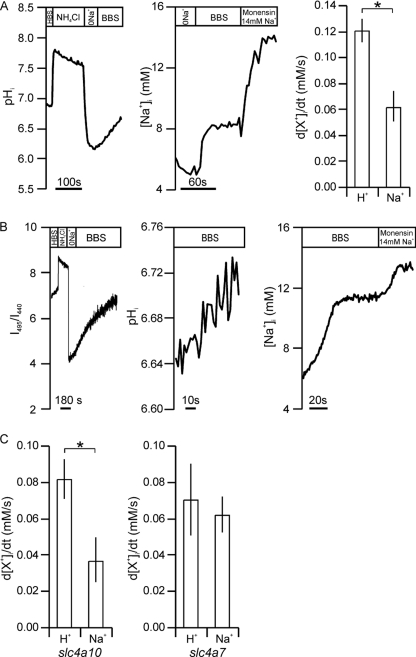
FIGURE 3.
Estimations of Na+HCO3− stoichiometry for slc4a10- and slc4a7-transfected cells. A, pHi recording of BCECF-loaded slc4a10-transfected cells by fluorescence microscopy using fast exchange perfusion chambers (left panel). Center panel, corresponding [Na+]i recordings in acidified CoroNa Green-loaded slc4a10-transfected cells. Right panel, mean d[X+]i/dt values ± S.E. (error bars), where X+ refers to H+ and Na+, as indicated (n = 6). B, left panel, cuvette fluorometry trace with the coverslip placed in a cuvette throughout the experiment. Center and right panels, traces of Na+-dependent pHi recoveries (center panel) and [Na+]i changes (right panel) upon acidification of slc4a10-transfected cells where acidification was performed before transfer to the cuvette. C, mean d[Na+]i/dt values and d[H+]i/dt values ± S.E. of slc4a10-transfected cells (left panel, n = 6) and slc4a7-transfected cells (Nbcn1, right panel, n = 6) corrected for the corresponding values from dsRed-transfected cells. *, statistical significance. Error bars = S.E.
Na+HCO3− Transport in slc4a10-transfected Cells Is Dependent on the Presence of Cl−
The Na+:HCO3− stoichiometry estimates suggest that a negative counterion or a positive cotransported ion is needed to maintain electroneutrality. Cl− has previously been suggested as the negative counter ion for the slc4a10 gene product. To remove both intracellular and extracellular Cl−, the slc4a10-transfected cells were incubated in the absence of extracellular Cl− for 30 min prior to pHi measurements. Cells were acidified by the addition of 20 mm propionate/propionic acid to the BBS (Fig. 4A), as the cells did not acidify by NH4− prepulsing in the absence of Cl−. The Na+-dependent pHi recovery of the Cl−-depleted rat slc4a10-transfected cells was dramatically decreased compared with cells with normal Cl− content (Fig. 4B, p = 0.0002, n = 6, initial pH values for slc4a10 in the absence of Cl−: 6.75 ± 0.05 versus 6.74 ± 0.05 in the presence of Cl−). The Na+:HCO3− transport activity was decreased to the level observed in dsRed-transfected control cells. Depleting dsRed-transfected cells of Cl− prior to pHi measurements did not further inhibit Na+-dependent pHi recovery (n = 6, not significant; initial pH values in Cl−-depleted dsRed-transfected cells: 6.78 ± 0.05 versus 6.75 ± 0.06 in the presence of Cl−). Cl− depletion was assessed by cellular loss of 36Cl−. After Cl− loading, cells gradually lost intracellular Cl− tracer to a level close to background count after 30 min (Fig. 4C). These experiments indicate that the 30-min Cl−-free incubation was sufficient to deplete the cells of Cl.
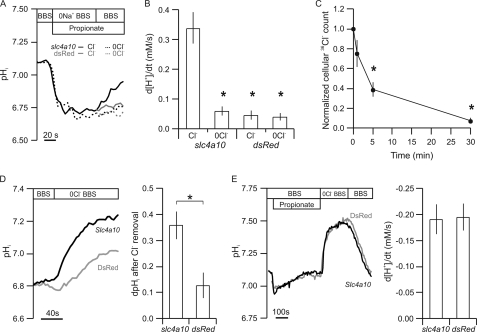
FIGURE 4.
The slc4a10 polypeptide is Cl−-dependent and sensitive to the Cl− gradient. A, pHi recordings from slc4a10-transfected (black) and negative control cells (gray) by fluorescence microscopy. Cells were depleted of Cl− for 30 min prior to the experiment by incubation in Cl−-free solution. Cells were acidified by addition of 20 mm propionate/propionic acid to a Na+-free solution. The Na+-dependent pHi recovery rate was determined after the readdition of Na+, as indicated. B, bar graph showing mean values ± S.E. (error bars) of d[H+]i/dt (n = 6). C, time course of cellular Cl− depletion from 36Cl−-loaded cells upon removal of the ion from the extracellular medium. D, pHi recordings of slc4a10-transfected (black) and negative control cells (gray) by fluorescence microscopy, where Cl− was removed from the extracellular medium as indicated. Bar graph shows the mean values ± S.E. of the maximal change in pHi (n = 6). E, rat slc4a10-transfected and control cells alkalized by propionic acid (20 mm added to solution) prepulsing and the Cl−-dependent pHi recovery rate determined. Bar graph shows the mean values ± S.E. of the pHi recovery rates. Error bars = S.E.
Na+ HCO3− Transport in slc4a10-transfected Cells Is Sensitive to the Transmembrane Cl− Gradient
The Cl− requirement for Na+ and HCO3− import by the rat slc4a10 gene product was further investigated by creating an outward Cl− gradient from the cells. An outward chemical Cl− gradient is expected to induce Na+:HCO3− import and thereby to induce intracellular alkalization in the case Cl− is exported by the transporter in slc4a10-transfected cells. The alkalization resulting from the acute removal of Cl− was significantly greater in slc4a10-transfected cells than the dsRed-transfected cells (Fig. 4D, initial pHi values in the rat slc4a10-transfected cells: 6.81 ± 0.08 versus 6.71 ± 0.04 in dsRed-transfected cells, p = 0.28, n = 6). The same pattern was observed in mouse slc4a10-expressing cells (dpHi in mouse slc4a10-expressing cells: 0.36 ± 0.05 versus dsRed-transfected cells: 0.07 ± 0.07, p = 0.012, n = 5). The alkalization observed in the dsRed-transfected cells most likely results from the reversed activity of an endogenous anion exchanger (i.e. HCO3− import and Cl− extrusion). Thus, the anion exchange capacity of the cell lines was compared to ascertain that the observed difference in alkalization between the slc4a10- and dsRed-transfected cells represents slc4a10-induced HCO3− import. The anion exchanger activity was investigated at high pHi at which forward anion exchange is favored and Na+:HCO3− import is minimal. The alkalization was achieved by prepulsing the cells for 5 min by adding propionate/propionic acid followed by a shift to Cl−-free solution in the continued presence of CO2/HCO3−. The Cl− was reintroduced after a steady-state pHi value had been reached, and the following pHi change was determined as a measure for anion exchange activity. There was no statistical difference in anion exchange activity between the slc4a10- and dsRed-transfected cells (Fig. 4E; for anion exchange: p = 0.914, n = 5; initial pH values for slc4a10: 7.35 ± 0.03 versus 7.42 ± 0.05 for dsRed, p = 0.28).
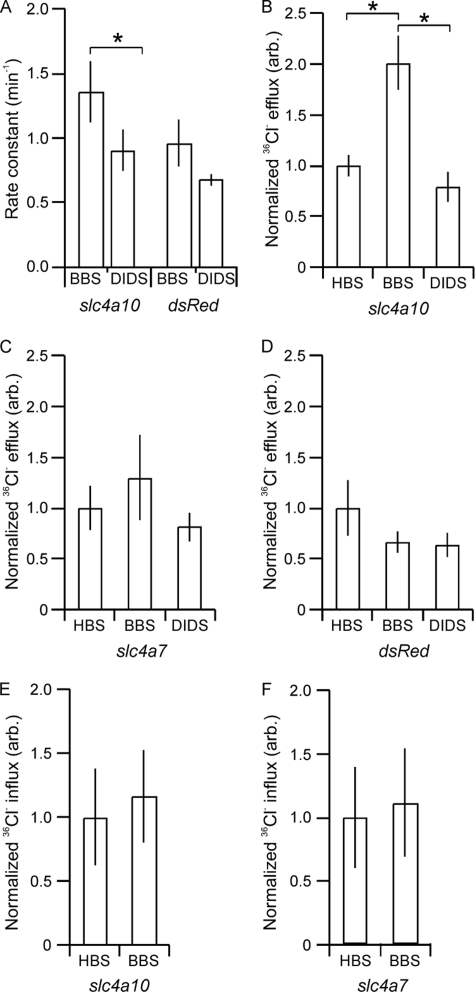
slc4a10-transfected Cells Exhibit DIDS-sensitive 36Cl− Efflux after Acidification
Direct evidence for Cl− transport by membrane proteins has classically been obtained by measuring 36Cl− transport. Cl− efflux rate constants were determined during recovery from NH4Cl-induced acidification to assess Cl− extrusion during high rate Na+ and HCO3− import. The 36Cl− efflux was computed before and after the readdition of Na+ in the presence of the Cl− channel blocker NPPB with and without DIDS. The Na+-dependent 36Cl− efflux was significantly lower in the presence than in the absence of DIDS in the mouse slc4a10-transfected cells (Fig. 5A; p = 0.02, n = 5), but not in dsRed-transfected cells (p = 0.21, n = 4). The Na+-dependent DIDS-sensitive dCl−/dt can be calculated to 0.13 mm/s by multiplying the DIDS-sensitive rate constant of 0.4 min−1 by an estimated [Cl−]i of 20 mm. Although this number may be inaccurate, it is in the same order of magnitude as the dNa+/dt and dH+/dt determined above. DIDS sensitivity of the Na+-dependent Cl− efflux during pHi recovery from acidification was verified in another series of experiments (Fig. 5, B–D). In the absence of CO2/HCO3−, the rate constant of the slc4a10 polypeptide equaled the value in the presence of DIDS and CO2/HCO3− (Fig. 5B). This indicated that the Cl− efflux not only depends on Na+, but is dependent on HCO3− as well. The values were corrected for the corresponding Na+-independent Cl− efflux rate constant from the same experiment (0.70 ± 0.13 in Na+-free HBS, 0.67 ± 0.15 in Na+-free BBS, n = 5, not significant). As a control, the corrected Cl− efflux rate constants were also estimated for cells transfected with the Cl−-independent Nbcn1 or dsRed under identical circumstances (Fig. 5, C and D, respectively). We did not observe statistically significant differences in rate constants under the three experimental conditions for Nbcn1- or dsRed-transfected cells. Under identical experimental conditions, the Cl− influx in slc4a10-transfected cells was not increased by the addition of CO2/HCO3−. The values were practically identical to those of slc4a7-transfected cells (Fig. 5, E and F; baseline cpm over background in HBS slc4a10: 35.57 ± 13.56; slc4a7: 42.40 ± 16.72, not significant). These values most likely corresponds to the baseline Cl− influx in HBS, as this influx was tripled in the same cell lines when the Na+,K+,2Cl− cotransport inhibitor furosemide was omitted from the solution (mean baseline cpm: 44 with furosemide and 138 without furosemide).
FIGURE 5.
The slc4a10 polypeptide mediates Cl− efflux but not Cl− influx. A, mean rate constants for the 36Cl− efflux of mouse slc4a10-transfected and control cells during the first 2 min after readdition of Na+ to acidified cells in the absence (BBS) and the presence of DIDS. The difference in apparent rate constants between the columns for each cell line represents the Na+-dependent DIDS-sensitive change in Cl− per time unit (n = 5). B, similar experiments performed on cells transfected with slc4a10 in presence/absence of CO2/HCO3− and DIDS. Rate constants were normalized to the value obtained in the absence of HCO3−. These 36Cl− efflux measurements were run in parallel with slc4a7-transfected cells (C) and dsRed-transfected cells (D). In separate experiments, 36Cl− influx was measured for the first 2 min of pHi recovery after acidification in cells transfected with slc4a10 (E) or slc4a7 (F) in the presence/absence of CO2/HCO3−. *, statistical significance.
Specific Charged Amino Acids Are Necessary for Normal slc4a10-induced Transport
Mutations in mouse slc4a10 were introduced at putative cation and anion attracting sites, selected as detailed under “Experimental Procedures.” In brief, all charged amino acids in the proposed hinge-like loops of slc4a10 that vary systematically among the gene family members were exchanged for uncharged amino acids (Glu890, Glu892, His976, and His980). In addition, a negatively charged amino acid outside these domains was mutated as well (Glu584). The Na+-dependent pHi recovery profiles of cell lines stably transfected with mutant slc4a10 forms were compared with the wild type slc4a10-transfected cell line (Fig. 6A). The pHi recovery rate was reduced by 62–76% (range) in E890A, E892A, H976L, and H980G single amino acid mutations compared with wild type slc4a10 cells (Fig. 6B, p < 0.001, n = 5), whereas transport was unaffected in the E584A cell line (p = 0.70, n = 5). We note that the Na+-dependent pHi recovery in wild type mouse slc4a10-transfected cells was similar to that of rat slc4a10-transfected cells (Fig. 6B). Quadruple E890A/E892A/H976L/H980G mutation as well as double E890A/E892A and H976L/H980G mutation eliminated pHi recovery (quadruple: p = 0.0002, n = 5, E890A/E892A: p = 0.002, n = 5; H976L/H980G: p = 0.002, n = 5; Fig. 6C). For each mutant cell line, immunoblotting analysis of surface biotinylated slc4a10 gene product was normalized to actin content of cell homogenate (to correct for cell density in culture flasks). There was no significant difference in surface slc4a10 protein to actin ratio among the mutants and wt cells (Fig. 6D, range 0.7 to 1.7 of wt slc4a10, n = 4, p > 0.05). Fig. 5E shows representative micrographs of slc4a10-transfected NIH-3T3 cells used in the above experiments.
FIGURE 6.
Na+-dependent HCO3− transport by the mouse slc4a10 gene product after selective mutagenesis. Mutations of slc4a10 were introduced by site-directed mutagenesis in positions E584A, E890A, E892A, H976L, and H980G. Cell lines expressing each of these mutant forms were generated, and the Na+-dependent HCO3− transport was assessed. A, pHi recordings from wild type and mutated slc4a10-transfected and negative control cells (dsRed) by fluorescence microscopy. Cells were acidified by a 20 mm NH4Cl prepulse followed by a wash in Na+-free solution as indicated. The pHi recovery rates (d[H+]i/dt) were determined from the slope after readdition of Na+. B, mean d[H+]i/dt values ± S.E. (error bars) after acidification of wild type slc4a10 and formation of a single mutant slc4a10 (n = 5). C, mean values ± S.E. of the Na+-dependent pHi recovery rate after acidification in double E890A/E892A, double H976L/H980G, and quadruple E890A/E892A/H976L/H980G slc4a10-mutants. *, statistical significance. D, immunoblot analysis of slc4a10 protein expression after surface biotinylation. Cells were biotinylated on ice and surface proteins isolated on a neutravidin column. Immunoblotting these samples for slc4a10 protein was corrected for cell density by actin immunoblots of the corresponding cell homogenates. Bar graph shows the mean values ± S.E. of the surface slc4a10 protein/cell actin relation after normalization to mouse-slc4a10 (n = 4). E, immunocytochemical micrographs of dsRed-transfected cells, wild type, and mutant slc4a10-transfected cells, as indicated. Error bars = S.E.
DISCUSSION
Na+-driven HCO3− import is necessary for multiple cell types to maintain a suitable intracellular milieu and to mediate a transcellular transport of base equivalents in specialized epithelia. The integral membrane proteins responsible for this Na+:HCO3− cotransport all belong to the solute carrier family 4, slc4a. Some controversy exists regarding the transport mode of one of these Na+-dependent HCO3− transporters, the slc4a10 gene product. Originally, the protein was characterized as a Na+-dependent Cl−/HCO3− exchanger, Ncbe (2), but a recent report suggested renaming the protein to Nbcn2 because Cl− transport was not linked to the HCO3− transport (7). slc4a10 disruption leads to (i) impaired regulation of intracellular pH upon acidification in the choroid plexus and hippocampal neurons, (ii) decreased brain ventricle volume, indicating abnormal cerebrospinal fluid formation, and (iii) increased seizure threshold (4). Because a human mutation in the SLC4A10 has been reported to induce a severe phenotype (5), we believe it crucial to explore the nature of this gene product further.
There is agreement in the literature on the most basic transport characteristics of the slc4a10 gene product. The Na+ dependence of the slc4a10-induced pHi regulation presented here is in accordance with all three previous studies characterizing the gene product (7, 2, 6). We show that intracellular [Na+] actually increases during the pHi recovery, providing direct evidence for the involvement of Na+ import by the transporter. This concurs with the 22Na+ uptake into untagged SLC4A10-injected Xenopus oocytes observed only in the presence of HCO3− (2). To propose amino residues that may be involved in Na+ attraction and translocation, we mutated both negative amino acids in the loops that are shared by Na+-transporting slc4a-derived proteins. By homology to the Ae1 protein, the Glu584 is located in the end of TM1, which is most unlikely to represent a flexible part of the protein. This mutation served as a negative control. In contrast, Glu890 and Glu892 are situated in the first putative hinge-like loop between the proposed TM9 and TM10, previously suggested to be involved in ion translocation in Ae1 (8). No other candidate amino acids could be deduced from the primary structures of slc4a genes. We found that mutation of only the two latter decreased slc4a10-induced Na+-dependent pHi recovery, which makes us speculate whether the amino acids may be involved in attracting or even translocating Na+. Double mutation of these sites prevented pHi recovery. To our knowledge, these amino acids have not been mutated in studies of other slc4a polypeptides, including Ae1. Apart from actual transport of Na+, the present study also verifies the previous three investigations in the absolute requirement for CO2/HCO3− and places the slc4a10 product within the group of DIDS-sensitive Na+-dependent HCO3− transporters.
The major discrepancies regarding the transport mediated by the slc4a10 gene product seems to be the possible dependence and/or transport of Cl−. Wang et al. found that HCO3− import after acidification in transiently SLC4A10-transfected HEK cells was virtually absent after 1-h Cl− depletion (i.e. internal and external Cl−-free) (2). Giffard et al. (6) found the same using stably FLAG-tagged rat slc4a10-transfected NIH-3T3 cells after 20-min Cl− depletion. This demonstrates either that the protein transports Cl− or that it is merely sensitive to the presence of Cl−. Evidence for direct Cl− transport by the protein came from the demonstration of SLC4A10-induced Cl− efflux which was DIDS-sensitive and dependent on Na+, Cl−, and HCO3− (2). Furthermore, long term equilibration at various extracellular [Cl−] yielded a linear correlation between [Cl−] and Na+ uptake in SLC4A10-injected Xenopus oocytes in the same study. In contrast to this and the present work, Parker et al. (7) found that pHi recovery is independent of Cl− even after 20 min under Cl−-free conditions and that enhanced GFP-tagged SLC4A10-induced Cl− efflux was independent of extracellular Na+. However, the authors do observe Na+-driven Cl−/HCO3− exchange in the absence of extracellular Cl−. Finally, the lack of net Cl− extrusion induced by SLC4A10 was demonstrated in the same study comparing the surface [Cl−] by microelectrodes to oocytes injected with known Cl−-dependent and -independent slc4a-derived transporters. The data pattern of the SLC4A10-injected oocytes was similar to the Cl−-independent transporters and different from known Cl− extruders. Some of the inconsistencies may rely on differences between mammalian cells and Xenopus oocytes, temperature, and composition of solutions, the transfection/injection efficiency or molecular tagging of the transport proteins. Discrepancy in transport mode caused by the expression systems has previously been described for other members of the slc4a family, Nbce2 and Nbce1 (14–18). To circumvent these putative sources of error, we use stable transfection with incorporation of a single copy of untagged slc4a10 into a predefined locus in a mammalian fibroblast cell line without native Na+:HCO3− transport and performed all experiments at a temperature and in buffers that are appropriate for the mammalian protein.
In agreement with the studies by Wang et al. (2) and Giffard et al. (6), we find that slc4a10-induced transport is Cl−-dependent because as depletion of internal and external Cl− completely inhibits pHi recovery from acid values. We also find that acutely creating an outward chemical gradient for Cl− enhances slc4a10-induced transport. This is indicative of Cl− transport but does not rule out that the protein somehow senses a Cl− gradient without transporting the ion. Finally, we found evidence for a Na+-dependent, CO2/HCO3−-dependent, DIDS-sensitive Cl− efflux in slc4a10-transfected cells in agreement with Wang et al. Importantly, this Cl− efflux during pHi recovery was not accompanied by detectable Cl− influx. Analysis of the apparent Na+:HCO3− stoichiometry was performed to support the functional coupling of Na+:HCO3− to Cl− efflux. The Na+:HCO3− stoichiometry is supposed to be 1:2 or greater when slc4a10 exchanges Cl− for HCO3− to maintain the electroneutrality. It is noted that electroneutrality has been demonstrated by microelectrode recording of SLC4A10-injected Xenopus oocytes in just one study (7). We estimated a Na+:HCO3− stoichiometry of 1:2 in slc4a10-transfected cells, whereas the apparent ratio in slc4a7-transfected cells was 1:1 (encoding the Cl−-independent electroneutral Na+:HCO3− cotransporter Nbcn1).
There are more ways to explain the apparent transport of two HCO3−/Na+ molecules: (i) two molecules of HCO3−, (ii) one molecule of CO32−, or (iii) countertransport of H+. The first two possibilities would require more than one positive amino acid in a given domain to attract and translocate the negative ion(s). Interestingly, the second large hinge-like loop between proposed TM11 and TM12 of slc4a10 contained two such amino acids (His976 and His980) and varies among slc4a10-derived proteins of varying transport mode. Mutations in each of these significantly reduced slc4a10-induced Na+-dependent pHi recovery, and double mutation of these amino acids hindered any transport. Mutations of the corresponding amino acid residues in Ae1 (Arg827 and Trp831) led to identical degree of transport inhibition (8). Quadruple mutation of the specific charged amino acids in the hinge-like regions also completely prevented slc4a10-induced Na+-dependent pHi recovery. The reason for the observed decreases in slc4a10 polypeptide function may be that the four charged amino acids are involved in the attraction or even the translocation of Na+ and HCO3−. Certainly, the changes are not explained by variations in membrane abundance as judged from the surface biotinylation analysis. Major changes in protein folding are usually detected by the cellular quality control systems and are not likely to reach the plasma membrane. Nevertheless, smaller changes in tertiary structure may have been induced by these mutations. It is evident that many more residues of these polypeptides are involved in ion transport. As an example, Ser731 and His734 in the first hinge-like loop of Ae1 are conserved among more types of transporters. Single mutations of these residues deleted anion exchange but introduced cation leak (19). In this study, we focus on the amino acids that may underlie the variations in ionic requirement among the gene family members. The omission of charged amino acids, which are shared by the group of slc4a proteins known to transport more than one HCO3− molecule/Na+, greatly affects transport by the slc4a10-derived protein. Thus, the mutational analysis would support that the slc4a10 gene product belongs to this group of transporters.
In conclusion, we find that the slc4a10 expression induces Na+-dependent Cl−/HCO3− exchange in a mammalian cell system with our experimental conditions, and we propose the protein name remains Ncbe. We speculate that the first hinge-like region is necessary for the Na+ translocation process and the second for the HCO3− or CO32− translocation process. Further structural analyses including crystallization of the transporter are necessary to determine fully the transport mode of Ncbe.
Acknowledgments
We thank Robert A. Fenton for guidance concerning transfection strategy and surface biotinylation and Christian Valdemar Westberg, Tina Drejer, and Helle Hoyer for expert technical assistance.
This work was supported by the Danish Medical Research Council, Karen Elise Jensens Fond, the Lundbeck Foundation, Aarhus University Research Foundation, and the Novo Nordisk Foundation. The Water and Salt Research Center at Aarhus University was established and funded by the Danish National Research Foundation (Danmarks Grundforskningsfond).
- pHi
- intracellular pH
- BBS
- bicarbonate-buffered saline
- BCECF
- 2′,7′-bis(carboxyethyl)-5,6-carboxyfluorescein
- DIDS
- 4,4′-diisothiocyanatostilbene-2,2′-disulfonate
- FRT
- flipase recognition target
- HBS
- HEPES-buffered salt solution
- Nbcn
- electroneutral Na+;HCO3− cotransporter
- Ncbe
- Na+-dependent Cl−/HCO3− exchanger
- Nbce
- electrogenic Na+;HCO3− cotransporter
- Ndcbe
- Na+-dependent Cl−/HCO3− exchanger
- NPPB
- 5-nitro-2-(3-phenylpropylamino)-benzoate
- Slc4a
- solute carrier family 4.
REFERENCES
- 1.Boron W., Boulpaep E. L. (2009) Medical Physiology, 2nd Ed., pp. 667–671, W. B. Saunders Co., Philadelphia, PA [Google Scholar]
- 2.Wang C. Z., Yano H., Nagashima K., Seino S. (2000) J. Biol. Chem. 275, 35486–35490 [DOI] [PubMed] [Google Scholar]
- 3.Praetorius J., Nejsum L. N., Nielsen S. (2004) Am. J. Physiol. Cell Physiol. 286, C601–C610 [DOI] [PubMed] [Google Scholar]
- 4.Jacobs S., Ruusuvuori E., Sipilä S. T., Haapanen A., Damkier H. H., Kurth I., Hentschke M., Schweizer M., Rudhard Y., Laatikainen L. M., Tyynelä J., Praetorius J., Voipio J., Hübner C. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 311–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gurnett C. A., Veile R., Zempel J., Blackburn L., Lovett M., Bowcock A. (2008) Arch. Neurol. 65, 550–553 [DOI] [PubMed] [Google Scholar]
- 6.Giffard R. G., Lee Y. S., Ouyang Y. B., Murphy S. L., Monyer H. (2003) Eur. J. Neurosci. 18, 2935–2945 [DOI] [PubMed] [Google Scholar]
- 7.Parker M. D., Musa-Aziz R., Rojas J. D., Choi I., Daly C. M., Boron W. F. (2008) J. Biol. Chem. 283, 12777–12788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu Q., Lee D. W., Casey J. R. (2003) J. Biol. Chem. 278, 3112–3120 [DOI] [PubMed] [Google Scholar]
- 9.Praetorius J., Nielsen S. (2006) Am. J. Physiol. Cell Physiol. 291, C59–C67 [DOI] [PubMed] [Google Scholar]
- 10.Boyarsky G., Ganz M. B., Sterzel R. B., Boron W. F. (1988) Am. J. Physiol. Cell Physiol. 255, C844–C856 [DOI] [PubMed] [Google Scholar]
- 11.Damkier H. H., Prasad V., Hübner C. A., Praetorius J. (2009) Am. J. Physiol. Cell Physiol. 296, C1291–C1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Damkier H. H., Nielsen S., Praetorius J. (2007) Am. J. Physiol. Regul. Integr. Comp. Physiol. 293, R2136–R2146 [DOI] [PubMed] [Google Scholar]
- 13.Moeller H. B., Praetorius J., Rützler M. R., Fenton R. A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 424–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romero M. F., Fong P., Berger U. V., Hediger M. A., Boron W. F. (1998) Am. J. Physiol. Renal Physiol. 274, F425–F432 [DOI] [PubMed] [Google Scholar]
- 15.Gross E., Hawkins K., Abuladze N., Pushkin A., Cotton C. U., Hopfer U., Kurtz I. (2001) J. Physiol. 531, 597–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sassani P., Pushkin A., Gross E., Gomer A., Abuladze N., Dukkipati R., Carpenito G., Kurtz I. (2002) Am. J. Physiol. Cell Physiol. 282, C408–C416 [DOI] [PubMed] [Google Scholar]
- 17.Millar I. D., Brown P. D. (2008) Biochem. Biophys. Res. Commun. 373, 550–554 [DOI] [PubMed] [Google Scholar]
- 18.Virkki L. V., Wilson D. A., Vaughan-Jones R. D., Boron W. F. (2002) Am. J. Physiol. Cell Physiol. 282, C1278–C1289 [DOI] [PubMed] [Google Scholar]
- 19.Guizouarn H., Martial S., Gabillat N., Borgese F. (2007) Blood 110, 2158–2165 [DOI] [PubMed] [Google Scholar]