Abstract
Impairment of the cystic fibrosis transmembrane conductance regulator (CFTR) Cl− channel causes cystic fibrosis, a fatal genetic disease. Here, to gain insight into CFTR structure and function, we exploited interspecies differences between CFTR homologues using human (h)-murine (m) CFTR chimeras containing murine nucleotide-binding domains (NBDs) or regulatory domain on an hCFTR backbone. Among 15 hmCFTR chimeras analyzed, all but two were correctly processed, one containing part of mNBD1 and another containing part of mNBD2. Based on physicochemical distance analysis of divergent residues between human and murine CFTR in the two misprocessed hmCFTR chimeras, we generated point mutations for analysis of respective CFTR processing and functional properties. We identified one amino acid substitution (K584E-CFTR) that disrupts CFTR processing in NBD1. No single mutation was identified in NBD2 that disrupts protein processing. However, a number of NBD2 mutants altered channel function. Analysis of structural models of CFTR identified that although Lys584 interacts with residue Leu581 in human CFTR Glu584 interacts with Phe581 in mouse CFTR. Introduction of the murine residue (Phe581) in cis with K584E in human CFTR rescued the processing and trafficking defects of K584E-CFTR. Our data demonstrate that human-murine CFTR chimeras may be used to validate structural models of full-length CFTR. We also conclude that hmCFTR chimeras are a valuable tool to elucidate interactions between different domains of CFTR.
Keywords: ABC Transporter, Chloride Channels, Protein Chimeras, Protein Folding, Trafficking, Cystic Fibrosis, F508del, NBD1 Structure, Human-Murine CFTR Chimeras
Introduction
Cystic fibrosis (CF)4 is the most common lethal genetic disease in the Caucasian population, resulting from the dysfunction of the cystic fibrosis transmembrane conductance regulator (CFTR) (1). CFTR is a multidomain protein containing 1480 amino acid residues located at the apical membrane of epithelial cells where it functions as a chloride (Cl−) ion channel regulated by cAMP-dependent phosphorylation and cycles of ATP binding and hydrolysis (2, 3). Based on its structure and function, CFTR (ABCC7) is a member of the ATP-binding cassette (ABC) transporter superfamily. It contains two membrane-spanning domains, two nucleotide-binding domains (NBDs), and a unique regulatory domain (RD) with multiple consensus phosphorylation sites and many charged amino acids (2, 4). The most common disease-causing mutation, occurring in ∼90% of CF patients worldwide on at least one CFTR allele, is deletion of phenylalanine 508 (F508del) located in NBD1.
A critical aspect of CF research is to understand how the F508del mutation disrupts CFTR function at the molecular and cellular levels. Depending on the cell type examined, the maturation efficiency of wild-type (WT)-CFTR protein is 25–70% (5–7). By contrast, very little or virtually no functional F508del-CFTR reaches the cell surface because it is retained in the endoplasmic reticulum (ER) by the ER quality control (ERQC) machinery and targeted for degradation (8). The exact nature of the structural divergence imposed on CFTR by absence of F508del, which is recognized by the ERQC (9, 10), is currently unknown. To understand better how F508del perturbs CFTR architecture, high resolution structures of murine (m) and human (h) NBD1 as well as of human F508del-NBD1 (11, 12) have been resolved. However, the structure of human NBD1 incorporates a series of mutations, which rescue in vivo the cell surface expression and function of F508del-CFTR (13). Thus, the available crystal structure of F508del-NBD1 plausibly corresponds to a partially corrected conformation of this domain. Based on these NBD1 structures and atomic resolution structures of other ABC transporters (14), molecular models of the CFTR protein have been developed (15, 16). These molecular models are a valuable guide for studies of CFTR structure and function.
Another powerful approach to investigate the structure and function of hCFTR is to examine interspecies differences to identify conserved and divergent regions. Since the identification and cloning of the hCFTR gene (4), various homologues have been isolated from different species (e.g. mouse, sheep, pig, Xenopus, macaque, rabbit, bovine, killifish, and salmon); all possess a domain organization similar to that of hCFTR (17). Nevertheless, significant interspecies differences in CFTR processing and function have been reported previously. For example, human and murine CFTR Cl− channels exhibit different patterns of channel gating (18, 19), which are specified, in part, by amino acid sequence differences in the NBDs (20). Moreover, Ostedgaard et al. (21) identified differences in the processing of the F508del mutation between human, porcine, and murine CFTR. In that study, both porcine and murine F508del-CFTR proteins were found to be at least partially processed like their WT counterparts, and this processing was unaffected by the origin of the cell line where the mutant proteins were expressed. Thus, cross-species CFTR chimeras appear to be an excellent resource to investigate the folding and functional consequences of interdomain interactions in CFTR. Indeed, insights into the biochemical properties of mCFTR subdomains in the context of hCFTR, such as those provided by the present study, will help to further understanding of the structural and functional effects of disease-causing mutations in both NBDs and validate existing structural models (15, 16, 22).
In the present work, we aimed to identify the structural features of hCFTR (mis)folding using a human-murine comparative approach by investigating whether CFTR domains (NBDs and RD) are structurally interchangeable between human and murine CFTR. Our data reveal that two chimeras (one in NBD1 and another in NBD2) failed to be processed into their mature forms. Systematic mutagenesis of the divergent murine residues present in these chimeras followed by biochemical and functional studies revealed that K584E is responsible for the maturation defect of the hmNBD1 chimera. Furthermore, replacement of Leu581 (interacting with Lys584 in the structure of hNBD1) by the corresponding murine amino acid (Phe581) rescued the maturation of K584E-CFTR. Thus, our results demonstrate that hmCFTR chimeras can be used to identify critical residues responsible for both structural and functional differences between human and murine CFTR. Furthermore, the CFTR mutants generated here constitute a valuable resource to characterize the possible binding sites and mechanism of action of small molecules used in CFTR assist therapies (23).
EXPERIMENTAL PROCEDURES
Construction of Human-Murine CFTR Chimeras
Human-murine CFTR chimeras containing sequences from murine NBD1, NBD2, and RD were constructed by homologous recombination as described previously (20).
Cells, Site-directed Mutagenesis, and CFTR Expression
The QuikChange® mutagenesis kit (Stratagene) was used to introduce mutations into the pNUT WT-CFTR cDNA. Each mutation was verified by sequencing. For a list of primers used, see supplemental Table 2. We transiently expressed wild-type and chimeric CFTRs in HEK-293 cells using Lipofectin® (Invitrogen). Cells were transfected with 1 μg of cDNA for all CFTR variants. 48 h after transfection, we extracted total protein from HEK-293 cells expressing wild-type and chimeric CFTRs and assayed for CFTR expression by Western blotting. To generate cell lines stably expressing high levels of CFTR variants, baby hamster kidney (BHK) cells were transfected with CFTR cDNAs (3 μg) using Lipofectin (Invitrogen) and selected for stable transfectants using methotrexate (500 μm). For each CFTR variant, 10 BHK clones were analyzed by Western blotting. Of these, the clone expressing the highest level of CFTR protein was selected for further analyses, ensuring that CFTR expression levels among the different CFTR variants were equivalent. Cells were cultured, seeded, and used as described previously (24). In some experiments (see figure legends), cell surface expression of F508del-CFTR was enhanced by incubating cells at 26 °C for 24 h.
Western Blotting, Pulse-Chase, and Immunoprecipitation
To assay for CFTR protein expression by Western blot (WB), cells expressing CFTR variants were lysed, and extracts were analyzed as described (25) using the anti-CFTR antibody 596 (CF Foundation). Densitometry was performed as described previously (25). Cells were starved for 30 min in methionine-free α-modified Eagle's medium (Invitrogen) before being radiolabeled for 25 min in the same medium supplemented with 150 μCi/ml [35S]methionine (ICN Biomedicals). For the chase (0, 0.5, 1, 2, and 3 h), the labeling medium was replaced by α-modified Eagle's medium supplemented with fetal bovine serum (8%, v/v; Invitrogen) and non-radioactive methionine (1 mm; Sigma-Aldrich). Cells were then lysed in radioimmunoprecipitation assay buffer (1 ml) containing deoxycholic acid (1%, w/v; Sigma-Aldrich), Triton X-100 (1%, v/v; GE Healthcare Bio-Sciences), SDS (0.1%, w/v; Invitrogen), Tris (50 mm, pH 7.4; Sigma-Aldrich), and NaCl (150 mm). CFTR protein was immunoprecipitated as described by Farinha et al. (26) after centrifugation of samples at 14,000 × g for 30 min at 4 °C. To detect specifically CFTR, the supernatant was incubated overnight with 1.5 μg of the anti-CFTR monoclonal antibody M3A7, which recognizes NBD2 and the C terminus of CFTR (residues 1197–1480; Chemicon), at 4 °C, and then protein G-agarose beads (25 μg; Roche Applied Science) were added for a further 4 h at 4% (v/v). Beads were washed four times using radioimmunoprecipitation assay buffer (1 ml), and protein was eluted for 1 h at room temperature (RT) with cracking buffer (80 μl) containing dithiothreitol (0.5 mm; Sigma-Aldrich), bromphenol blue (0.001%, w/v), glycerol (5%, v/v), SDS (1.5%, w/v), and Tris (31.25 mm), pH 6.8. Samples were separated electrophoretically on 7% (v/v) polyacrylamide gels. Then, gels were prefixed (methanol (30%, v/v) and acetic acid (10%, v/v)) for 30 min, washed thoroughly in water, and soaked in salicylic acid (1 m) for 1 h. After drying at 80 °C under vacuum for 2 h, gels were exposed to x-ray film (Fujifilm Medical Systems). Fluorograms of gels were digitized (Sharp JX-330, Sharp Europe), and integrated peak areas were determined using ImageMaster® software (GE Healthcare).
Iodide Efflux
CFTR-mediated iodide efflux was measured at room temperature as described (19) using the cAMP agonist forskolin (10 μm) and the CFTR potentiator genistein (50 μm; Sigma-Aldrich). Prior to commencing experiments, BHK cells expressing CFTR variants were incubated for 1 h in loading buffer containing 136 mm NaI, 3 mm KNO3, 2 mm Ca(NO3)2, 20 mm Hepes, and 11 mm glucose, pH 7.4 with 1 m NaOH and then washed thoroughly with efflux buffer (136 mm NaNO3 replacing 136 mm NaI in the loading buffer). The amount of iodide in each sample of efflux buffer was determined using an iodide-selective electrode (MP225, Thermo Electron Corp., Waltham, MA). BHK cells were loaded, and experiments were performed at RT (∼23 °C).
Immunocytochemistry
Immunocytochemistry was performed as described previously (27). In brief, BHK cells expressing CFTR variants were rinsed twice with cold PBS and fixed with ice-cold 4% p-formaldehyde (Fluka BioChemica) in PBS for 20 min. After four washes with cold PBS at RT, nonspecific staining was prevented by blocking with BSA (1%, w/v) in PBS for 30 min. Cells were stained with anti-wheat germ agglutinin for 1 h, coupled with Texas Red, and then permeabilized with Triton (0.2%, w/v) for 20 min. Finally, cells were washed three times with PBS (10 min each at RT) and incubated with (i) monoclonal anti-CFTR antibody 570 and then (ii) secondary antibody, Alexa Fluor 488 anti-mouse, for 1 h each. Cells were again washed three times with PBS (10 min each at RT), and slides were mounted with Vectashield (Vector Laboratories) containing 4′,6-diamidino-2-phenylindole dihydrochloride (Sigma-Aldrich) to stain nuclei and covered with a glass coverslip. Immunofluorescence staining was investigated using a confocal microscope (Leica TCS SPE). No background staining or autofluorescence was observed with untransfected BHK cells (data not shown).
Electrophysiology
CFTR Cl− currents were recorded in excised inside-out membrane patches using an Axopatch 200B patch clamp amplifier (MDS Analytical Technologies) and pCLAMP data acquisition and analysis software (versions 6.0.4 and 9.2, MDS Analytical Technologies) as described previously (18). The pipette (extracellular) solution contained 140 mm N-methyl-d-glucamine, 140 mm aspartic acid, 5 mm CaCl2, 2 mm MgSO4, and 10 mm TES, pH 7.3 with Tris ([Cl−], 10 mm). The bath (intracellular) solution contained 140 mm N-methyl-d-glucamine, 3 mm MgCl2, 1 mm CsEGTA, and 10 mm TES, pH 7.3 with HCl ([Cl−], 147 mm; [Ca2+]free, <10−8 m) and was maintained at 37 °C. After their excision, membrane patches were voltage-clamped at −50 mV, and CFTR Cl− channels were activated by the addition of ATP (1 mm; Sigma-Aldrich) and the catalytic subunit of protein kinase A (75 nm; Promega UK) to the intracellular solution within 5 min of patch excision. In this study, we used membrane patches containing small numbers of active channels (≤5). We determined the number of channels in a membrane patch from the maximum number of simultaneous channel openings observed during the course of an experiment. To minimize errors when counting the number of active channels, we used the strategies described by Cai et al. (28). We recorded, filtered, and digitized data as described (18). To measure single channel current amplitude (i), Gaussian distributions were fit to current amplitude histograms. For open probability (Po) analyses, lists of open and closed times were created using a half-amplitude crossing criterion for event detection. Transitions <1 ms in duration were excluded from the analyses, and Po was calculated as described previously (28).
Statistics
Results are expressed as means ± S.E. of n observations. To compare sets of data, we used Student's t test for iodide efflux and single channel data. Pulse-chase data were analyzed by comparing degradation rates (slopes of regression lines) by Student's t test. On a regression modeling procedure, the slope is known to follow a t distribution (29, 30). Therefore, slopes of two straight lines can be compared using a t distribution with n1 + n2 − 4 degrees of freedom where n1 and n2 are the number of points used on the regression procedure in groups 1 and 2, respectively. Differences between groups of data were considered statistically significant when p < 0.05.
RESULTS
Human-Murine CFTR Chimeras
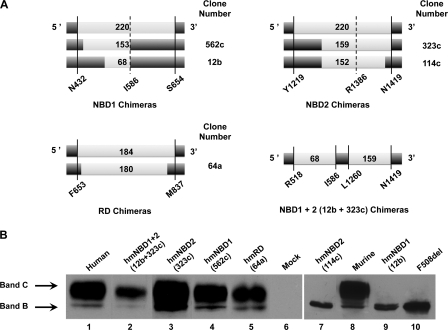
A previously described recombination strategy was used to generate hmCFTR chimeras (20). Because of the high degree of homology between hCFTR and mCFTR sequences, homologous recombination frequently occurred within the murine domains rather than at the chimeric junction defined by the PCR primers. From the library of chimeras generated by Scott-Ward et al. (20), in the present work, we focused on the following constructs (Fig. 1A): 562c-NBD1 (limits of inserted mCFTR sequence, Leu433–Val586), 12b-NBD1 (Lys518–Val586), 323c-NBD2 (Met1260–Ser1419), 114c-NBD2 (Met1260–Arg1412), 64a-RD (Ser654–Leu834), and NBD1+2 (Lys518–Val586; Met1260–Ser1419). The latter NBD1+2 refers to chimera 12b+323c, which is not the same as that previously described (20); hence, we term it hereafter 12b-NBD1+323c-NBD2. As controls, we also analyzed WT- and F508del-hCFTR as well as mCFTR.
FIGURE 1.
Biochemical analyses of hmCFTR chimeras in which one or two domains of full-length hCFTR (dark gray rectangles) were replaced by an equivalent one(s) from mCFTR (light gray rectangles). A, schematic representations of the hmCFTR chimeras. The light gray segments represent mCFTR inserts (numbers designate length in mCFTR amino acid residues) into hCFTR (dark gray rectangles). Note that the extent of protein sequence classified here as mCFTR is depicted as the largest contiguous protein sequence inserted into each construct that is flanked at each end by hCFTR residues (i.e. residues that differ between mCFTR and hCFTR sequences). Therefore, sequences of residues that are common to both mCFTR and hCFTR and may extend beyond the mCFTR blocks are denoted here as hCFTR because they do not represent deviations from the human CFTR sequence. The continuous lines and coordinates indicate the chimeric junctions defined by the PCR primers and confirmed here by sequencing; the dashed lines and coordinates show the original C termini of NBD1 and NBD2 (4). For further information, see “Results.” B, Western blot analysis of hmCFTR chimeras transiently expressed in HEK-293 cells using the anti-CFTR antibody 596 (CF Foundation). The amount of total protein applied was 60 μg. The positions of bands B and C are indicated by arrows. Data shown are representative of multiple experiments (n = 10).
Processing Efficiency of Human-Murine CFTR Chimeras
The traffic of CFTR protein to the cell membrane can be assessed indirectly by its maturation. Indeed, WT-CFTR synthesized in the ER as a core-glycosylated, immature form undergoes additional glycosylation in the Golgi apparatus to generate its mature form (band C; ∼170–190 kDa) (9, 31). By contrast, due to its ER retention, F508del-CFTR can only be detected as the core-glycosylated, ER-specific immature form of CFTR (band B; ∼150 kDa) (31). Quantification of these immature and mature forms at steady state by WB provides a measure of the extent of maturation of a given CFTR variant and also, indirectly, of whether its in vivo folding has been achieved by ERQC protein conformation criteria. Thus, such data can also be interpreted as indicative of structural divergence between WT- and mutant CFTR.
When we evaluated the maturation efficiency of the hmCFTR chimeras by assessing their production of immature and mature CFTR protein at steady state by WB analysis in the same cell line, we found that two chimeras failed to generate the mature form of CFTR (band C): clone 12b-NBD1 (Fig. 1B, lane 9) and clone 114c-NBD2 (Fig. 1B, lane 7). Indeed, both these chimeras were only detected as their immature core-glycosylated ER-specific forms (150 kDa; band B) (31). These results contrast with all the other chimeras analyzed: 12b-NBD1+323c-NBD2 (Fig. 1B, lane 2), 323c-NBD2 (lane 3), 562c-NBD1 (lane 4), 64a-RD (lane 5), human CFTR (lane 1), and murine CFTR (lane 8). For each of these CFTR constructs, the mature form was detected, albeit at varying levels. After quantification of independent experiments (n = 10), the rank order of processing efficiency was as follows (WT-CFTR taken as 100%): 323c-NBD2 (129 ± 6%) ≫ murine (86 ± 5%) = human (83 ± 3%) = 562c-NBD1 (80 ± 4%) = 64a-RD (77 ± 4%) > 12b-NBD1+323c-NBD2 (67 ± 5%).
Additional chimeric constructs analyzed by WB in transiently transfected HEK-293 cells included the following: 20b-NBD1 (Thr531–Tyr647), 966c-NBD1 (Ile481–Val546), 158a-NBD2 (Met1220–Asp1409), 189b-NBD2 (Met1220–Tyr1381), 207c-NBD2 (Val1226–Asn1420), 51a-RD (Phe653–Met837), 52a-RD (Thr654–Val693), 71a-RD (Thr654–Leu834), and 85a-RD (Thr654–Val837). Like hCFTR and mCFTR, each of these hmCFTR chimeras was processed normally (data not shown).
When we compared the murine amino acid sequences of the two non-processed chimeric proteins (12b-NBD1 and 114c-NBD2) with their corresponding regions in human CFTR, we found that 12 and 27 amino acid residues diverge between the human and murine sequences of 12b-NBD1 and 114c-NBD2, respectively (see also supplemental Fig. 1). To identify which of these residues could account for the failure of these chimeric proteins to mature, we selected as most probable those residues with greatest sequence divergence between human and murine CFTR based on their values for physicochemical distance (32). Thus, we identified six residues in 12b-NBD1 (E527Q, E528Q, S531T, K536Q, I539T, and K584E) and 12 residues in 114c-NBD2 (T1263I, P1290T, K1302Q, Y1307N, Q1309K, S1311K, R1325K, V1338T, C1344Y, L1367I, D1394G, and E1409D) (see supplemental Fig. 1 and supplemental Table 1, A and B).
Processing of Point Mutants (Murine Residues) into Human CFTR
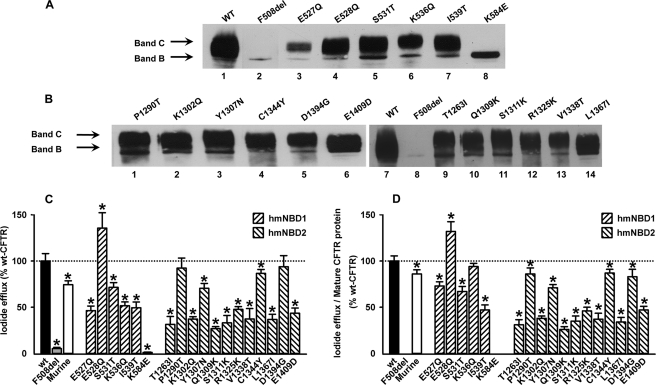
The above six NBD1 and 12 NBD2 point mutants (mCFTR residues) were introduced into full-length human CFTR, the constructs were used to generate stable BHK cell lines, and CFTR protein expression was assessed by WB analysis (Fig. 2).
FIGURE 2.
Biochemical and functional analyses of CFTR variants in NBD1 and NBD2. Western blot analysis of total protein extracts from BHK cells stably expressing the indicated CFTR variants in NBD1 (A) and NBD2 (B) using the anti-CFTR antibody 596 (CF Foundation) is shown. The amounts of total protein applied were 20 μg for WT and F508del and 40 μg for all other CFTR variants. The positions of bands B and C are indicated by arrows. Quantification of these data is provided in Table 1. Data shown are representative of multiple experiments (n = 18). C, functional analysis of CFTR variants in NBD1 and NBD2. Data are the magnitude of CFTR-mediated iodide efflux elicited by the indicated CFTR constructs expressed in BHK cells and cultured at 37 °C. For each construct, the peak value of iodide efflux stimulated by forskolin (10 μm) and genistein (50 μm) in the time courses shown in supplemental Fig. 2 are expressed as a percentage of that of WT-CFTR. For comparison, the magnitude of CFTR-mediated iodide efflux elicited by F508del-CFTR and wild-type murine CFTR are shown. Data are means ± S.E. (n = 6). Asterisks indicate values that are significantly different from WT-CFTR (p < 0.05). D, magnitude of CFTR-mediated iodide efflux normalized to the percentage of mature protein produced by each CFTR construct relative to WT-CFTR (for further information, see Table 1). Error bars correspond to S.E.
With one exception (K584E; Fig. 2A, lane 8), all point mutants derived from 12b-NBD1 were processed (lanes 3–7) similarly to WT-hCFTR (Fig. 2A, lane 1). Like F508del (Fig. 2A, lane 2), K584E (lane 8) only generated immature CFTR protein (band B; 150-kDa). By contrast, all point mutants derived from 114c-NBD2 were processed to the fully glycosylated (band C; 170–190-kDa) form (Fig. 2B, lanes 1–6 and 9–14) like WT-hCFTR (lane 7). These data suggest that all CFTR constructs except K584E are delivered to the cell surface. Results from quantification of the mature form (as percentage of total expressed) for each of the CFTR point mutants studied (Table 1) show that physicochemical distance (supplemental Table 1, A and B) is not inversely correlated with extent of protein processing.
TABLE 1.
Summary information of CFTR point mutants analyzed in present study
| CFTR variants | Clinical dataa | Band C/band Bb (±S.E., n = 5) | Processingc | Normalized processingd | Normalized iodide efflux functione (±S.E., n = 6) | Iodide efflux to processed proteinf |
|---|---|---|---|---|---|---|
| % | % | % | % peak intensity | % | ||
| WT-CFTR | —g | 83 ± 3 | 77 | 100 | 100 ± 8 | — |
| Murine | — | 86 ± 5 | 66 | 86 | 74 ± 4 | 86 ± 4 |
| E527Q | Mild CF | 64 ± 5 | 49 | 63 | 46 ± 4 | 73 ± 4 |
| E528Q | — | 86 ± 5 | 79 | 102 | 135 ± 16 | 132 ± 10 |
| S531T | — | 87 ± 6 | 81 | 105 | 71 ± 5 | 67 ± 5 |
| K536Q | — | 69 ± 3 | 42 | 54 | 51 ± 4 | 94 ± 3 |
| I539T | Revertant | 112 ± 5 | 81 | 105 | 49 ± 6 | 46 ± 5 |
| L581F | — | 118 ± 3 | 83 | 107 | 72 ± 5 | 67 ± 3 |
| L581F/K584E | — | 125 ± 2 | 77 | 100 | 100 ± 12 | 100 ± 8 |
| T1263I | Mild CF | 75 ± 3 | 76 | 98 | 31 ± 8 | 31 ± 5 |
| P1290T | Asymptomatic | 87 ± 3 | 82 | 106 | 92 ± 10 | 86 ± 6 |
| K1302Q | — | 72 ± 3 | 77 | 100 | 37 ± 2 | 37 ± 2 |
| Y1307N | — | 82 ± 2 | 76 | 98 | 70 ± 5 | 71 ± 3 |
| Q1309K | — | 79 ± 4 | 77 | 100 | 26 ± 2 | 26 ± 3 |
| S1311K | — | 73 ± 4 | 72 | 93 | 33 ± 7 | 35 ± 5 |
| R1325K | — | 64 ± 6 | 78 | 101 | 47 ± 2 | 46 ± 4 |
| V1338T | — | 88 ± 2 | 77 | 100 | 37 ± 11 | 37 ± 6 |
| C1344Y | — | 71 ± 4 | 76 | 98 | 86 ± 4 | 87 ± 4 |
| L1367I | — | 72 ± 5 | 80 | 103 | 36 ± 5 | 34 ± 5 |
| D1394G | — | 78 ± 4 | 86 | 111 | 93 ± 12 | 83 ± 8 |
| E1409D | — | 70 ± 3 | 70 | 90 | 43 ± 5 | 47 ± 4 |
a Data from the CFTR mutation database.
b Percentage of processing given by (band C/band B) × 100.
c Percentage of processing given by (band C/(band B + band C) × 100).
d Ratio of the “percent band C” value for each mutant (from previous column) over the same “percentage for WT-CFTR.”
e Iodide efflux expressed as percentage of peak intensity relative to that of WT-CFTR.
f Ratio of “Normalized iodide efflux function” values (from previous column) normalized to “Normalized processing” values (from column 5).
g —, not applicable/no information.
Functional Characterization of Point Mutants at Residues Divergent between Human and Murine CFTR in NBD1 and NBD2
To investigate the function of the above six NBD1 and 12 NBD2 point mutants, we used the iodide efflux technique, which is a convenient method to assay the function of a large population of CFTR Cl− channels in intact cells. When BHK cells expressing WT-CFTR were treated with 10 μm forskolin, a cAMP agonist, and 50 μm genistein, a CFTR potentiator, a robust efflux of I− was obtained (Fig. 2C; see also supplemental Fig. 2).
Like BHK cells expressing F508del-CFTR (Fig. 2C and supplemental Fig. 2A), those expressing K584E-CFTR failed to elicit an efflux of I− when treated with forskolin and genistein (Fig. 2C and supplemental Fig. 2C). The likely explanation for this result is the lack of K584E-CFTR expression at the cell membrane as suggested by the absence of mature CFTR protein (Fig. 2A). All other NBD1 and NBD2 mutants that generated mature CFTR protein (Fig. 2, A and B) elicited an efflux of I−, albeit with varying intensity compared with that of WT-CFTR (Fig. 2C). For most NBD1 and NBD2 mutants, the magnitude of I− efflux was less than that of WT-CFTR; for two mutants in NBD2 (P1290T- and D1394D-CFTR), it was the same; and for one NBD1 mutant (E528Q-CFTR), the magnitude of I− efflux was greater than that of WT-CFTR (Fig. 2C; supplemental Fig. 2, B, D, and F; and Table 1).
To normalize these functional data, the iodide efflux values were divided by the amount of protein at the plasma membrane relative to WT-CFTR, i.e. normalized band C (Table 1 and Fig. 2D). Although for most cases the amount of band C produced correlates well with the extent of CFTR activity, these data show that this is not always the case. For example, compare the data for S531T and K536Q in Fig. 2, C and D.
Taken together and consistent with the WB data (Fig. 2, A and B, and Table 1), our iodide efflux data suggest that most of the NBD1 and NBD2 mutants exhibited channel activity, arguing that they reach the cell surface. These data also show that physicochemical distance is not a good predictive parameter for either the extent of CFTR processing or activity (Table 1 and supplemental Table 1, A and B). By contrast, the agonists had barely any effect on BHK cells expressing K584E-CFTR (Fig. 2C), a result that is consistent with the trafficking defect of this mutant (Fig. 2A).
Characterization of Processing Defect of K584E-CFTR
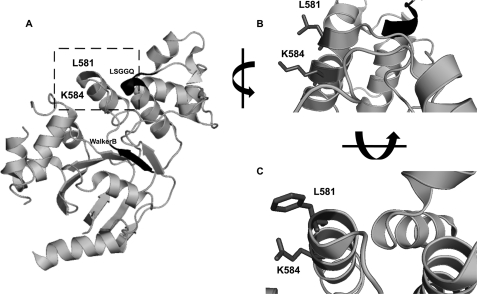
The lack of maturation and of detectable activity for the K584E-CFTR mutant led us to investigate the surrounding environment of Lys584 in the published structure of hNBD1 (12). The rationale behind this approach was to uncover which residues near or interacting with Lys584 (hNBD1) might contribute to the putative conformational change caused by the K584E mutation. We found that in the hNBD1 structure Lys584 interacts with Leu581 (Fig. 3, A and B) and that predictively in K584E the glutamic acid residue at position 584 becomes solvent-exposed (Fig. 3B). Interestingly, in mNBD1, Glu584 interacts with Phe581, preventing exposure of this acidic residue (Fig. 3C).
FIGURE 3.
A, stereo ribbon diagram of human F508del-NBD1 structure (Protein Data Bank code 2BBT) revealing the positions of Leu581 and Lys584 (highlighted by the square). The LSGGQ motif and the Walker B motif are also shown (black ribbon and arrow, respectively). B, close-up view of the same structure as in A (human NBD1) in the vicinity of Lys584 (dark gray) showing the interaction of its side chain with that of Leu581 (dark gray). C, similar close-up view but in the mouse NBD1 structure (Protein Data Bank code 1RXO) showing the interaction between the side chains of the corresponding mouse residues, namely Glu584 and Phe581 (both in dark gray). In B and C, the side chains of residues 581 and 584 are shown (dark gray).
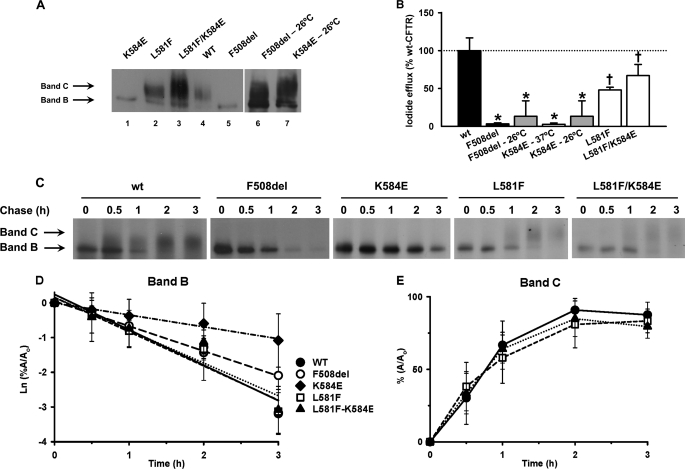
Thus, we postulated that replacing Leu581 by a phenylalanine residue (as in mNBD1) might improve CFTR folding and rescue the processing defect of K584E-CFTR. To test this hypothesis, we introduced L581F in cis with K584E-CFTR by site-directed mutagenesis (L581F/K584E-CFTR) and generated the corresponding stable BHK cell line as well as one expressing L581F-CFTR. We then analyzed these cell lines in parallel with that expressing K584E-CFTR by biochemical (Fig. 4, A and C–E), cell biology (Fig. 5), and functional approaches (Figs. 4B and 6).
FIGURE 4.
Rescue of trafficking defect of K584E-CFTR by L581F-CFTR. A, biochemical analysis by Western blot of total protein extract from BHK cells stably expressing K584E (lane 1), L581F (lane 2), L581F/K584E (lane 3), WT (lane 4), F508del (lane 5), F508del at 26 °C (lane 6), and K584E at 26 °C (lane 7). The amounts of total protein applied were 40 μg for F508del and K584E at 26 °C and 30 μg for the other CFTR variants. The positions of bands B and C are indicated by arrows. B, magnitude of peak iodide efflux elicited by the indicated CFTR constructs expressed in BHK cells and cultured at either 37 or 26 °C. For each construct, the peak value of iodide efflux stimulated by forskolin (10 μm) and genistein (50 μm) in the time courses shown in supplemental Fig. 3 are expressed as a percentage of that of WT-CFTR. Data are means ± S.E. (n = 6). Asterisks indicate values that are significantly different from WT-CFTR incubated at 37 °C (p < 0.05); crosses indicate significant differences between both CFTR variants incubated at 37 °C (p < 0.05). C, turnover and processing of WT-, F508del-, K584E-, L581F-, and L581F/K584E-CFTR determined in BHK cells stably expressing these CFTR variants by pulse-chase experiments followed by immunoprecipitation. Cells were radiolabeled with [35S]methionine for 20 min and then chased for the indicated times (0, 0.5, 1, 2, and 3 h) before lysis with radioimmunoprecipitation assay buffer and immunoprecipitation with the anti-CFTR antibody M3A7 (Chemicon) (see “Experimental Procedures”). After electrophoresis and fluorography, images were analyzed by densitometry. The positions of bands B and C are indicated by arrows. D, rate of turnover of the immature form of CFTR (band B) shown as the natural logarithm of the amount of band B at a given time of the chase (A) relative to the amount at the beginning of the experiment (Ao). E, efficiency of conversion into mature CFTR (band C) shown as the percentage of band C detected at a given time of the chase (A) relative to the amount at the beginning of the experiment (Ao). In D and E, data are mean ± S.E. (n = 4). In D, the lines are the fit of first-order regressions to the data. Error bars in B, D, and E correspond to S.E.
FIGURE 5.
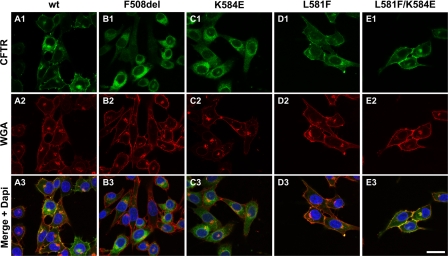
Immunolocalization of CFTR constructs in BHK cells. BHK cells expressing the indicated CFTR variants grown at 37 °C were analyzed by immunofluorescence using the anti-CFTR antibody 570 (green; upper panels) and the anti-wheat germ agglutinin (WGA) coupled with Texas Red (red; middle panels). The lower panels show the merged images with nuclei stained blue with DAPI. Data shown are representative of n = 4 experiments. Bar, 25 μm.
FIGURE 6.
Single channel analysis of CFTR constructs K584E-, L581F-, and L581F/K584E-CFTR. A, representative recordings of CFTR Cl− channels in excised inside-out membrane patches from BHK cells expressing the indicated CFTR variants cultured at 37 °C. ATP (1 mm) and protein kinase A (75 nm) were continuously present in the intracellular solution, voltage was −50 mV, and there was a large Cl− concentration gradient across the membrane patch ([Cl−]int = 147 mm; [Cl−]ext = 10 mm). Dotted lines indicate where channels are closed, and downward deflections correspond to channel openings. For WT-, F508del-, K584E-, and L581F/K584E-CFTR, membrane patches contained a single active channel, but for L581F-CFTR, the membrane patch contained two active channels. B and C, single channel current amplitude (i) and Po of the indicated CFTR variants recorded using the conditions described in A. Data are means ± S.E. (n = 4–5 except F508del-CFTR where n = 10). Asterisks indicate values that are significantly different from those of wild-type CFTR (p < 0.05). In C, Po values for F508del-CFTR are from Ref. 24. Error bars in B and C correspond to S.E.
Fig. 4A demonstrates that both L581F- (lane 2) and L581F/K584E- (lane 3) were detected as their fully glycosylated forms (band C) in contrast to K584E-CFTR (lane 1) for which only immature protein could be detected. We interpret these data to suggest that K584E is rescued by L581F, possibly mediated by the side chain of phenylalanine 581 that most probably fills the empty space previously occupied by the side chain of Lys584 (Fig. 3B) as occurs in the structure of mNBD1 (Fig. 3C) (11). Data from WB also show that the trafficking defect of K584E-CFTR was rescued by incubation of cells at 26 °C (Fig. 4A, lane 7), similarly to F508del-CFTR (lane 6).
The magnitude of I− efflux elicited by L581F-CFTR (Fig. 4B and supplemental Fig. 3A) was smaller than that of WT-CFTR. However, that of L581F/K584E-CFTR was intermediate between L581F- and WT-CFTR (Fig. 4B and supplemental Fig. 3A). Furthermore, when cells expressing K584E-CFTR were incubated at 26 °C for 24 h (Fig. 4B and supplemental Fig. 3B), they generated an efflux of I− similar to that of F508del-CFTR-expressing cells incubated at 26 °C for 24 h; albeit this response of K584E-CFTR was 5-fold smaller than that of WT-CFTR at 37 °C. These data together with those of Fig. 4A (lane 6) indicate that, like F508del-CFTR (Fig. 4A, lane 6) (33), the processing of K584E-CFTR is temperature-sensitive. At 26 °C, some K584E-CFTR protein is delivered to the cell membrane because the trafficking defect of this mutant was (at least partially) reverted at this permissive temperature.
To learn more about the stability of the K584E-, L581F-, and L581F/K584E-CFTR variants, we examined the turnover rate of their immature forms (band B) and their efficiency of conversion into their mature forms (band C). Pulse-chase analyses followed by CFTR immunoprecipitation (Fig. 4C) demonstrate that the turnover rates of band B of both L581F- (Fig. 4D, open squares) and L581F/K584E-CFTR (Fig. 4D, filled triangles) were the same as those of WT-CFTR (Fig. 4D, filled circles). However, the turnover rate of K584E-CFTR (Fig. 4D, filled diamonds) was significantly (p < 0.05) reduced compared with those of L581F-, L581F/K584E-, WT-, and F508del-CFTR (Fig. 4D, open squares, filled triangles, filled circles, and open circles, respectively). Consistent with the data in Fig. 4D, the processing efficiencies of L581F- (Fig. 4E, open squares) and L581F/K584E-CFTR (Fig. 4E, filled triangles) were not significantly different from those of WT-CFTR (Fig. 4D, filled circles). Taken together, these data suggest that the immature form of K584E-CFTR is significantly stabilized compared with those of L581F-, L581F/K584E-, WT-, and even F508del-CFTR.
Localization of K584E-, L581F-, and L581F/K584E-CFTR by Immunofluorescence
Our biochemical studies demonstrated that K584E-CFTR is only detected in its immature form, suggesting that it is misfolded and retained in the ER. To test this possibility and learn whether the biochemically detected rescue of K584E-CFTR by L581F-CFTR (mature form) indeed corresponds to protein present at the cell surface, we used immunofluorescence and confocal microscopy.
Immunodetection of CFTR showed that the K584E-CFTR mutant (Fig. 5, C1) was predominately located intracellularly like F508del-CFTR (B1). By contrast, L581F- (D1), L581F/K584E- (E1), and WT-CFTR (A1) were mostly found at the plasma membrane co-localizing with wheat germ agglutinin (Fig. 5, D3, E3, and A3). When applied without permeabilization, wheat germ agglutinin is a marker of the cell surface that recognizes sialic acid and N-acetylglucosaminyl sugars highly prevalent at the plasma membrane (34). We interpret these data to suggest that L581F rescues the cell surface expression of K584E, confirming our biochemical and functional data, which argue that L581F-CFTR is a revertant mutant for K584E-CFTR.
Single Channel Behavior of Processing Mutant K584E-CFTR
In previous research, we demonstrated that revertant (e.g. G550E-CFTR (24)) and solubilizing mutations (e.g. F429S/F494N/Q637R (13)) rescue defects in CFTR channel gating in addition to promoting the cell surface expression of F508del-CFTR. We therefore speculated that L581F-CFTR might augment the single channel activity of K584E-CFTR. To test this possibility, we used the excised inside-out configuration of the patch clamp technique.
Fig. 6A demonstrates that addition of ATP (1 mm) and protein kinase A (75 nm) to the intracellular solution bathing excised inside-out membrane patches from cells expressing K584E-, L581F-, and L581F/K584E-CFTR cultured at 37 °C activated single channels with properties and regulation characteristic of wild-type CFTR. By contrast, culture of cells expressing F508del-CFTR at reduced temperature is required to observe channel activity, and these Cl− channels possess properties distinct from those of wild-type CFTR (24, 33) (Fig. 6A). To quantify single channel activity, we measured single channel current amplitude (i) and Po. Consistent with the behavior of other CFTR variants containing point mutations in the NBDs (2), K584E-, L581F-, and L581F/K584E-CFTR had values of i similar to that of WT-CFTR (Fig. 6B).
The pattern of channel gating of WT-CFTR is characterized by bursts of channel openings interrupted by brief flickery closures and separated by longer closures between bursts (Fig. 6A). The F508del mutation perturbs the rate of channel opening with the result that the closed time interval separating channel openings is increased greatly and Po is attenuated markedly (35) (Fig. 6, A and C). Visual inspection of single channel records suggested that the gating behavior of the CFTR variants K584E-, L581F-, and L581F/K584E-CFTR all resembled that of WT-CFTR (Fig. 6A). Consistent with this idea, the Po of L581F/K584E-CFTR was the same as that of WT-CFTR, whereas those of K584E- and L581F-CFTR were slightly, albeit significantly (p < 0.05), reduced (Fig. 6C). Thus, K584E-CFTR profoundly disrupts CFTR processing but only modestly affects the Cl− channel function of CFTR. Of note, these effects of K584E-CFTR were rescued by the revertant mutation L581F-CFTR.
DISCUSSION
In this study, we used human-murine CFTR chimeras and point mutants to explore the structural basis of differences in the function of human and murine CFTR (18, 19).
Maturation of Chimeras
With two exceptions, our data demonstrate that replacement of different protein regions in the NBDs and RD of hCFTR with the equivalent regions of mCFTR were without effect on CFTR processing. These data indicate that interchanging sections of human domains with the equivalent murine regions had no apparent consequence on the folding of the CFTR protein. However, two CFTR chimeras failed to mature: 12b-NBD1 and 114c-NBD2 containing part of NBD1 (Lys518–Val586) and NBD2 (Met1260–Arg1412), respectively. The hmCFTR chimera 12b-NBD1 contains a conserved region of mNBD1 that includes the LSGGQ and Walker B motifs, whereas the hmCFTR chimera 114c-NBD2 includes the LSHGH and Walker B motifs of mNBD2. Because these two regions of CFTR are very much conserved between human and murine CFTR, differences in amino acid residues can highlight how processing is abolished in these two mutants. Of note, the processing of the NBD1+NBD2 chimera (12b-NBD1+323c-NBD2) was less efficient (67 ± 5%) than either human or murine CFTR. This result implies that the two murine NBDs acquire better folding when they are together in the murine rather than the human background and emphasizes the relevance of interdomain associations for folding of full-length CFTR.
It is interesting that the hmCFTR chimera 12b-NBD1 failed to be processed because the hmCFTR chimera 966c-NBD1 containing a wider portion of NBD1 at the N terminus but shorter at the C terminus (Ile481–Val546) was correctly processed (data not shown). Thus, it would seem likely that the region present in 12b-NBD1 and absent in 966c-NBD1 (Val546–Val586) contains the residue(s) causing the maturation defect of 12b-NBD1. Indeed, the point mutant that we found to cause a failure in CFTR maturation (K584E-CFTR) lies exactly in this region. Remarkably, the 12b-NBD1+323c-NBD2 chimera (Lys518–Val586; Met1260–Ser1419), which contains the same murine portion of NBD1 as 12b-NBD1, was found to be correctly processed. This result suggests that the presence of this murine NBD2 region (Met1260–Ser1419) is able to mask the structural cue of 12b-NBD1 that is recognized by the ERQC, leading to its maturation defect. Alternatively or in addition, this NBD2 region may also increase the folding rate and/or the overall stability of the 12b-NBD1 chimera.
The hmCFTR chimera 114c-NBD2 also failed to be processed. Remarkably, another NBD2 chimera 323c-NBD2 (Met1260–Ser1419) with the same N-terminal boundary and just seven additional residues at its C terminus (1413–1419) was found to be fully processed. When comparing this seven-amino acid extension of 323c-NBD2, which is absent in 114c-NBD2, only one residue (at position 1419) was found to diverge between murine (Ser1419) and human (Asn1419) CFTR. It thus becomes apparent that this residue alone could rescue the trafficking defect of the 114c-NBD2 chimera. Interestingly, 323c-NBD2 was the hmCFTR chimera, which demonstrated higher processing efficiency than either human or murine CFTR.
As for the RD hmCFTR chimeras analyzed (data for 64a-RD shown here and for 52a-RD, 71a-RD, and 85a-RD not shown), we did not find any effects on CFTR processing. Consistent with these results, transfer of the RD of mCFTR to hCFTR was without effect on CFTR channel gating (20). Taken together, these data likely reflect the high degree of structural plasticity of the RD, which has been proposed to be intrinsically disordered and highly dynamic following phosphorylation at multiple sites (36).
Our present and previous (20) data suggest that the structural requirements for protein processing and channel function are distinct. Previously, we demonstrated that the hmCFTR chimeras 562c-NBD1 (Leu433–Val586) and 323c-NBD2 (Met1260–Ser1419) exhibited patterns of channel gating with only subtle differences from those of WT-hCFTR (20). Only by simultaneous transfer of the mCFTR sequences Leu433–Val586 and Met1260–Ser1419 to hCFTR did the latter Cl− channel acquire the dramatically prolonged openings of the subconductance state of murine CFTR (20). These data argue that optimal channel gating requires precise interactions between NBD1 and NBD2, a fundamental tenet of the ATP-driven NBD dimerization model of CFTR channel gating (37). Our previous data (20) also raise the interesting possibility that channel gating, albeit suboptimal, is possible when NBDs from two CFTR homologues are mixed. In contrast, increased protein processing levels can result from improved trafficking efficiency, which is not necessarily coupled to improved folding. This is the case when ER retention motifs are masked, such as by removal of the arginine-framed tripeptides, as we showed before for 4RK-F508del-CFTR (24).
Processing and Activity of Point Mutants in NBD1 and NBD2
Biochemical studies of CFTR variants identified by physicochemical distance analysis of residues in 12b-NBD1 revealed that the mutation K584E disrupts the maturation of CFTR protein. By contrast, a similar analysis of 114c-NBD2 failed to identify CFTR variants that disrupt CFTR processing; mutation of the 12 residues with highest physicochemical distance between human and murine CFTR was without effect on CFTR maturation. This suggests that one or more of the remaining 15 conserved sequence variations between human and murine CFTR in 114c-NBD2 might account for the defective processing of this CFTR chimera. Alternatively, multiple sequence changes might be required to account for the defective processing of this chimera. Nevertheless, as discussed above, N1419S alone likely rescues the processing defect of 114c-NBD2. This argues that the interacting residues of N1419S cause the trafficking defect of the hmCFTR chimera 114c-NBD2.
Our data also suggest that mutations in NBD1 have a more severe effect on CFTR processing than those in NBD2. Previous work supports this idea that NDB2 might play a less important role in the folding of full-length CFTR. For example, CFTR constructs lacking NBD2 exit the ER (38), and deletion of NBD1, but not NBD2, prevented CFTR trafficking to the cell surface (39, 40). Furthermore, CF-associated mutations are more prevalent in NBD1 than NBD2 (CFTR mutation database). Consistent with this idea, Gregory et al. (42) showed that the increased susceptibility of NBD1 mutations to misprocessing might be a result of the surrounding domains or the greater susceptibility of NBD1 mutants to detection by the ERQC.
Among the 20 point mutants described in this study (the above 18 point mutants plus L581F and L581F/K584E), we found four that are listed in the CFTR mutation database (CFTR mutation database), namely E527Q, T1263I, and P1290T, which are described in association with mild CF, and I539T, which is described as an F508del-revertant mutation (Table 1). Our own data suggest that P1290T (associated with asymptomatic CF) is worthy of further study because it does not affect CFTR processing or function. This raises the interesting possibility that P1290T might be a sequence variation (polymorphism) like V562I (24).
Regarding the CFTR channel activity (as determined by the iodide efflux technique) of these point mutants, there were some that, although correctly processed, exhibited significantly reduced activity. The most striking differences for the NBD1 mutants (Table 1 and Fig. 2, compare C with D) were (Iodide efflux to processed protein (%) far right column) E527Q (64/46), I539T (112/49), and L581F (118/72), whereas for the NBD2 mutants, they were T1263I (75/31), K1302Q (72/37), Q1309K (79/26), S1311K (73/33), V1338T (88/37), L1367I (72/36), and E1409D (70/43). Curiously, the point mutant with the highest discrepancy was I539T, which rescues the trafficking defect of F508del-CFTR (43). These data suggest that this revertant mutation greatly improves trafficking but slows channel gating, which can be interpreted as evidence for some structural divergence from the native conformation of the WT channel. By contrast, the revertant mutation G550E, which enhances both CFTR trafficking and gating, was proposed to correct the defective folding of F508del-CFTR (24).
Interestingly, by quantifying the maturation or function of these point mutants (Table 1), we found no inverse correlation between physicochemical distance values and extent of processing or levels of CFTR activity. This argues that the context of the amino acid change overrides its nature. These data also emphasize the difficulty of making predictions about the functional consequences of missense mutations (and even more of patient phenotypes based on genotypes), thus demonstrating the necessity of experimental analyses.
Characterization of K584E and Rescuing by Leu581
Like F508del-CFTR (33), the trafficking defect of K584E-CFTR is temperature-sensitive. However, in contrast to F508del-CFTR, active K584E-CFTR Cl− channels could be detected in cells cultured at 37 °C using the single channel patch clamp, although this mutant could not be detected at the cell surface by immunocytochemistry or in its processed form by WB. To investigate this discrepancy, we used our processing (Table 1) and single channel data (Fig. 6) to calculate predicted macroscopic currents for K584E-, L581F-, and L581F/K584E-CFTR and compared the values obtained with the magnitude of iodide efflux generated by these different CFTR constructs (Fig. 2C). Macroscopic CFTR Cl− current or CFTR-mediated iodide efflux (ICFTR) is determined from the product of the number of CFTR Cl− channels in the cell membrane (N), the current amplitude (i) of an individual CFTR Cl− channel, and the probability (Po) that a single CFTR Cl− channel is open.
If we set each of these variables to 100% for wild-type CFTR, we can compare the CFTR-mediated iodide efflux generated by wild-type and mutant CFTRs. Table 2 presents values of each variable, the predicted value of ICFTR determined by calculating N × i × Po and the observed value of ICFTR. Differences between observed and predicted values might principally reflect errors resulting from the calculation of N based on the amount of CFTR protein as band C and on values of Po determined from excised inside-out membrane patches. Nevertheless, the predicted and observed values agree well. Thus, these data provide a molecular explanation for the quantitative decrease in CFTR-mediated iodide efflux caused by K584E and for the rescuing action of L581F.
TABLE 2.
Comparison of predicted and measured CFTR-mediated iodide efflux for K584E-, L581F-, and L581F/K584E-CFTR
N, the number of Cl− channels in the cell membrane; i, single-channel current; N × i × Po, the predicted CFTR-mediated iodide efflux based on CFTR processing and single channel data; ICFTR, measured CFTR-mediated iodide efflux. To estimate N relative to wild-type CFTR, we used the percent processing data from Table 1; i and Po data were from Fig. 6, B and C, with data for wild-type CFTR assigned values of 100%; ICFTR data were from Fig. 2C. The predicted value for F508del-CFTR concurs with our previous data (44) and values calculated by other investigators (e.g. Haws et al. (48), 2%; Wang et al. (41), 0.4%).
| CFTR | N | i | Po | N × i × Po | ICFTR |
|---|---|---|---|---|---|
| % | % | % | % | % | |
| Wild type | 100 | 100 | 100 | 100 | 100 |
| F508del | 5 | 104 | 12 | 1 | 0 |
| K584E | 1 | 90 | 73 | 1 | 0 |
| L581F | 107 | 88 | 81 | 75 | 72 |
| L581F/K584E | 100 | 91 | 103 | 93 | 100 |
The different effects of K584E on CFTR processing and Cl− channel function are reminiscent of A455E and P574H, two CF mutations associated with a milder clinical phenotype (44). Although production of the mature forms of A455E and P574H was reduced and very much delayed, A455E had channel activity similar to and P574H had channel activity greater than that of WT-CFTR (44). Conversely, the NBD1 CF mutant G551D is processed correctly but forms a Cl− channel with a profound gating defect that is not regulated by intracellular ATP (28, 45).
A further explanation for the discrepancy between the processing (WB) and single channel patch clamp data of K584E-CFTR is that this trafficking mutant might escape the ER via a non-conventional route (46). But this explanation has to be discarded because, by immunofluorescence, K584E-CFTR could not be detected at the plasma membrane. It thus seems likely that only very little (below biochemical/immunofluorescence detection levels) of the K584E-CFTR reaches the cell surface, but once correctly inserted, this CFTR variant has a significant capacity to transport Cl−.
To understand the structural basis by which K584E disrupts the processing and function of CFTR, we used a model of full-length CFTR.5 This trafficking mutant lies on the highly conserved region of NBD1 where Glu584 is solvent-exposed and interacts with Leu581. Accordingly, we changed the interacting residue in human CFTR (Leu581) to the corresponding residue in murine CFTR (Phe581) and found that it rescued the trafficking defect of K584E. In the crystal structure of NBD1 (11, 12), it is likely that K584E disrupts the interaction of Lys584 with neighboring residues and, hence, the folding of NBD1. The introduction of the L581F mutation likely restores the processing defect of K584E possibly mediated by the side chain of Phe581 that fills the empty space left by the removal of Lys584. Moreover, confirmation that both L581F and L581F/K584E are present at the cell surface was provided by functional studies (both iodide efflux and patch clamp).
A further dimension of the present work is to use hmCFTR chimeras and point mutants in the NBDs to elucidate the protein motifs responsible for the differential efficiencies with which some small molecules modulate human and murine CFTR Cl− channels. For example, Lansdell et al. (18) demonstrated that the inorganic phosphate analogue PPi potentiates robustly the gating behavior of hCFTR but is without effect on mCFTR. Using hmCFTR chimeras, we previously localized the binding site with which PPi interacts to enhance CFTR channel gating to NBD2 (20). Moreover, de Jonge et al. (47) tested a battery of CFTR correctors and potentiators for their effects on F508del-murine CFTR and demonstrated that all CFTR correctors tested rescued the cell surface expression of F508del-murine CFTR, whereas the majority of CFTR potentiators were without effect on F508del-mCFTR channel gating. These data argue that hmCFTR chimeras and point mutants in the NBDs are a valuable resource to elucidate the mode of action of CFTR potentiators.
With this study of hmCFTR chimeras, we gained further insight into the structural and functional properties of CFTR domains. We identified an NBD1 trafficking mutant (K584E) that, despite being inefficiently processed, exhibits some activity as a regulated Cl− channel. Moreover, by using the available structure of NBD1, we predicted that substitution of another NBD1 residue (L581F) should restore the processing defect of K584E. By experimentally confirming this prediction, we demonstrate here that residues 581 and 584 are close to each other in the H6 α-helix. Finally, our data show how hmCFTR chimeras can be used to validate structural models of full-length CFTR.
Supplementary Material
Acknowledgments
We thank Prof. R. C. Ford (University of Manchester) for valuable discussions of CFTR structure and our laboratory colleagues for essential assistance with data acquisition, analysis, and interpretation. We also thank the CF Foundation (United States) for the anti-CFTR antibodies 596 and 570 used in this study.
This work was supported in part by Fundação para a Ciência ea Tecnologia (FCT), Portugal Grants PIC/IC/83103/2007; and POCTI/MGI/47382/2002 and pluriannual funding of BioFIG from FCT, Portugal (Fonds Européen de Développement Régional-European Union) (to M. D. A.) and by Biotechnology and Biological Sciences Research Council Grant BB/C517517/1 and the CF Trust (to D. N. S. and A. C. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3 and Tables 1 and 2.
R. Ford, personal communication.
- CF
- cystic fibrosis
- CFTR
- cystic fibrosis transmembrane conductance regulator
- NBD
- nucleotide-binding domain
- RD
- regulatory domain
- WB
- Western blot
- ABC
- ATP-binding cassette
- ER
- endoplasmic reticulum
- ERQC
- ER quality control
- m
- murine
- h
- human
- BHK
- baby hamster kidney
- TES
- N-tris[hydroxymethyl]methyl-2-aminoethanesulfonic acid
- Po
- open probability.
REFERENCES
- 1.Welsh M., Ramsey B., Accurso F., Cutting G. (2001) in The Metabolic and Molecular Bases of Inherited Disease (Scriver C., Beaudet A., Sly W. eds), 8th Ed., pp. 5121–5188, McGraw Hill, New York [Google Scholar]
- 2.Sheppard D. N., Welsh M. J. (1999) Physiol. Rev. 79, S23–S45 [DOI] [PubMed] [Google Scholar]
- 3.Gadsby D. C., Vergani P., Csanády L. (2006) Nature 440, 477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riordan J. R., Rommens J. M., Kerem B., Alon N., Rozmahel R., Grzelczak Z., Zielenski J., Lok S., Plavsic N., Chou J. L. (1989) Science 245, 1066–1073 [DOI] [PubMed] [Google Scholar]
- 5.Lukacs G. L., Chang X. B., Bear C., Kartner N., Mohamed A., Riordan J. R., Grinstein S. (1993) J. Biol. Chem. 268, 21592–21598 [PubMed] [Google Scholar]
- 6.Ward C. L., Kopito R. R. (1994) J. Biol. Chem. 269, 25710–25718 [PubMed] [Google Scholar]
- 7.Varga K., Jurkuvenaite A., Wakefield J., Hong J. S., Guimbellot J. S., Venglarik C. J., Niraj A., Mazur M., Sorscher E. J., Collawn J. F., Bebök Z. (2004) J. Biol. Chem. 279, 22578–22584 [DOI] [PubMed] [Google Scholar]
- 8.Amaral M. D. (2004) J. Mol. Neurosci. 23, 41–48 [DOI] [PubMed] [Google Scholar]
- 9.Lukacs G. L., Mohamed A., Kartner N., Chang X. B., Riordan J. R., Grinstein S. (1994) EMBO J. 13, 6076–6086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qu B. H., Thomas P. J. (1996) J. Biol. Chem. 271, 7261–7264 [DOI] [PubMed] [Google Scholar]
- 11.Lewis H. A., Buchanan S. G., Burley S. K., Conners K., Dickey M., Dorwart M., Fowler R., Gao X., Guggino W. B., Hendrickson W. A., Hunt J. F., Kearins M. C., Lorimer D., Maloney P. C., Post K. W., Rajashankar K. R., Rutter M. E., Sauder J. M., Shriver S., Thibodeau P. H., Thomas P. J., Zhang M., Zhao X., Emtage S. (2004) EMBO J. 23, 282–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis H. A., Zhao X., Wang C., Sauder J. M., Rooney I., Noland B. W., Lorimer D., Kearins M. C., Conners K., Condon B., Maloney P. C., Guggino W. B., Hunt J. F., Emtage S. (2005) J. Biol. Chem. 280, 1346–1353 [DOI] [PubMed] [Google Scholar]
- 13.Pissarra L. S., Farinha C. M., Xu Z., Schmidt A., Thibodeau P. H., Cai Z., Thomas P. J., Sheppard D. N., Amaral M. D. (2008) Chem. Biol. 15, 62–69 [DOI] [PubMed] [Google Scholar]
- 14.Hollenstein K., Frei D. C., Locher K. P. (2007) Nature 446, 213–216 [DOI] [PubMed] [Google Scholar]
- 15.Serohijos A. W., Hegedus T., Aleksandrov A. A., He L., Cui L., Dokholyan N. V., Riordan J. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3256–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mornon J. P., Lehn P., Callebaut I. (2008) Cell. Mol. Life Sci. 65, 2594–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J. M., Cutler C., Jacques C., Boeuf G., Denamur E., Lecointre G., Mercier B., Cramb G., Férec C. (2001) Mol. Biol. Evol. 18, 1771–1788 [DOI] [PubMed] [Google Scholar]
- 18.Lansdell K. A., Delaney S. J., Lunn D. P., Thomson S. A., Sheppard D. N., Wainwright B. J. (1998) J. Physiol. 508, 379–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lansdell K. A., Kidd J. F., Delaney S. J., Wainwright B. J., Sheppard D. N. (1998) J. Physiol. 512, 751–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scott-Ward T. S., Cai Z., Dawson E. S., Doherty A., Da Paula A. C., Davidson H., Porteous D. J., Wainwright B. J., Amaral M. D., Sheppard D. N., Boyd A. C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 16365–16370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ostedgaard L. S., Rogers C. S., Dong Q., Randak C. O., Vermeer D. W., Rokhlina T., Karp P. H., Welsh M. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 15370–15375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenberg M. F., Kamis A. B., Aleksandrov L. A., Ford R. C., Riordan J. R. (2004) J. Biol. Chem. 279, 39051–39057 [DOI] [PubMed] [Google Scholar]
- 23.Amaral M. D. (2010) Curr. Drug Targets, in press [DOI] [PubMed] [Google Scholar]
- 24.Roxo-Rosa M., Xu Z., Schmidt A., Neto M., Cai Z., Soares C. M., Sheppard D. N., Amaral M. D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17891–17896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farinha C. M., Mendes F., Roxo-Rosa M., Penque D., Amaral M. D. (2004) Mol. Cell. Probes 18, 235–242 [DOI] [PubMed] [Google Scholar]
- 26.Farinha C. M., Penque D., Roxo-Rosa M., Lukacs G., Dormer R., McPherson M., Pereira M., Bot A. G., Jorna H., Willemsen R., Dejonge H., Heda G. D., Marino C. R., Fanen P., Hinzpeter A., Lipecka J., Fritsch J., Gentzsch M., Edelman A., Amaral M. D. (2004) J. Cyst. Fibros. 3, Suppl. 2, 73–77 [DOI] [PubMed] [Google Scholar]
- 27.Mendes F., Wakefield J., Bachhuber T., Barroso M., Bebok Z., Penque D., Kunzelmann K., Amaral M. D. (2005) Cell. Physiol. Biochem. 16, 281–290 [DOI] [PubMed] [Google Scholar]
- 28.Cai Z., Taddei A., Sheppard D. N. (2006) J. Biol. Chem. 281, 1970–1977 [DOI] [PubMed] [Google Scholar]
- 29.Kleinbaum D. G., Kupper L. L., Muller K. E. (2008) Applied Regression Analysis and Other Multivariate Methods, 4th Ed., pp. 114–138, Duxbury Press, Pacific Grove, CA [Google Scholar]
- 30.Helliwell P. S., Jackson S. (1994) Ann. Rheum. Dis. 53, 726–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng S. H., Gregory R. J., Marshall J., Paul S., Souza D. W., White G. A., O'Riordan C. R., Smith A. E. (1990) Cell 63, 827–834 [DOI] [PubMed] [Google Scholar]
- 32.Grantham R. (1974) Science 185, 862–864 [DOI] [PubMed] [Google Scholar]
- 33.Denning G. M., Anderson M. P., Amara J. F., Marshall J., Smith A. E., Welsh M. J. (1992) Nature 358, 761–764 [DOI] [PubMed] [Google Scholar]
- 34.Glozman R., Okiyoneda T., Mulvihill C. M., Rini J. M., Barriere H., Lukacs G. L. (2009) J. Cell Biol. 184, 847–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dalemans W., Barbry P., Champigny G., Jallat S., Dott K., Dreyer D., Crystal R. G., Pavirani A., Lecocq J. P., Lazdunski M. (1991) Nature 354, 526–528 [DOI] [PubMed] [Google Scholar]
- 36.Baker J. M., Hudson R. P., Kanelis V., Choy W. Y., Thibodeau P. H., Thomas P. J., Forman-Kay J. D. (2007) Nat. Struct. Mol. Biol. 14, 738–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vergani P., Lockless S. W., Nairn A. C., Gadsby D. C. (2005) Nature 433, 876–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cui L., Aleksandrov L., Chang X. B., Hou Y. X., He L., Hegedus T., Gentzsch M., Aleksandrov A., Balch W. E., Riordan J. R. (2007) J. Mol. Biol. 365, 981–994 [DOI] [PubMed] [Google Scholar]
- 39.Pollet J. F., Van Geffel J., Van Stevens E., Van Geffel R., Beauwens R., Bollen A., Jacobs P. (2000) Biochim. Biophys. Acta 1500, 59–69 [DOI] [PubMed] [Google Scholar]
- 40.Rich D. P., Gregory R. J., Cheng S. H., Smith A. E., Welsh M. J. (1993) Receptors Channels 1, 221–232 [PubMed] [Google Scholar]
- 41.Wang F., Zeltwanger S., Hu S., Hwang T. C. (2000) J. Physiol. 524, 637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gregory R. J., Rich D. P., Cheng S. H., Souza D. W., Paul S., Manavalan P., Anderson M. P., Welsh M. J., Smith A. E. (1991) Mol. Cell. Biol. 11, 3886–3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeCarvalho A. C., Gansheroff L. J., Teem J. L. (2002) J. Biol. Chem. 277, 35896–35905 [DOI] [PubMed] [Google Scholar]
- 44.Sheppard D. N., Ostedgaard L. S., Winter M. C., Welsh M. J. (1995) EMBO J. 14, 876–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bompadre S. G., Sohma Y., Li M., Hwang T. C. (2007) J. Gen. Physiol. 129, 285–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bannykh S. I., Bannykh G. I., Fish K. N., Moyer B. D., Riordan J. R., Balch W. E. (2000) Traffic 1, 852–870 [DOI] [PubMed] [Google Scholar]
- 47.de Jonge H., Wilke M., Bot A., Sheppard D. N. (2009) Pediatr. Pulmonol. Suppl. 32, 291–292 [Google Scholar]
- 48.Haws C. M., Nepomuceno I. B., Krouse M. E., Wakelee H., Law T., Xia Y., Nguyen H., Wine J. J. (1996) Am. J. Physiol. Cell Physiol. 270, C1544–C1555 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.