Abstract
Telomeres are terminal repetitive DNA sequences whose stability requires the coordinated actions of telomere-binding proteins and the DNA replication and repair machinery. Recently, we demonstrated that the DNA replication and repair protein Flap endonuclease 1 (FEN1) is required for replication of lagging strand telomeres. Here, we demonstrate for the first time that FEN1 is required for efficient re-initiation of stalled replication forks. At the telomere, we find that FEN1 depletion results in replicative stress as evidenced by fragile telomere expression and sister telomere loss. We show that FEN1 participation in Okazaki fragment processing is not required for efficient telomere replication. Instead we find that FEN1 gap endonuclease activity, which processes DNA structures resembling stalled replication forks, and the FEN1 interaction with the RecQ helicases are vital for telomere stability. Finally, we find that FEN1 depletion neither impacts cell cycle progression nor in vitro DNA replication through non-telomeric sequences. Our finding that FEN1 is required for efficient replication fork re-initiation strongly suggests that the fragile telomere expression and sister telomere losses observed upon FEN1 depletion are the direct result of replication fork collapse. Together, these findings suggest that other nucleases compensate for FEN1 loss throughout the genome during DNA replication but fail to do so at the telomere. We propose that FEN1 maintains stable telomeres by facilitating replication through the G-rich lagging strand telomere, thereby ensuring high fidelity telomere replication.
Keywords: Aging, Cancer Tumor Promoter, DNA Repair, DNA Replication, Telomere
Introduction
High fidelity DNA replication is critical for genome stability and continued cellular proliferation. Given the importance of high fidelity DNA replication to genomic stability, it is not surprising that numerous redundant mechanisms of DNA replication exist. Inherited syndromes in which DNA replication/repair proteins are mutated or lost but overall DNA replication continues relatively unabated (1–3) best illustrate the compensatory nature of these mechanisms. However, in some cases this compensation is incomplete, and thus, patients with these mutations manifest replication defects and genomic instability (2).
Deficiencies in various DNA replication/repair mechanisms become particularly detrimental in highly repetitive DNA sequences that present unique challenges to the DNA replication machinery (4, 5). For example, triplet repeats can lead to replication fork slippage, resulting in deleterious expansions and deletions (6). Similarly, replication fork pausing and stalling occur within telomeric repeats (7–10), and telomeres were recently identified as fragile sites (11, 12). Because fragile sites are thought to arise in response to replication stress, these data support the hypothesis that telomeric DNA presents a challenging template for the DNA replication machinery that requires the actions of specialized replication complexes, including a replication fork re-initiation complex (4, 13, 14). Recent work has shown that telomeres are highly sensitive to the loss of DNA replication/repair proteins shown to localize to stalled replication forks, including the Werner (WRN)2 and Flap endonuclease 1 (FEN1) proteins (15). Indeed, cells from WRN patients display overt telomere dysfunction, whereas only minor defects in genomic replication are observed (2, 16, 17), suggesting that other proteins compensate for WRN throughout the genome but are insufficient at the telomere.
DNA replication mechanisms at the telomere are coordinated by the six-protein Shelterin complex (including TRF1, TRF2, TIN2, POT1, RAP1, and TPP1) (4, 5, 18). For example, TRF2 interacts with and modulates the activities of numerous DNA replication and repair proteins (18). These interactions include TRF2 binding to the WRN and BLM helicases, which stimulates their activity in vitro, suggesting that TRF2 recruits them to replicate or repair telomeric DNA (19). In Schizosaccharomyces pombe, the TRF1/2 homolog Taz1 is essential for DNA replication through the telomeres (20). Upon Taz1 deletion, replication forks stall within telomeric repeats, and telomeres are rapidly lost (20). TRF1 plays a similar role in mammalian cells (11, 12). After deletion of TRF1, stalled replication forks accumulate within the telomeric repeats, resulting in a replication stress response characterized by an ATR (ataxia telangiectasia mutated (ATM)- and Rad3-related)-dependent DNA damage response and expression of fragile sites within telomeric DNA (11, 12). Together, these data underscore the importance of the coordinated actions of the Shelterin components and the DNA replication and repair machinery to efficiently complete telomere replication.
FEN1 is a structure-specific endonuclease that plays an important role in DNA metabolism. FEN1 participates in Okazaki fragment processing during lagging strand DNA replication (21) and is important for several DNA repair processes (22, 23). FEN1 co-localizes to stalled replication forks where it interacts with the RecQ helicase, WRN, and is postulated to re-initiate stalled DNA replication forks (15, 24). Recently, we demonstrated that FEN1 is vital for telomere stability (25). Indeed, FEN1 depletion in telomerase-deficient cells leads to a DNA damage response at telomeres and telomere dysfunction characterized by loss of lagging strand-replicated sister telomeres (STLs) (25, 26). Furthermore, genetic rescue experiments demonstrate that the nuclease activity and the C-terminal WRN-interacting domain of FEN1 are important for telomere stability (25).
The above findings prompted us to investigate how FEN1 contributes to telomere stability. Here, for the first time we demonstrate that FEN1 promotes efficient re-initiation of stalled replication forks. The C-terminal domain of FEN1 and its gap endonuclease (GEN) activity are critical for its ability to re-initiate stalled replication forks. However, FEN1 depletion does not affect progression through S phase or SV40 large T antigen-dependent in vitro DNA replication of non-repetitive sequences. Instead, FEN1 depletion leads to replicative stress within telomeric sequences as evidenced by expression of fragile sites. Finally, we demonstrate that the PCNA-interacting domain of FEN1 is dispensable for its telomere function and that the GEN activity is critical for its ability to prevent STLs. We propose that FEN1 maintains stable telomeres through efficient re-initiation of stalled replication forks that occur in the G-rich telomere, ensuring high fidelity telomere replication.
EXPERIMENTAL PROCEDURES
Cell Culture
All cells were grown as reported (25–28). Briefly, cells were grown at 37 °C in 5% CO2. HeLa and 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% heat-inactivated fetal calf serum (FCS) and 1% penicillin/streptomycin. BJ fibroblasts were cultured in Dulbecco's modified Eagle's medium with 15% medium 199 (Sigma), 15% heat-inactivated FCS, and 1% penicillin/streptomycin.
Virus Production and Infection
Lentiviral production and cell infections were carried out as described (25, 26, 29). Briefly, 293T cells were transfected with TransIT-LT1 (Mirius, Madison, WI). Virus was harvested 48 h post-transfection, and infections were carried out overnight in the presence of 10 μg/ml of protamine sulfate. After infection, transduced cells were selected with 1 μg/ml puromycin.
For adenovirus production, FEN1 cDNAs were cloned into the pShuttle vector (Stratagene, La Jolla, CA) at the EcoRV site. The human WT (hWT), D181A (DA), and ΔC cDNAs were previously described (25); the ΔP cDNA was previously described (30); the ΔPΔC cDNA was constructed using a forward primer complementary to the FLAG epitope, 5′-GGTACCATGGACTACAAAGACCATGACGG-3′, and the reverse primer 5′-CTCGAGTTATTAGGTGCTGCCTTGGCGGCTCTTAC-3′ and was cloned into the pShuttle plasmid; the murine WT (mWT) and mutant E160D (mED) cDNAs were previously described (24). After subcloning, the FEN1 cDNAs were recombined into the pAdEasy-1 plasmid (Stratagene), and the resultant DNA was transfected into HEK293 cells to produce infectious adenovirus. Adenovirus production and concentration were carried out according to the manufacturer's protocol using the AdEasy XL Adenoviral Vector System (Stratagene). Adenovirus was titered before use with the AdEasy Viral Titer kit (Stratagene, La Jolla, CA) according to the manufacturer's protocol.
Western Blot Analyses
All Western blots were conducted as described (25). Antibodies used: rabbit polyclonal anti-FEN1 (#586, Bethyl Laboratories, Montgomery, TX), mouse monoclonal anti-Actin (ABCAM, Cambridge, MA), rabbit polyclonal anti-TRF2 (H-300; Santa Cruz Biotechnology, Santa Cruz, CA), mouse monoclonal anti-FLAG M2 (Sigma), rabbit polyclonal anti-cyclophilin A (Cell Signaling Technology, Danvers, MA).
S Phase Progression Assay
HeLa cells were cultured for 1 h in the presence of 50 μm 5-bromo-2-deoxyuridine (BrdU) in the dark. The cells were then washed in phosphate-buffered saline (PBS), placed in culture medium, and harvested at the indicated times. The harvested cells were washed with PBS and fixed in 4% paraformaldehyde and 0.1% Triton X-100 in PBS for 20 min at room temperature. Cells were further permeabilized with 0.1% Triton X-100 for 10 min on ice and fixed for an additional 5 min in 4% paraformaldehyde and 0.1% Triton X-100 in PBS. The DNA was denatured with 30 μg of DNase I (Sigma) at 37 °C for 1 h. BrdU was detected with an Alexa Fluor 488-conjugated anti-BrdU antibody (A21303, Invitrogen), and the DNA content of the cells was determined by 7-amino-actinomycin D (BD Biosciences) staining. The stained cells were analyzed on a FACSCalibur machine (BD Biosciences).
SV-40 Large-T Antigen-dependent in Vitro DNA Replication Assay
The crude cell extracts for this assay were prepared using HeLa cells as described (31). Briefly, HeLa cells were harvested and washed in cold isotonic buffer (20 mm HEPES, pH 7.8, 1.5 mm MgCl2, 5 mm KCl, 250 mm sucrose, 1 mm dithiothreitol (DTT), 0.1 mm phenylmethylsulfonyl fluoride) and then with cold hypotonic buffer (isotonic buffer without sucrose). The cells were then swollen on ice for 15 min in hypotonic buffer and lysed with 10 strokes of the Dounce homogenizer (pestle B). The cell lysate suspension was incubated on ice for another 60 min. After this incubation, the lysate was centrifuged at 1700 × g at 4 °C for 10 min to remove the nuclei and then centrifuged again at 12,000 × g for 10 min at 4 °C to clarify the lysate. The resulting lysate was flash-frozen in liquid nitrogen and stored at −80 °C. Linear plasmid DNA (pSVO.11–2K (7)) used in the replication reactions was prepared by equilibrium centrifugation in cesium chloride-ethidium bromide gradients and then digested with BbsI (New England Biolabs, Ipswich, MA). The in vitro replication reactions were carried out as described (31). Briefly, each 25-μl reaction contained 30 mm HEPES-HCl, pH 7.8, 7 mm MgCl2, 4 mm ATP, 200 μm each of CTP, UTP, and GTP, 100 μm each of dATP, dGTP, and dTTP, 0.5 mm DTT, 40 mm creatine phosphate, 0.625 units of creatine phosphokinase, 50 μm (2.5 μCi) [α-32P]dCTP (PerkinElmer Life Sciences), 50 ng of linearized plasmid DNA, 1 μg of Large-T antigen (Chimerx, Madison, WI), and 100 μg of cytoplasmic extract. The reaction was incubated for 10 min on ice and then at 37 °C for the indicated time. To stop the reaction, an equal volume of stop solution (2% SDS, 50 mm EDTA, 1 mg/ml Proteinase K) was added, and the reaction was incubated for an additional 30 min at 37 °C. The reactions were subject to a phenol/chloroform/isoamyl alcohol extraction, and the DNA was precipitated with isopropanol followed by a 70% ethanol wash. To verify that the products were generated by semi-conservative replication, additional samples were digested after precipitation with 10 units of DpnI (New England Biolabs) for 5 min at 37 °C, which completely degraded the methylated plasmid template. The isolated DNA was separated on an agarose electrophoresis gel to determine replication products that were quantified using a PhosphorImager (Amersham Biosciences).
Replication Re-initiation Assay
The protocol was adapted from Kennedy et al. (32) and Sengupta et al. (33). Briefly, cells were cultured with 1.5 mm hydroxyurea for 16 h. The cells were then released from hydroxyurea inhibition into medium containing 150 μm BrdU for 10 min in the dark. The cells were fixed immediately, permeabilized with 0.5% Triton X-100, and treated with 10 units of DNase I at 37 °C for 1 h to denature the DNA. The antibodies used for staining were mouse anti-BrdU (BD Biosciences), rabbit anti-FLAG M2 (Sigma), Alexa Fluor 488 goat anti-mouse, and Alexa Fluor 546 goat anti-rabbit (Invitrogen).
Chromatin Immunoprecipitation (ChIP)
ChIP was conducted as described (25).
Metaphase Preparation, FISH, and Chromosome Orientation in Situ Hybridization (CO-FISH)
Metaphase preparation, FISH, and CO-FISH were conducted as described (25). Aphidicolin treatments were conducted as described (11).
Statistical Analysis
Student's t test (two-tailed distribution with equal variance) was used for BrdU foci, CO-FISH, and fragile telomere analyses.
RESULTS
FEN1 Depletion Does Not impact S Phase Progression
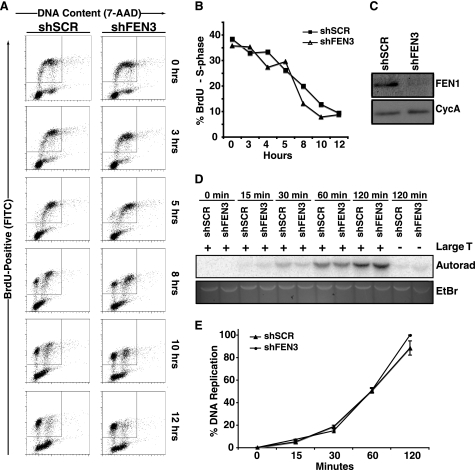
Previously, we demonstrated that FEN1 depletion in telomerase-negative cells resulted in telomere dysfunction (25). However, in telomerase-positive cells neither telomere dysfunction nor cytogenetic abnormalities were observed upon FEN1 depletion (25). Although this observation suggested that FEN1 is dispensable for genomic replication at large, we wished to more directly assess the impact of FEN1 depletion on genomic replication by measuring S phase progression. Because telomere dysfunction might impact S phase progression and this defect is rescued in telomerase-positive cells (26), telomerase-positive HeLa cells were transduced with a lentiviral construct encoding a short hairpin RNA (shRNA) targeting the FEN1 3′-UTR (shFEN3) or a control hairpin (shSCR). Expression of shFEN3 led to a significant reduction in FEN1 protein compared with control cells expressing shSCR. To follow cells through the cell cycle, cells were pulsed with BrdU for 1 h to label the S phase population, and cells were followed as they continued through the cell cycle. As expected from our previous work, in the absence of telomere dysfunction, there was no difference in S phase progression when cells were transduced with shFEN3 or shSCR. As shown in Fig. 1A, regardless of the status of FEN1, ∼35% of the cells were in S phase after a 1-hour BrdU pulse. Both control and FEN1-depleted cells exited S phase and progressed through the cell cycle with similar kinetics (Fig. 1B). These data indicate that FEN1 depletion does not significantly impact cell cycle progression and suggests that it is not essential for DNA replication in vivo.
FIGURE 1.
FEN1 depletion does not affect S phase progression or in vitro DNA replication. A, cell cycle progression of HeLa cells expressing shSCR or shFEN3 is shown. HeLa cells were labeled with BrdU for 1 h and analyzed at the indicated times using an anti-BrdU antibody (FITC-conjugated) and 7-amino-actinomysin D (7-AAD) to label DNA content. BrdU-positive cells are displayed on the y axis and represent cells that transit through S phase during BrdU labeling. The x axis displays the DNA content of the cells as indicated by incorporation of 7-amino-actinomycin D (G1 and G2/M cells have a 2 n and 4 n content of DNA, respectively). B, quantification of the percent of BrdU-positive cells in S phase after BrdU pulse (representative experiment is shown) is shown. The cells present in the inset boxes in A are BrdU-positive and consist of cells in G1, S, and G2/M phases of the cell cycle. Only the S phase cells (those that are present between G1 and G2 (2n and 4n DNA content, respectively) within the BrdU-positive population are plotted on the graph. Error bars represent S.E. C, shown are Western blots of S100 lysates from control and FEN1-depleted HeLa cells. Cyclophilin A (CycA, lower panel) is shown as a loading control. D, an SV40 Large T antigen-dependent in vitro DNA replication assay was conducted using lysates from control (shSCR) and FEN1-depleted (shFEN3) HeLa cells as described under “Experimental Procedures.” The assay was stopped at the indicated times, and the replication products were separated via gel electrophoresis. The replication products were detected via autoradiography (Autorad), and the input DNA was observed via ethidium bromide (EtBr) staining. E, shown is quantification of the replication products at the indicated times in D. Two independent experiments were conducted in duplicate, and the average of the four experiments is shown. The error bars represent S.E.
FEN1 Depletion Does Not Impact DNA Replication Kinetics in Vitro
Above, we showed that FEN1 depletion does not impact S phase progression, suggesting that other nucleases compensate for FEN1 loss during Okazaki fragment processing. However, because minor effects on DNA replication might be missed by the S phase progression assay, we next examined the impact of FEN1 depletion on DNA replication kinetics through non-telomeric DNA sequences. To measure replication kinetics in the presence or absence of FEN1, we conducted an SV40 Large T antigen-dependent in vitro DNA replication assay (31) using cell lysates isolated from control or FEN1-depleted HeLa cells (Fig. 1C). The DNA replication reaction was reconstituted with lysates from control or FEN1-depleted cells and carried out for 0, 15, 30, 60, and 120 min using a linearized plasmid containing and SV40 origin of replication. We compared the kinetics of replication by measuring the formation of newly synthesized full-length linear DNA. As shown in Figs. 1, D and E, there was no difference in DNA replication efficiency when lysates from control versus FEN1-depleted cells were used. In addition, we found that the product was insensitive to DpnI treatment (data not shown), indicating that DNA replication was semi-conservative and proceeded with the same efficiency in control and FEN1-depleted cells. Previous work (31) utilizing a defined, reconstituted system indicated that FEN1 is required for SV40-dependent DNA replication. However, our data clearly show that replication continues unabated upon FEN1 depletion. These results are in agreement with our S phase progression data and suggest that other nucleases (e.g. Dna2 and/or ExoI) present in the cell lysate compensate for FEN1 function during DNA replication in non-telomeric sequences.
FEN1 Depletion Leads to Inefficient Replication Fork Restart
Recently, we demonstrated that in human cells, FEN1 depletion results in telomere dysfunction while having little impact on total genome stability (25). Above, we showed that FEN1 depletion has no impact on S phase progression or DNA replication kinetics in vitro. These results were intriguing as they suggested that other proteins compensate for FEN1 depletion during genomic replication and/or repair, but these same proteins are ineffective within telomeric sequences. Interestingly, the FEN1ΔC mutant that does not interact with the WRN is unable to rescue telomere dysfunction upon depletion of endogenous FEN1 (25, 35). Given the data implicating FEN1 and WRN in replication fork re-initiation (15, 24) and the perceived need for this complex for efficient telomere replication, we hypothesized that FEN1 is required for the re-initiation of stalled replication forks in telomeric sequences. Therefore, we first addressed how FEN1 depletion impacts DNA replication fork re-initiation after hydroxyurea treatment.
We have hypothesized that failure to rescue stalled replication forks results in STLs. Therefore, we created an experimental paradigm that allowed us to examine the impact of FEN1 depletion on the efficiency of re-initiation of stalled replication forks. Because telomerase rescues the STL phenotype (25) and we wished to first examine the impact of FEN1 loss on re-initiation of stalled replication forks in the absence of telomere dysfunction, we carried out our analysis in telomerase-positive HeLa cells.
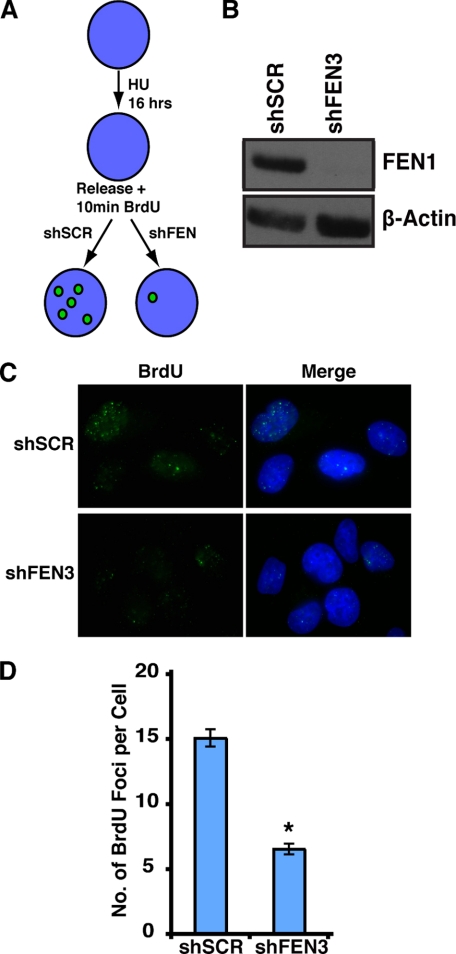
Hydroxyurea treatment causes nucleotide depletion, resulting in DNA replication fork stalling. Upon removal of hydroxyurea, nucleotide pools recover and stalled DNA replication forks re-initiate, allowing S phase to proceed. To investigate whether FEN1 contributes to DNA replication fork re-initiation, we induced stalled DNA replication forks in HeLa cells by treating them with hydroxyurea for 16 h and then releasing them in the presence of BrdU for 10 min. Because BrdU is only incorporated where DNA replication forks re-initiate, the efficiency of fork re-initiation can be determined by quantifying BrdU foci (Fig. 2A) (32). We hypothesized that if FEN1 participates in the stabilization or restart of stalled DNA replication forks, its depletion would result in fewer re-initiation events, and thus, fewer BrdU foci would be observed.
FIGURE 2.
FEN1 depletion decreases re-initiation of stalled replication forks. A, shown is a schematic of the stalled replication fork re-initiation assay and the expected results. HU, hydroxyurea. B, a Western blot analysis shows FEN1 depletion. Short hairpins against FEN1 (shFEN3) or a scrambled sequence (shSCR) were expressed in HeLa cells. FEN1 (upper panel) and β-actin (lower panel) protein levels were assessed by Western blot analysis. C, representative images show that FEN1 depletion decreases BrdU incorporation in hydroxyurea-treated cells. Immunofluorescence was conducted using an anti-BrdU antibody (green) and 4′,6-diamidino-2-phenylindole (DAPI, blue). D, quantification of the number of BrdU foci per cell in HeLa cells transduced with the indicated shRNA is shown. BrdU foci in no fewer than 100 cells were counted for each condition, and the experiment was conducted twice (a representative experiment is presented). Error bars represent S.E. (*, p < 0.0001).
HeLa cells were transduced with a lentiviral construct encoding shFEN3 or shSCR. Expression of shFEN3 led to a significant reduction in FEN1 protein compared with cells expressing a control hairpin (shSCR) (Fig. 2B). Control cells and FEN1-depleted cells were cultured for 16 h in the presence of hydroxyurea and then released from hydroxyurea inhibition in the presence of BrdU for 10 min (Fig. 2A). BrdU foci were observed by immunofluorescence. As expected, FEN1 depletion resulted in a striking decrease in the number of BrdU foci, indicating that FEN1 is important for efficient re-initiation of stalled DNA replication forks in vivo (Fig. 2C). In cells expressing the control hairpin, there was an average of 15 BrdU foci per cell. In contrast, FEN1 depletion led to an average of 6.5 BrdU foci per cell, a greater than 50% decrease (p < 0.0001; Fig. 2D). Importantly, upon FEN1 depletion, cells retained the ability to re-initiate stalled DNA replication forks, albeit less efficiently. Together these results demonstrate that FEN1 is important for efficient restart and/or stabilization of stalled DNA replication forks.
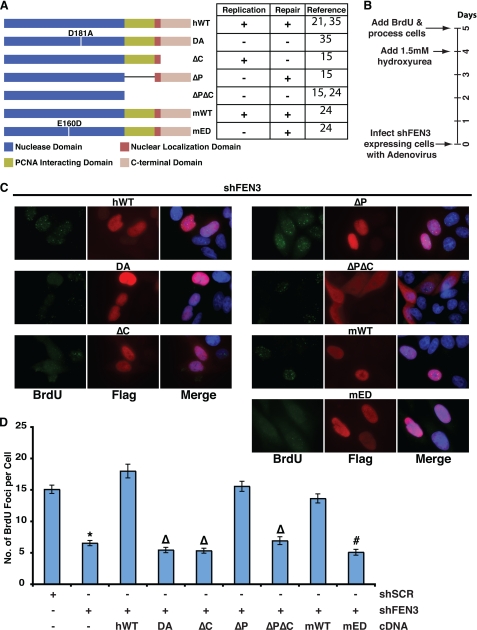
To date, the biochemical properties of FEN1 critical to the restart of stalled DNA replication forks have not been determined. Therefore, we carried out the re-initiation assay described above in cells depleted of endogenous FEN1 and expressing various FEN1 mutants as outlined in Fig. 3A. The different FEN1 alleles used in this study included 1) human wild-type (hWT), which is competent for both replication and repair functions, 2) DA, which lacks nuclease activity (36), thus representing a loss-of-function allele, 3) delta C (ΔC; a 20-amino acid deletion of the C terminus), which retains near wild-type ability to process flap structures together with the replication clamp (30, 37), PCNA, and is, therefore, competent for Okazaki fragment processing but is unable to bind the BLM and WRN helicases and participate in FEN1 DNA repair functions (30, 35), 4) delta P (ΔP; an eight-amino acid deletion), which retains the ability to interact with the RecQ helicases, BLM and WRN, but is unable to interact with PCNA (30, 37), thus rendering it replication-incompetent yet repair-competent, and 5) deltaP-deltaC (ΔPΔC; 44-amino acid deletion of the C terminus), which deletes the FEN1 nuclear localization signal and abrogates its ability to interact with PCNA, BLM, and WRN, thus creating a second loss-of-function allele that retains the nuclease domain. Finally, we expressed an mED mutant FEN1, which retains near wild-type levels of FEN activity and the ability to participate in DNA replication but is devoid of a GEN activity (38). The GEN activity has been shown to process DNA bubble structures reminiscent of stalled replication forks and is hypothesized to participate in re-initiation of stalled replication forks (24, 39).
FIGURE 3.
The gap endonuclease activity and C terminus of FEN1 are essential to re-initiate stalled replication forks. A, the schematic shows the different FEN1 alleles used in the study. Inferences on whether the different FEN1 alleles are replication competent or repair competent are shown on the right of the schematic with their associated references. These inferences were made based on nuclease activity and ability to interact with the WRN and PCNA proteins. The mutant proteins are as follows: ΔC (amino acids 360–380 deleted), ΔP (amino acids 337–344 deleted), and ΔPΔC (amino acids 337–380 deleted). B, the timeline of the experimental procedure is given in days. C, representative images show BrdU incorporation after hydroxyurea treatment in FEN1-depleted cells expressing wild-type or FEN1 mutants. Immunofluorescence was conducted using an anti-BrdU (green) antibody, anti-FLAG (red) antibody, and DAPI (blue). D, quantification of the number of BrdU foci per cell in FEN1-depleted HeLa cells with the indicated ectopic FEN1 expression (wild-type or mutant) is shown. Only cells expressing FLAG-tagged FEN1 (marked by red in C) was quantified. No fewer than 75 cells were counted for each condition, and the experiment was conducted twice (a representative experiment is presented). The error bars represent S.E. (*, p < 0.0001 compared with shSCR; Δ, p < 0.0001 compared with hWT; #, p < 0.0001 compared with mWT).
To facilitate our analysis, we depleted cells of endogenous FEN1. After depletion of FEN1, cells were infected with adenoviral constructs expressing a wild-type or mutant FEN1 allele. Transduced cells were allowed to grow for 4 days and then treated with hydroxyurea for 16 h followed by a 10-min BrdU pulse to label re-initiated DNA replication forks (Fig. 3B). To facilitate identification of successfully transduced cells, each of the FEN1 constructs was tagged with a FLAG epitope. Therefore, after the BrdU pulse, cells were fixed and stained with anti-BrdU and anti-FLAG antibodies, and BrdU foci were only quantified in FLAG-positive cells that expressed the transduced cDNAs. As expected, expression of hWT FEN1 recovered the number of BrdU foci lost in FEN1-depleted cells to numbers slightly higher than that observed in control cells. Indeed, expression of wild-type FEN1 led to an average of 18 BrdU foci per nucleus compared with 6.5 foci in FEN1-depleted cells, demonstrating that the phenotype observed was specific to FEN1 loss (Figs. 3, C and D). The significance of this slight increase in re-initiated replication forks is unclear but may be related to the level of FEN1 expression. In contrast, expression of the nuclease-deficient FEN1 mutant (also devoid of GEN activity), DA, did not rescue FEN1 depletion and resulted in an average of 5.5 foci per nucleus, indicating that the nuclease activity of FEN1 is critical for its function in the re-initiation of stalled DNA replication forks (Figs. 3, C and D). Similarly, expression of the ΔPΔC mutant, a functionally null allele, was unable to rescue the reduction in BrdU foci observed upon FEN1 depletion (Figs. 3, C and D). Expression of FEN1ΔC also failed to rescue the decreased number of BrdU foci observed in FEN1-depleted cells. Because this mutant is able to interact with PCNA and is competent for Okazaki fragment processing, this result suggests that the interactions between FEN1 and the RecQ helicases, BLM and WRN, are important for the role of FEN1 in the re-initiation of stalled DNA replication forks (Figs. 3, C and D). Finally, we found that expression of the ΔP mutant resulted in an average of 15.6 BrdU foci (Figs. 3, C and D), demonstrating that the FEN1 interaction with PCNA is not critical for its role in the re-initiation of stalled DNA replication forks.
Analysis of our FEN1 mutants indicates that FEN1 activity distinct from its ability to participate in Okazaki fragment processing is critical for the restart of stalled DNA replication forks. This result and the existence of the FEN1 GEN activity, which is stimulated by WRN (39) to cleave DNA bubble structures that resemble stalled replication forks, suggests that this activity is functionally important at stalled replication forks. To establish whether the GEN function is important for the restart of stalled replication forks, we next tested the impact of expression of a GEN-deficient FEN1 mED allele. Expression of the mED mutant failed to rescue the phenotype observed in FEN1-depleted cells, which displayed an average of 5.1 BrdU foci per nucleus (Figs. 3, C and D). As expected, the mouse wild-type protein, mWT, completely recovered the number of BrdU foci observed upon FEN1 depletion with an average of 17 foci per nucleus (Figs. 3, C and D). Because the mED mutant processes Okazaki fragments near wild type levels, these data indicate that FEN1 GEN activity is required to restart stalled DNA replication forks.
FEN1 Localizes to the Telomere
Our previous work supports the hypothesis that FEN1 activity at the telomere is critical for high fidelity DNA replication and that other nucleases compensate for FEN1 loss across the genome but fail to do so at the telomere (25). Given these results, we next wished to characterize the impact of the FEN1 mutants described above at the telomere. Because recent work demonstrates that FEN1 localizes to the mammalian telomere (9, 25, 40), we first examined the ability of the FEN1 mutants to localize to the telomere.
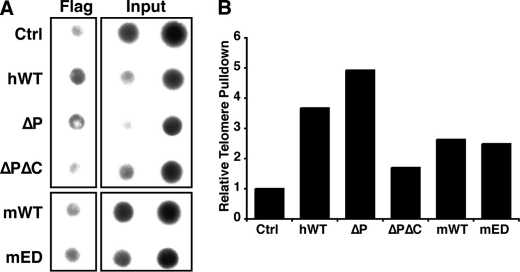
To determine whether the various FEN1 mutants retain the ability to localize to the telomere, we carried out ChIP experiments. As expected, the hWT FEN1 and FEN1ΔP mutant localized to the telomere (Figs. 4, A and B). In contrast, the FEN1ΔPΔC mutant was unable to precipitate telomeric DNA (Figs. 4, A and B). The latter result was expected because the ΔPΔC mutant lacks the nuclear localization domain and is unable to localize to the nucleus (Fig. 3A). Finally, both the mWT and mED proteins localized to the telomere. These data indicate that FEN1 mutants that retain the ability to participate in replication fork re-initiation also localize to the telomere.
FIGURE 4.
FEN1 mutants localize to the telomere. A, FEN1 alleles localize to the telomere. Representative ChIP analysis of 293T cells (Ctrl) or 293T cells transfected with wild-type FEN1 (hWT or mWT) or FEN1 mutants (ΔP, ΔPΔC, or mED), subjected to immunoprecipitation with the FLAG (M2) antibody. Precipitated DNA was probed for the presence of telomeric sequences as described under “Experimental Procedures.” The inputs indicate 0.1 and 0.2% of total. B, quantification of the representative ChIP assay is shown. Percent of telomeric DNA immunoprecipitated with the FLAG antibody was calculated using input DNA, and the control pulldown percentage was set to 1.
FEN1 Depletion Leads to the Induction of Fragile Telomeres
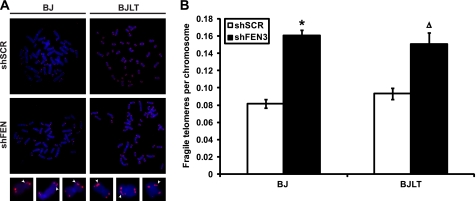
Telomeres are chromosome fragile sites as evidenced by the appearance of multiple telomeric signals after aphidicolin treatment (11, 12). Interestingly, the presence of these multiple telomere signals also arises upon depletion of Apollo, ATM, ATR, BLM, and TRF1, suggesting that several protein components repress telomere fragility (11, 12, 41, 42). Because fragile sites are thought to result from replication stress and an inability to resolve stalled replication forks (43), this observation raised the possibility that the STL observed upon FEN1 depletion (25) is the result of unresolved stalled replication forks and expression of fragile sites within telomeric sequences. Given our results above demonstrating that FEN1 facilitates re-initiation of stalled replication forks, we postulated that FEN1 depletion would lead to fragile telomere expression. Analysis of metaphase spreads prepared from aphidicolin-treated or FEN1-depleted BJ fibroblasts revealed an increase in fragile telomeres (data not shown and Fig. 5A). Indeed, 16% of the chromosomes from BJ fibroblasts depleted of FEN1 demonstrated the fragile telomere phenotype, significantly up from the control cells (8.2%; p < 0.0001). Surprisingly, this increase in fragile telomere expression was also observed upon FEN1 depletion in BJ fibroblasts expressing SV40 Large T antigen and telomerase (BJLT). FEN1 depletion in BJLT cells resulted in 15.1% of chromosomes exhibiting multiple telomere signals, significantly higher than the 9.3% observed in the control samples (Fig. 5B). These results indicate that FEN1 plays a role in the repression of fragile site expression at mammalian telomeres. Furthermore, because telomerase expression rescues the STL phenotype (25) but not expression of telomeric fragile sites, these results suggest that fragile telomere expression is either upstream of STLs or represents a second form of telomere dysfunction independent of STLs.
FIGURE 5.
FEN1 depletion results in fragile site expression at telomeres. A, representative FISH of metaphases obtained from BJ fibroblasts (BJ) or BJ fibroblasts expressing SV40 Large T antigen and telomerase (BJLT). Cells expressing a control hairpin (shSCR) or depleted of FEN1 (shFEN3) are indicated. Chromosomes were hybridized with the PNA telomere probe Cy3-(CCCTAA)3 (red) and stained with DAPI (blue). Magnified images show representative fragile telomeres (arrowheads). B, quantification of telomere fragility, depicted as the number of fragile telomeres observed per chromosome. No fewer than 60 metaphases from two independent experiments were analyzed for each condition, and an average of the two experiments is shown (*, p < 0.0001; Δ, p < 0.001). Error bars represent S.E.
FEN1 DNA Replication Fork Re-initiation Activity Is Critical to Telomere Stability
The telomere consists of G-rich repetitive DNA that has the propensity to form secondary structures, including G-quadruplexes that can impede the movement of the DNA replication fork (4, 7, 9, 14, 20). Indeed, it has been hypothesized that stalled DNA replication forks frequently occur at the telomere (4, 9). Failure to resolve a stalled DNA replication fork within the telomere would lead to fork collapse, formation of a double strand DNA break, and telomere loss (44). In support of this, recent studies suggest that collapsed replication forks at telomeres lead to the formation of very short telomeres (10, 16, 45) and, as discussed above, the expression of fragile telomeres (11, 12). We recently demonstrated that FEN1 depletion results in telomere dysfunction characterized by STLs (25), indicating that FEN1 functions in telomere maintenance through DNA replication or repair. Given our observation that FEN1 contributes to efficient re-initiation of stalled DNA replication forks, we next wished to determine whether it was the role of FEN1 in Okazaki fragment processing or the restart of stalled DNA replication forks that contributes to telomere stability. Because telomerase expression compensates for FEN1 loss at the telomere, thus masking the STL phenotype (25, 26), we utilized BJ fibroblasts, which express insufficient telomerase to maintain telomere lengths (46) for these studies.
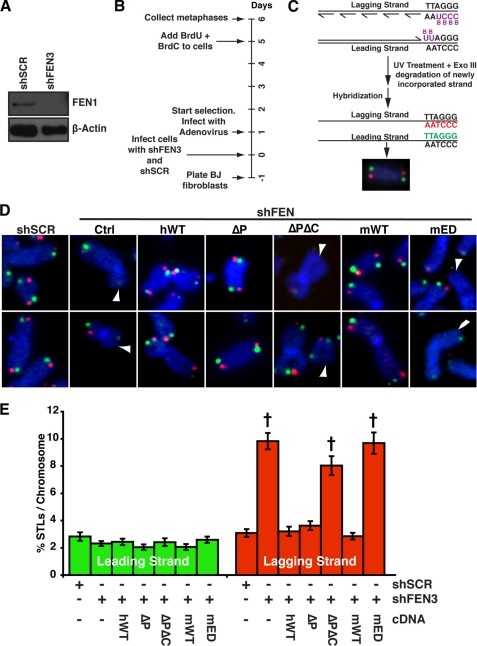
To determine the impact of FEN1 mutant expression on telomere stability, endogenous FEN1 was depleted from BJ fibroblasts (Fig. 6A). After shRNA-mediated FEN1 depletion, cells were infected with an adenovirus expressing a wild-type or mutant FEN1 allele (Fig. 6B, greater than 85% of the cells were infected; data not shown). Because FEN1 depletion leads to lagging strand-specific STL, we analyzed the strand-specific loss of telomeres in cells expressing different FEN1 alleles (25). To carry out this analysis, we utilized a technique referred to as chromosome CO-FISH, which takes advantage of the fact that the G- and C-rich strands of the telomere are exclusively replicated by lagging and leading strand DNA synthesis, respectively (Fig. 6C). In agreement with our previous results (25), FEN1 depletion led to specific loss of lagging strand-replicated telomeres (9.8% in shFEN3 cells compared with 3.1% in the control shSCR cells; p < 0.0001) while having no impact on telomeres replicated by the leading strand machinery (Figs. 6, D and 6E). Expression of wild-type FEN1 rescued the lagging strand STL phenotype (3.2% lagging strand STLs were observed, similar to that observed in shSCR control cells), indicating that the phenotype was specific to FEN1 depletion. Similarly, expression of the FEN1ΔP mutant resulted in 3.6% lagging strand STLs (p < 0.0001 compared with shFEN3), indicating that the FEN1 interaction with PCNA and, hence, its ability to participate in Okazaki fragment processing is not important for its function at the telomere. In contrast, expression of the ΔPΔC null allele led to 8% lagging strand STLs, indicating that it failed to rescue telomere dysfunction upon FEN1 depletion. Intriguingly, in contrast to the mWT protein, which rescued the lagging strand STL defect upon FEN1 depletion, the mED mutant failed to rescue FEN1 depletion at the telomere. Indeed, expression of mWT significantly decreased the number of lagging strand STLs upon FEN1 depletion to 2.8%, whereas expression of the mED mutant resulted in lagging strand STLs (9.7%, p < 0.0001) similar to those observed in ΔPΔC-expressing cells (Fig. 6) and in ΔC-expressing cells (25). However, the FEN1ΔC protein demonstrated reduced telomere localization, raising the possibility that the STL phenotype observed upon FENΔC expression was not because of reduced FEN1 re-initiation function but, rather, its reduced telomere localization. Because the mED mutant retains the ability to participate in Okazaki fragment processing and localizes to the telomere at the same efficiency as the wild-type protein (Fig. 4), the failure of mED to rescue the STL phenotype indicates that the FEN1 gap endonuclease activity is critical for its role at the mammalian telomere. Furthermore, these data demonstrate that FEN1 interaction with PCNA is dispensable for its role at the telomere.
FIGURE 6.
The gap endonuclease activity of FEN1 is essential for its function at the telomere. A, Western blot analysis of FEN1 (upper panel) from BJ fibroblasts expressing a control hairpin (shSCR) or depleted of FEN1 (shFEN3) is shown. β-Actin (lower panel) is shown as a loading control. B, shown is a timeline of experimental procedure given in days. C, a CO-FISH schematic is shown. Newly synthesized DNA strands incorporate BrdU and BrdC. UV and ExoIII treatment resulted in degradation of newly synthesized DNA containing BrdU and BrdC, and the template strands were hybridized with Cy3-(CCCTAA)3 (red, lagging strand) and fluorescein-(TTAGGG)3 (green, leading strand) PNA probes (25). D, representative CO-FISH of chromosomes from BJ fibroblasts expressing shSCR or shFEN3 and the indicated FEN1 alleles is shown. Ctrl refers to cells that do not express exogenous FEN1 protein. Color schemes are as described in C. DNA was stained with DAPI (blue). The arrowheads indicate missing telomeres. E, shown is quantification of STLs on metaphase chromosomes after depletion of endogenous FEN1 and expression of the indicated FEN1 allele, depicted as percentage of chromosomes with missing leading (green) and lagging (red) strand telomeres. A minimum of 60 metaphases from two independent experiments was analyzed per treatment in a blinded fashion, and an average of the two experiments is shown (*, p < 0.0001). The error bars represent S.E.
DISCUSSION
Telomeres perform a critical cellular function by distinguishing the chromosome end from a bona fide double-stranded DNA break. As such, mechanisms that modify the activities of DNA repair and replication proteins, presumably through interaction with the Shelterin complex, have evolved to protect the telomere and ensure its faithful replication. The need for telomere-specific replication mechanisms is likely due to the nature of the telomeric DNA sequence, which presents a number of challenges to the DNA replication machinery (4, 5). G-rich, repetitive, telomeric sequences have a high propensity to form secondary structures such as G-quadruplexes (G4) that impede the progressing replication fork, leading to the formation of stalled forks (4, 13, 14). Indeed, telomeres were recently identified as fragile sites (11), and several reports have indicated pausing or stalling of replication forks within telomeres (8–10, 47). Additionally, telomere replication is primarily initiated by the most centromere-distal origin of replication and continues unidirectionally toward the end of the telomere (11). If a replication fork stalls within the telomere and is not re-initiated, the absence of a converging replication fork would result in telomere loss. Therefore, mechanisms that facilitate replication fork movement through the telomere are critical to high fidelity telomere replication.
The importance of the Shelterin complex to telomere replication is underscored by several studies. For example, Taz1 in S. pombe and TRF1 in mice are required for efficient telomere replication. Loss of Taz1 results in replication fork stalling throughout telomeric sequences (20), whereas loss of TRF1 leads to expression of fragile telomeres (11). The ability of telomere-binding proteins to facilitate replication fork progression through the telomere is postulated to require recruitment of specialized proteins (4, 11). For example, TRF1 and TRF2 interact with and stimulate the RecQ helicases, BLM and WRN (11, 48, 49), suggesting that they recruit these proteins to enhance DNA replication and/or repair at the telomeres. Interestingly, a recent study demonstrated that TRF2 increases branch migration of Holliday Junction (HJ) intermediates, suggesting that this promotes the formation of chicken foot structures in the context of a stalled replication fork at telomeres (50). FEN1 also interacts with TRF2 (25, 40), and because FEN1 GEN activity is postulated to process chicken foot structures (24, 39), this raises the possibility that TRF2 engages the RecQ helicase-FEN1 complex coordinately at the telomere to resolve stalled replication forks and enable their efficient restart.
WRN participates in the re-initiation of stalled replication forks in vivo (51, 52). Interestingly, FEN1 was shown to localize with WRN, raising the possibility that it contributes to replication fork restart (15). Furthermore, FEN1 and WRN process branch migrating structures that resemble regressed replication forks in vitro (15). The present study demonstrates for the first time that FEN1 functionally participates in the re-initiation of stalled replication forks in vivo. Together with previous work (53), this indicates that the FEN1 role in S phase is 2-fold; first, in Okazaki fragment processing during DNA replication and, second, in the re-initiation of stalled replication forks. FEN1 localizes to mammalian telomeres during S phase (9, 25), so it could be involved in one or both of the functions outlined above. However, given that the PCNA-interacting domain of FEN1 is dispensable for telomere stability, our data indicate that the role of FEN1 in Okazaki fragment processing is non-essential for telomere stability. This result indicates that either sufficient FEN1 remains in FEN1-depleted cells to support continued replication or that other nucleases such as DNA2 or EXO1, which can also process Okazaki fragments (54–58), compensate for FEN1 loss during lagging strand DNA replication. However, these same nucleases are insufficient when replication forks stall within telomeric sequences. Indeed, we find that in the absence of the ability of FEN1 to re-initiate stalled replication forks, sister telomeres are lost despite the presence of other nucleases. Interestingly, other proteins involved in the re-initiation of stalled replication forks such as PARP1 and PARP2 have also been implicated in telomere maintenance (59–61), further indicating the importance of the re-initiation process for efficient telomere replication. An alternate hypothesis is that FEN1 is important for fork stabilization after hydroxyurea treatment. The assay we have conducted cannot differentiate between FEN1-dependent fork stabilization and fork re-initiation.
Intriguingly, the C-terminal region of FEN1 is essential for its function at the telomere and also mediates its interaction with another RecQ helicase, BLM (35). Similar to WRN, BLM is able to unwind G4 DNA, is critical for the re-initiation of stalled replication forks, and has recently been suggested to be important for efficient telomere replication (11, 33, 62, 63). This suggests that there is complicated interplay between WRN, BLM, and FEN1 at mammalian telomeres. Although the function of BLM at telomeres has not been well characterized, recent work suggests that it is important for repression of fragile telomeres (11). Interestingly, FEN1 depletion also leads to an increase fragile telomere expression, raising the possibility that these proteins work as a complex to repress telomere fragility. Together, these data are consistent with the hypothesis that FEN1 and the RecQ helicases play an important role in the maintenance of stable telomeres through re-initiation of stalled replication forks.
Here we demonstrate that FEN1 is important for efficient re-initiation of stalled replication forks in vivo. This function of FEN1 is dependent on its C-terminal domain and its GEN activity. However, despite the importance of FEN1 in re-initiation of stalled replication forks, FEN1 depletion in telomerase-positive cells did not affect S phase progression or SV40 Large T antigen-dependent in vitro DNA replication, suggesting that other nucleases compensate for FEN1-dependent replication function throughout the genome. However, these same proteins fail to compensate for FEN1 at the telomere. Indeed, FEN1 depletion leads to increased telomere fragility and lagging strand STLs. As with the re-initiation of stalled replication forks, both the FEN1 C terminus and GEN activity are essential for its function at telomeres, whereas its ability to interact with PCNA is dispensable. Collectively, these data demonstrate that FEN1 is necessary for efficient replication of telomeres, and we propose that FEN1 promotes replication fork re-initiation within telomeric sequences.
Acknowledgments
We are grateful to Dr. Ulrich Hübscher for providing the FEN1ΔP cDNA, Dr. Binghui Shen for providing the mWT and mED FEN1 cDNAs, and Dr. Fuyuki Ishikawa for providing the pSVO.11–2K plasmid. We are also grateful to Dr. Marc Wold, Dr. Peter Burgers, and members of the Stewart Laboratory for valuable discussions and to Ying Jie Lock, Ermira Pazolli, and Dr. Susana Gonzalo for critical reviews.
This work was supported in part by the Children's Discovery Institute (to S. A. S.) and Cellular, Biochemical, and Molecular Sciences Predoctoral Training Grant T32 GM007067-35 (to D. C. T.).
- WRN
- Werner
- FEN1
- Flap endonuclease 1
- STL
- sister telomere loss
- GEN
- gap endonuclease
- CO-FISH
- orientation-FISH
- hWT
- human WT
- mWT
- mutant WT
- mED
- mutant E160D
- DA
- D181A
- HJ
- Holliday junction
- PCNA
- proliferating cell nuclear antigen
- BrdU
- 5-bromo-2-deoxyuridine
- BLM
- Bloom
- PNA
- peptide nucleic acid.
REFERENCES
- 1.Wu L., Hickson I. D. (2002) Mutat. Res. 509, 35–47 [DOI] [PubMed] [Google Scholar]
- 2.Sidorova J. M. (2008) DNA Repair 7, 1776–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh D. K., Ahn B., Bohr V. A. (2009) Biogerontology 10, 235–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilson E., Géli V. (2007) Nat. Rev. Mol. Cell Biol. 8, 825–838 [DOI] [PubMed] [Google Scholar]
- 5.Verdun R. E., Karlseder J. (2007) Nature 447, 924–931 [DOI] [PubMed] [Google Scholar]
- 6.Kovtun I. V., McMurray C. T. (2008) Cell Res 18, 198–213 [DOI] [PubMed] [Google Scholar]
- 7.Ohki R., Ishikawa F. (2004) Nucleic Acids Res. 32, 1627–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makovets S., Herskowitz I., Blackburn E. H. (2004) Mol. Cell. Biol. 24, 4019–4031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verdun R. E., Karlseder J. (2006) Cell 127, 709–720 [DOI] [PubMed] [Google Scholar]
- 10.Khadaroo B., Teixeira M. T., Luciano P., Eckert-Boulet N., Germann S. M., Simon M. N., Gallina I., Abdallah P., Gilson E., Géli V., Lisby M. (2009) Nat. Cell Biol. 11, 980–987 [DOI] [PubMed] [Google Scholar]
- 11.Sfeir A., Kosiyatrakul S. T., Hockemeyer D., MacRae S. L., Karlseder J., Schildkraut C. L., de Lange T. (2009) Cell 138, 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martínez P., Thanasoula M., Muñoz P., Liao C., Tejera A., McNees C., Flores J. M., Fernández-Capetillo O., Tarsounas M., Blasco M. A. (2009) Genes Dev. 23, 2060–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parkinson G. N., Lee M. P., Neidle S. (2002) Nature 417, 876–880 [DOI] [PubMed] [Google Scholar]
- 14.Maizels N. (2006) Nat. Struct. Mol. Biol. 13, 1055–1059 [DOI] [PubMed] [Google Scholar]
- 15.Sharma S., Otterlei M., Sommers J. A., Driscoll H. C., Dianov G. L., Kao H. I., Bambara R. A., Brosh R. M., Jr. (2004) Mol. Biol. Cell 15, 734–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crabbe L., Verdun R. E., Haggblom C. I., Karlseder J. (2004) Science 306, 1951–1953 [DOI] [PubMed] [Google Scholar]
- 17.Crabbe L., Jauch A., Naeger C. M., Holtgreve-Grez H., Karlseder J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 2205–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Lange T. (2005) Genes Dev. 19, 2100–2110 [DOI] [PubMed] [Google Scholar]
- 19.Opresko P. L., von Kobbe C., Laine J. P., Harrigan J., Hickson I. D., Bohr V. A. (2002) J. Biol. Chem. 277, 41110–41119 [DOI] [PubMed] [Google Scholar]
- 20.Miller K. M., Rog O., Cooper J. P. (2006) Nature 440, 824–828 [DOI] [PubMed] [Google Scholar]
- 21.Li X., Li J., Harrington J., Lieber M. R., Burgers P. M. (1995) J. Biol. Chem. 270, 22109–22112 [DOI] [PubMed] [Google Scholar]
- 22.Liu Y., Kao H. I., Bambara R. A. (2004) Annu. Rev. Biochem. 73, 589–615 [DOI] [PubMed] [Google Scholar]
- 23.Shen B., Singh P., Liu R., Qiu J., Zheng L., Finger L. D., Alas S. (2005) BioEssays 27, 717–729 [DOI] [PubMed] [Google Scholar]
- 24.Zheng L., Zhou M., Chai Q., Parrish J., Xue D., Patrick S. M., Turchi J. J., Yannone S. M., Chen D., Shen B. (2005) EMBO Rep 6, 83–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saharia A., Guittat L., Crocker S., Lim A., Steffen M., Kulkarni S., Stewart S. A. (2008) Curr. Biol. 18, 496–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saharia A., Stewart S. A. (2009) Oncogene 28, 1162–1167 [DOI] [PubMed] [Google Scholar]
- 27.Stewart S. A., Hahn W. C., O'Connor B. F., Banner E. N., Lundberg A. S., Modha P., Mizuno H., Brooks M. W., Fleming M., Zimonjic D. B., Popescu N. C., Weinberg R. A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12606–12611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stewart S. A., Ben-Porath I., Carey V. J., O'Connor B. F., Hahn W. C., Weinberg R. A. (2003) Nat. Genet. 33, 492–496 [DOI] [PubMed] [Google Scholar]
- 29.Stewart S. A., Dykxhoorn D. M., Palliser D., Mizuno H., Yu E. Y., An D. S., Sabatini D. M., Chen I. S., Hahn W. C., Sharp P. A., Weinberg R. A., Novina C. D. (2003) RNA 9, 493–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stucki M., Jónsson Z. O., Hübscher U. (2001) J. Biol. Chem. 276, 7843–7849 [DOI] [PubMed] [Google Scholar]
- 31.Brush G. S., Kelly T. J., Stillman B. (1995) Methods Enzymol. 262, 522–548 [DOI] [PubMed] [Google Scholar]
- 32.Kennedy B. K., Barbie D. A., Classon M., Dyson N., Harlow E. (2000) Genes Dev. 14, 2855–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sengupta S., Linke S. P., Pedeux R., Yang Q., Farnsworth J., Garfield S. H., Valerie K., Shay J. W., Ellis N. A., Wasylyk B., Harris C. C. (2003) EMBO J. 22, 1210–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma S., Sommers J. A., Gary R. K., Friedrich-Heineken E., Hübscher U., Brosh R. M., Jr. (2005) Nucleic Acids Res. 33, 6769–6781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen B., Nolan J. P., Sklar L. A., Park M. S. (1996) J. Biol. Chem. 271, 9173–9176 [DOI] [PubMed] [Google Scholar]
- 37.Stucki M., Stagljar I., Jónsson Z. O., Hübscher U. (2001) Prog. Nucleic Acid Res. Mol. Biol. 65, 261–298 [DOI] [PubMed] [Google Scholar]
- 38.Zheng L., Dai H., Zhou M., Li M., Singh P., Qiu J., Tsark W., Huang Q., Kernstine K., Zhang X., Lin D., Shen B. (2007) Nat. Med. 13, 812–819 [DOI] [PubMed] [Google Scholar]
- 39.Liu R., Qiu J., Finger L. D., Zheng L., Shen B. (2006) Nucleic Acids Res. 34, 1772–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muftuoglu M., Wong H. K., Imam S. Z., Wilson D. M., 3rd, Bohr V. A., Opresko P. L. (2006) Cancer Res. 66, 113–124 [DOI] [PubMed] [Google Scholar]
- 41.van Overbeek M., de Lange T. (2006) Curr. Biol. 16, 1295–1302 [DOI] [PubMed] [Google Scholar]
- 42.Undarmaa B., Kodama S., Suzuki K., Niwa O., Watanabe M. (2004) Biochem. Biophys. Res. Commun. 315, 51–58 [DOI] [PubMed] [Google Scholar]
- 43.Durkin S. G., Glover T. W. (2007) Annu. Rev. Genet. 41, 169–192 [DOI] [PubMed] [Google Scholar]
- 44.Branzei D., Foiani M. (2005) Curr. Opin. Cell Biol. 17, 568–575 [DOI] [PubMed] [Google Scholar]
- 45.Xu L., Blackburn E. H. (2007) Mol. Cell 28, 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Masutomi K., Yu E. Y., Khurts S., Ben-Porath I., Currier J. L., Metz G. B., Brooks M. W., Kaneko S., Murakami S., DeCaprio J. A., Weinberg R. A., Stewart S. A., Hahn W. C. (2003) Cell 114, 241–253 [DOI] [PubMed] [Google Scholar]
- 47.Ivessa A. S., Zhou J. Q., Schulz V. P., Monson E. K., Zakian V. A. (2002) Genes Dev. 16, 1383–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lillard-Wetherell K., Machwe A., Langland G. T., Combs K. A., Behbehani G. K., Schonberg S. A., German J., Turchi J. J., Orren D. K., Groden J. (2004) Hum. Mol. Genet. 13, 1919–1932 [DOI] [PubMed] [Google Scholar]
- 49.Opresko P. L., Otterlei M., Graakjaer J., Bruheim P., Dawut L., Kølvraa S., May A., Seidman M. M., Bohr V. A. (2004) Mol. Cell 14, 763–774 [DOI] [PubMed] [Google Scholar]
- 50.Poulet A., Buisson R., Faivre-Moskalenko C., Koelblen M., Amiard S., Montel F., Cuesta-Lopez S., Bornet O., Guerlesquin F., Godet T., Moukhtar J., Argoul F., Déclais A. C., Lilley D. M., Ip S. C., West S. C., Gilson E., Giraud-Panis M. J. (2009) EMBO J. 28, 641–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dhillon K. K., Sidorova J., Saintigny Y., Poot M., Gollahon K., Rabinovitch P. S., Monnat R. J., Jr. (2007) Aging Cell 6, 53–61 [DOI] [PubMed] [Google Scholar]
- 52.Sidorova J. M., Li N., Folch A., Monnat R. J., Jr. (2008) Cell Cycle 7, 796–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nikolova T., Christmann M., Kaina B. (2009) Anticancer Res. 29, 2453–2459 [PubMed] [Google Scholar]
- 54.Moreau S., Morgan E. A., Symington L. S. (2001) Genetics 159, 1423–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kang H. Y., Choi E., Bae S. H., Lee K. H., Gim B. S., Kim H. D., Park C., MacNeill S. A., Seo Y. S. (2000) Genetics 155, 1055–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bae S. H., Seo Y. S. (2000) J. Biol. Chem. 275, 38022–38031 [DOI] [PubMed] [Google Scholar]
- 57.Ayyagari R., Gomes X. V., Gordenin D. A., Burgers P. M. (2003) J. Biol. Chem. 278, 1618–1625 [DOI] [PubMed] [Google Scholar]
- 58.Kao H. I., Campbell J. L., Bambara R. A. (2004) J. Biol. Chem. 279, 50840–50849 [DOI] [PubMed] [Google Scholar]
- 59.Dantzer F., Giraud-Panis M. J., Jaco I., Amé J. C., Schultz I., Blasco M., Koering C. E., Gilson E., Ménissier-de Murcia J., de Murcia G., Schreiber V. (2004) Mol. Cell. Biol. 24, 1595–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ye J. Z., de Lange T. (2004) Nat. Genet. 36, 618–623 [DOI] [PubMed] [Google Scholar]
- 61.Bryant H. E., Petermann E., Schultz N., Jemth A. S., Loseva O., Issaeva N., Johansson F., Fernandez S., McGlynn P., Helleday T. (2009) EMBO J. 28, 2601–2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun H., Karow J. K., Hickson I. D., Maizels N. (1998) J. Biol. Chem. 273, 27587–27592 [DOI] [PubMed] [Google Scholar]
- 63.Davies S. L., North P. S., Hickson I. D. (2007) Nat. Struct. Mol. Biol. 14, 677–679 [DOI] [PubMed] [Google Scholar]