Abstract
In the large intestine organic cation transporter type-2 (OCTN2) is recognized as a transporter of compounds such as carnitine and colony sporulation factor, promoting health of the colon intestinal epithelium. Recent reports suggest that OCTN2 expression in small intestine is under control of peroxisome proliferator-activated receptor-α (PPARα). However, PPARα contribution to colonic OCTN2 expression remains controversial. Here we examined the transcriptional regulation of colon OCTN2 gene by PPARγ. To exclude any additional modulation of other PPAR to OCTN2 expression, we used both in vivo and in vitro PPAR-null models and specific PPAR inhibitors. The PPARγ agonists thiazolidinediones increased both OCTN2 mRNA and protein expression in colonic epithelial cell lines independently by PPARα expression. The induction was blocked only by PPARγ antagonists or by γORF4, a PPARγ isoform with dominant negative activity, suggesting a PPARγ-dependent mechanism. A conserved noncanonical PPAR-responsive element was found by computational analysis in the first intron of human OCTN2 gene and validated by EMSA assay. Promoter-reporter assays further confirmed transcriptional functionality of the putative PPAR response element, whereas selective mutation caused complete loss of responsiveness to PPARγ activation. Finally, adenovirus-mediated overexpression of constitutively active PPARγ mutant increased colon OCTN2 expression in PPARα−/− mice. Interestingly, animals overexpressing colon PPARγ showed a significant increase in plasma carnitine, thus demonstrating the functional contribution of large intestine to systemic carnitine homeostasis. This study reveals a PPARγ-dependent absorption machinery in colon that is likely involved in the health of colon epithelium, in the microbiota-host interactions and in the absorption of nutraceuticals and drugs.
Keywords: Carnitine, Gene Expression, Intestine, Membrane Proteins, Mouse, Organic Cation Transporters (OCTN), Peroxisome Proliferator-activated Receptors (PPAR)
Introduction
Carnitine (l-3-hydroxy-4-N,N,N-trimethylamino-butyrate) is an essential metabolite that plays a fundamental role in intermediary metabolism (1–3). Carnitine homeostasis in mammals is maintained by acquisition of carnitine from dietary sources, a modest rate of endogenous carnitine biosynthesis, and efficient reabsorption of carnitine (4). Intestinal absorption and distribution of carnitine and its derivatives within the body and maintenance of substantial concentration gradients between the cells and the extracellular compartment are regulated by organic cation transporters (OCTNs)2 that belong to the solute carrier 22A family (5–7). Three OCTNs have been identified so far: OCTN1 (SLC22A4), OCTN2 (SLC22A5), and OCTN3 (SLC22A21) (8–10). OCTN1 and OCTN2 are highly expressed in several tissues, such as kidney, intestine, skeletal muscle, heart, liver, and brain (8, 11–14), whereas OCTN3 is almost exclusively expressed in testes (10, 15). Because of its high binding affinity for carnitine and its wide expression, OCTN2 is the most important carnitine transporter. OCTN1 contributes less to carnitine transport than OCTN2 because of its low carnitine transport activity. The fact that mutations of OCTN2 in human diseases or in juvenile visceral steatosis mice results in systemic carnitine deficiency demonstrates the essential role of this carrier in carnitine homeostasis (5, 16). OCTN2 has been also demonstrated to regulate the tissue distribution of carnitine, as juvenile visceral steatosis mice showed not only a decreased intestinal and renal absorption of carnitine but also a concomitant altered distribution of this substrate to target tissues (6).
Despite the fundamental role played by intestine in carnitine absorption, to date little has been known about the transcriptional mechanisms underlying the regulation of the OCTN2 gene expression in the gastrointestinal tract. Previous studies showed that the levels of OCTN2 mRNA were increased in the liver and small intestine of rats fed clofibrate or supplemented with WY-14643 (17, 18). Clofibrate, a hypolipidemic drug, and WY-14643 are ligand and activators of peroxisome proliferator-activated receptor α (PPARα), a transcription factor belonging to the nuclear hormone receptor superfamily. PPARα has been shown to be expressed at a high level in the small intestine, but its expression decreases from jejunum to colon, where its transcripts are barely detectable (19). The site-specific distribution of PPAR in the bowel may provide an explanation for the findings that treatment with PPARα agonists significantly increased OCTN2 expression only in small intestine.
Independently of PPARα expression and/or activation, OCTN2 exhibits the strongest expression in the colon where it contributes to the preservation of colonocyte metabolic function (20). Recently, we have shown that carnitine is a rate-limiting factor for the maintenance of physiological butyrate oxidation in colon epithelium (21). Using an animal model of experimental colitis, we have established a causal relationship between carnitine depletion, due to OCTN2 decrease, and colonocyte damage, due to the inability of mitochondria to maintain normal butyrate β-oxidation. Moreover, only in the colon it has been demonstrated an additional function of OCTN2; quorum-sensing molecules produced by colonic microbiota, such as the pentapetide secreted by Bacillus subtilis, is transported through OCTN2 into colon epithelia, where it induces cytoprotective heat shock proteins that prevent oxidant-induced intestinal epithelial cell injury and loss of barrier function (22).
Therefore, the identification of the factors regulating OCTN2 expression in the colon is an important topic from a biological and pharmacological point of view. Because PPARγ rather than PPARα predominates in colonocytes (10, 23), we hypothesized that OCTN2 gene expression could be induced in the large intestine by PPARγ rather than PPARα activation. To test this hypothesis, we examined the regulation of OCTN2 expression by PPARγ in intestinal mucosa of WT and PPARα null mice and in suitable human and animal-derived cell models.
EXPERIMENTAL PROCEDURES
Reagents
Troglitazone, rosiglitazone, bisphenol A diglycidyl ether (BADGE), and GW9662 were purchased by Cayman Chemical (Inalco, Milan, Italy), whereas GSK0660 was obtained from Sigma. If necessary, the compounds were dissolved in dimethyl sulfoxide (DMSO). The solvent was used as a control without significant effect.
Rabbit anti-OCTN2 antiserum was obtained from Vinci-Biochem (Florence, Italy). Reagents and secondary and nonspecific IgG antibody for immunoblotting analysis were from Dako (Milan, Italy). All other reagents and compounds used, if not specified, were obtained from Sigma.
Cell Line Preparation and Cell Culture
The young adult mouse colon cells and derivation of conditionally immortalized transgenic cell lines using the H-2Kb-tsA58 mouse (ImmortoMouse; Charles River Laboratories) were obtained as previously described (24). Briefly, PPARα−/− mice were mated with the ImmortoMouse. PPARα−/− mice carrying the heat-labile SV40 gene were sacrificed, and colon epithelium was prepared to derive PPARα−/− MCE cell lines as before (25). Cell lines were maintained in RPMI 1640 media with 5% FBS and 5 units/ml of murine IFNγ on collagen-coated plates and grown under permissive conditions at 33 °C with 5% CO2 (26). Before all experiments, cells were transferred to 37 °C (non-permissive conditions) with 0.5% FBS, IFNγ-free media overnight. NCM460 cells were obtained from In Cell (San Antonio, TX) and grown in M3:10 culture medium (In Cell). Human colonocytes were isolated and cultured as described previously (27).
Cell Carnitine Uptake
Carnitine uptake by MCE cell lines was analyzed following a methodology already described (21). Briefly, the cells and transport medium (125 mm NaCl, 4.8 mm KCl, 5.5 mm d-glucose, 1.2 mm CaCl2, 1.2 mm KH2PO4, 1.2 mm MgSO4, and 25 mm HEPES, pH 7.4) containing radiolabeled [3H]carnitine (0.5 μm, 0.5 mCi/ml) were preincubated separately and then mixed to start the carnitine uptake reaction. At appropriate times, the transport reaction was stopped by rapidly washing the cells 4 times with ice-cold 0.1 m MgCl2. The resultant pellets were solubilized in 3 n KOH and neutralized with HCl, and the associated radioactivity was measured in a liquid scintillation counter.
Human Colonic Tissue and Cellular Transfection
Human colonic tissue was obtained from individuals undergoing colonic resection at the University of Naples, Faculty of Medicine, for benign or malignant tumors. Donors had not received preoperative irradiation or chemotherapy, and use of human tissue was approved by the Institutional Ethical Committee. Where requested, colonocyte cell lines were transfected with γORF4 cDNAs cloned in pcDNA3 expression vectors or with PPARα-specific short interfering RNA (siRNA; Euroclone, Milan, Italy) using Lipofectamine reagent (Invitrogen). The siRNA used was able to reduce the expression level of the endogenous PPARα by ∼91% in PPARα-expressing hepatocytes.
Animals
Male PPARα-null mice (129S4/SvJae-Pparatm1Gonz/J; PPARα−/−) and corresponding wild-type (PPARαWT) control mice (129S1/SvImJ) were purchased from The Jackson Laboratory. Four-month-old male wild-type and PPARα-null mice were used in this study (n = 4–5 per group), with an average initial body weight of 25.1 g (±2.7 S.D.). Previous studies have shown that the biological effects of PPARγ occur independently of dietary composition (28). Troglitazone (150 mg/kg/day), rosiglitazone (15 mg/kg/day), and BADGE (10 mg/kg/day) were prepared as a suspension in distilled water and administered 2 h after the beginning of the light cycle by gavage for 4 days (0.2 ml per mouse), twice per day. The control group was administered an equal volume of distilled water by gavage. The volume of all doses (200 μl) equaled the half-maximum recommended volume for gastric gavages for mice (29).
The animals were housed on a regular dark-light cycle (light from 8:00 a.m. to 8:00 p.m.) with free access to standard rodent chow (∼10% kcal from fat) and water throughout the experimental period and cared for in compliance with the Italian Ministry of Health Guidelines (no. 86609 EEC). The experimental protocol was approved by the Bioethical Committee of the University of Naples.
Tissue Handling and Tissue and Cellular Lysate Preparations
Four days after the treatment and 6 h after the last gavage the mice were sacrificed, the colonic mucosa was scraped into homogenization buffer, and cells were lysed to extract the RNA and protein. Cell cultures were treated with or without GW9662 (5 μm), BADGE (10 μm), or GSK0660 (1 μm) for 1 h, exposed to DMSO or troglitazone (50 μm) for 12–24 h, and then scraped into cell lysis buffer.
Immunohistochemistry
Intestinal tissue was fixed in 10% neutral buffered formalin and embedded in paraffin before sectioning. Slides were immunostained with anti-OCTN2 antibody (Vinci-Biochem) followed by secondary polyclonal goat anti-rabbit immunoglobulins/AP (Dako). Slides were viewed by light microscopy.
Preparation of Nuclear Extracts of Treated Cells for Quantitative Measurement of PPARγ Activity
Levels of PPARγ activity after rosiglitazone (5 μm), GW9662 (10 μm), BADGE (10 μm), or combination treatment were measured in the nuclear fraction of cultured cells using the TransAM PPAR kits (Vinci-Biochem). TransAM PPAR kits include a 96-well plate with an immobilized oligonucleotide containing a PPAR response element (5′-AACTAGGTCAAAGGTCA-3′). The active form of PPAR contained in nuclear extract specifically binds to the oligonucleotide.
In Silico Screening of PPAR Response Elements (PPREs)
PPAR binding sites were searched by using PPAR targets finder, a tool written in PHP, specifically developed for the purpose of this prediction by one of us (A. Boccia). The ENSEMBL public repository Version 51.36 provided the genomic sequence containing the OCTN2 gene and relative annotations. Specifically, the TSS of the ENSEMBL transcript ENST00000245407 has been used as a reference to calculate gene coordinates for the predicted PPREs.
RNA Analyses
Total RNA was isolated from samples by using TRIzol reagent according to the manufacturer's protocol. The cDNA synthesis was carried out, and the mRNA expression of genes was measured by real-time detection PCR using specific primers on iCycler IQ5 Real-Time PCR detection system (Bio-Rad) with the SYBR Green PCR kit (Molecular Probes, Milan, Italy). To assess specific mRNA expression levels, quantitative PCR was used with the following designed gene-specific primers: human OCTN2-specific primer sequences 5′-CCATTGTGACCGAGTGGAACC-3′ (forward) and 5′-ACATTCTTCCGGCCAAACCTG-3′ (reverse); mouse OCTN2-specific primer sequences 5′-ACAGTATCCCGTTGGAGACG-3′ (forward) and 5′-ACACCAGGTCCCACTCTGTC-3′ (reverse); human β-actin-specific primer sequences 5′-TTGCCGACAGGATGCAGAAGGA-3′ (forward) and 5′-AGGTGGACAGCGAGGCCAGGAT-3′ (reverse); mouse β-actin-specific primer sequences 5′-ACGGCCAGGTCATCACTATTG-3′ (forward), 5′-CACAGGATTCCATACCCAAGAAG-3′ (reverse). Gene expression was quantified by using the comparative CT method, normalized to β-actin and expressed as-fold induction of control. The abundance of housekeeping gene β-actin mRNA, used for normalization, was not influenced by the treatment of mice and cells.
Promoter-Reporter Assay
To construct the OCTN-PPRE-luciferase reporter plasmids, we synthesized complementary oligonucleotides containing two repeats of the putative OCTN2 PPREs (Table 1) and flanking sites for KpnI and HindIII. The double strands were annealed and subcloned into the KpnI- and HindIII-digested pGL4.23 (luc2/minP) vector (Promega, Milan, Italy) that contains the minimal promoter minP followed by the luciferase reporter gene luc2 (Promega). After cloning, fragments were sequenced to confirm the integrity of the constructs. The 2× OCTN2-PPRE-Luc plasmids (1 mg/well) were transiently transfected together with pGL4.74 renilla luciferase (Rluc) (encoding the renilla luciferase reporter gene; Promega), which was used as an internal control reporter vector to normalize for differences in transfection efficiency into subconfluent target cells (PPARα−/−MCE). PPARα−/− MCE cells were also co-transfected with either human PPARγ expression plasmid (pCMX-hPPARγ) and human retinoid X receptor-α (RXRα) expression plasmid (pCMX-hRXRα) or empty vector (pCMX) using Lipofectamine 2000 (Invitrogen). Twenty-four hours later the cells were treated with DMSO or rosiglitazone in the presence or absence of GW9662. Cell lysates were harvested 24 h later to measure luciferase and β-galactosidase activities.
TABLE 1.
In silico screening of PPARγ binding sites
Putative PPREs are sorted on the basis of the position relative to the OCTN2 transcription start site. A prediction of the efficiency of each PPRE is provided in the form of average strength of PPARγ binding (Weak, Medium, Strong).
| PPRE sequence | TSS offset | Location | Predicted binding |
|---|---|---|---|
| GGGGAAAAGGGTA | −9413 | 5′ | Weak |
| GGGAAAAGGGTAA | −9412 | 5′ | Weak |
| AGGCCATGGGTCT | −8260 | 5′ | Weak |
| AGGAAAAAGGTGA | −6851 | 5′ | Weak |
| AGGGGAAAGGTGT | −4500 | 5′ | Weak |
| AGGAAAAAGGGAA | 1092 | Intron 1 | Weak |
| AGGTCATAGGGTG | 1638 | Intron 1 | Weak |
| AGGTGAAAGGGCA | 2885 | Intron 1 | Strong |
| AGGTCAGAGTTCA | 5090 | Intron 1 | Weak |
| AGGCCAAGCTTCA | 7018 | Intron 1 | Weak |
| AGGTAATATGCCA | 8977 | Intron 2 | Weak |
On the basis of the results obtained, a mutation construct of the putative PPRE selected was prepared by introducing a mutation in the PPRE sequence identified within the OCTN2 first intron with the site-directed mutagenesis kit (Agilent Technologies, Milan, Italy) using the oligonucleotides 5′-GACCTGTAAGTAGGTGTATGGGCATATAACTCTTA-3′ (forward) and 5′-TAAGAGTTATATGCCCATACACCTACTTACAGGTC-3′ (reverse). The mutant construct was controlled for the presence of the intended mutation and the absence of any unexpected mutations by DNS sequencing.
Electrophoretic Mobility Shift Assay (EMSA)
The human PPARγ and human RXR proteins were generated from the expression vectors by in vitro transcription/translation using the TNT1 Quick Coupled Transcription/Translation kit (Promega) according to the manufacturer's protocol. For EMSA, double-stranded oligonucleotides (100 ng) of the putative OCTN2 PPRE (wild type or mutated) were labeled with 0.05 mm DIG-ddUTP in labeling buffer (0.2 m potassium cacodylate, 25 mm Tris-HCl, 0.25 ng/ml bovine serum albumin, pH 6.6), 5 mm CoCl2, 20 units/ml terminal transferase (Roche Applied Science) and incubated for 15 min at 37 °C. Then 2 ml of each in vitro translated PPARγ and RXR proteins were incubated with 4 ng of DIG-labeled probes and a 5-, 50-, and 100-fold molar excess of unlabeled specific probes for competition in 1 mg of poly(dI-dC) and EMSA binding buffer (10 mm Tris-HCl, 120 mm KCl, 0.5 mm EDTA, 0.1% Triton-X-100, 12.5% glycerol, 0.2 mm DTT) for 30 min at RT. The protein-DNA complexes were subjected to electrophoresis on 6% native polyacrylamide gels and transferred to a positive-charged nylon membrane. The DIG-labeled DNA was detected by chemiluminescence using anti-digoxigenin-AP conjugate and Cold Shock Protein D (CSPD) (Roche Applied Science) according to the manufacturer's protocol (Roche Applied Science) and a Bio-Imaging Analysis (Bio-Rad).
Preparation of Recombinant Adenoviruses
A constitutively active PPARγ mutant VpPPARγ was generated by fusing the transcriptional activation domain of HSV Vp16 to the N terminus of mouse PPARγ1. The transactivation ability of VpPPARγ was assessed by transient transfection into PPARα−/− MCE and NCM460 cells. Transfections were performed multiple times in triplicate using CMV-β-gal to control for transfection efficiency. To generate the adenoviruses expressing VpPPARγ (AdPPARγ), the cDNA fragment containing VpPPARγ was subcloned into a pAdenoVator-CMV5-IRES-GFP (MP Biomedicals, Milan, Italy), and recombinant adenoviruses were generated by Adeno-QuestTM kit (MP Biomedicals) using QBI-HEK 293A cells. Adenoviruses expressing only GFP (AdGFP) were used as the control.
In Vivo Infusion of AdPPARγ or AdGFP into Mouse Colon
A solution of ∼1 × 1012 virions of AdPPARγ was infused into the colon of mice via a rectal approach. Briefly, animals were fasted for 48 h before viral infusion, and colonic fecal material was completely cleansed by feeding animals with GoLytely solution followed by three additional bowel irrigations with PBS buffer. To disrupt intestinal mucous, the mucolytic agent acetylcysteine was introduced into the rectum and maintained for 30 min before viral infusion. After being mildly sedated with halothane, animals were infused with a viral solution via the rectum by using a modified gavage and were kept in the Trendelenburg position for at least 30 min to enhance transduction of adenoviruses into the large intestine. Because the rapid turnover of intestinal epithelium may result in diminished PPARγ expression in newly formed epithelial cells, animals were killed on day 3 after the viral infusion. The entire colon was excised, cleaned, quickly frozen in liquid nitrogen, ground into fine powder, and used for RNA isolation.
Plasma Carnitine Profile of PPARα-null Mice
The plasma carnitine concentrations in PPARα−/− mice treated or not with rosiglitazone or rosiglitazone plus BADGE and in AdPPARγ- or AdGFP-infected PPARα−/− mice were determined before and after supplementation with carnitine (1 g/250 ml of drinking water; n = 6 for each group). Briefly, plasma samples were treated with perchloric acid (final concentration 3%), resulting in a supernatant and a pellet. Analysis of the supernatant yielded free carnitine and, after alkaline hydrolysis, total acid-soluble carnitine. The pellet yielded the long chain acylcarnitines (acyl group chain length ≥10 carbons) after alkaline hydrolysis. The short-chain acylcarnitine fraction (acyl group chain length <10 carbons) was calculated from the difference between total acid-soluble and free carnitine, and the sum of total acid-soluble and long-chain acylcarnitine represented total carnitine. Acylcarnitine measurements were made using flow injection tandem mass spectrometry. Data were acquired using a Micromass Quattro MicroTM system equipped with a model 2777 autosampler, a model 1525 HPLC solvent delivery system, and a data system controlled by the MassLynx 4.0 operating system (Waters, Milan, Italy).
All data are expressed as the mean ± S.E. Statistical analyses were performed with a t test or the non-parametric Mann-Whitney U test for significance using PRISM software (GraphPad, Inc., San Diego, CA). A p value of less than 0.05 indicated a significant difference between groups.
RESULTS
Increased Expression of OCTN2 mRNA by PPARγ Agonists
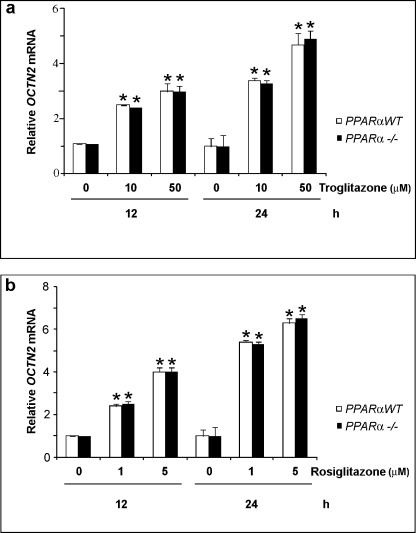
To determine the effect of PPARγ on the expression of OCTN2 gene, we treated adult mouse colon cells (MCE) expressing endogenous PPARα (26) with PPARγ agonists troglitazone and rosiglitazone, both being thiazolidinediones (TZDs), at various concentrations for different time periods. OCTN2 mRNA level was measured by quantitative reverse transcriptase-PCR (qRT-PCR). As shown in Fig. 1 (white bars), both agonists significantly increased the mRNA expression of OCTN2 gene in cells in a time- and dose-dependent manner. To ascertain whether PPARγ agonists induce OCTN2 gene expression in a PPARα-independent way, we generated conditionally immortalized MCE cell lines null for PPARα (PPARα−/− MCE cells). Quantitative RT-PCR analysis confirmed that OCTN2 gene was expressed to a similar extent in MCE and in PPARα−/− MCE cells and that its expression was similarly induced by PPARγ agonists (Fig. 1). We further examined the effect of PPARγ agonists in human colonocyte cell lines. Again, TDZs treatment was able to increase OCTN2 expression in human colonocytes (Fig. 2a). To confirm the pivotal role of only endogenous PPARγ on the expression of OCTN2 mRNA and to exclude any contribution of PPARα due to the stimulation procedure, human colonocytes were transfected with PPARα-specific siRNA. As expected, PPARα-specific siRNA transfection did not abrogate the TDZ-induced expression of OCTN2 in human colonocytes (Fig. 2a).
FIGURE 1.
TZDs increased OCTN2 expression in a dose- and time-dependent manner. MCE and PPARα−/− MCE cells were treated with the solvent DMSO (0.1%), and the TZDs troglitazone (a) or rosiglitazone (b) for the indicated time periods. Relative mRNA level of OCTN2 gene was determined by using qRT-PCR and, after normalization with β-actin, expressed as the -fold induction in relation to the control. Data represent the means ± S.E. of three independent experiments. *, p < 0.01 versus control.
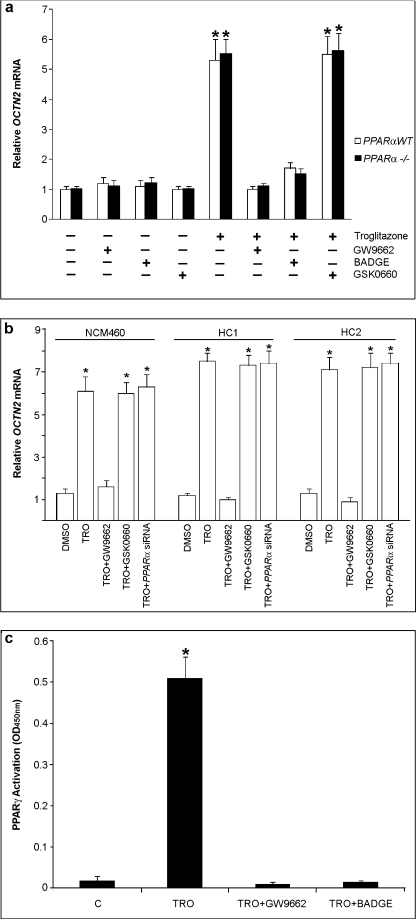
FIGURE 2.
TZD-induced OCTN2 expression is dependent on PPARγ activation. MCE and PPARα−/− MCE cells (a) or human colonocyte cell lines (NCM460, HC1, and HC2) (b) were treated with or without GW9662 (5 μm), BADGE (10 μm), or GSK0660 (1 μm) for 1 h and exposed to DMSO or troglitazone (TRO; 50 μm) for 24 h. Only human colonocyte cell lines were also transfected with siRNA specific for PPARα (40 nm) for 48 h before the exposure to troglitazone or DMSO. Relative mRNA levels of OCTN2 were determined by using qRT-PCR. *, p < 0,01 versus control. c, PPARα−/− MCE cells were treated with or without GW9662 (10 μm) or BADGE (10 μm) for 1 h and exposed to DMSO (C) or troglitazone (50 μm) for 4 h. The levels of PPARγ activation were measured by the PPARγ transfactor assay using nuclear protein extracts (see “Materials and Methods”). Results are the mean ± S.E. of five experiments. *, p < 0.01 versus control.
TDZs Induction of OCTN2 Expression and Its Inhibition by PPARγ Antagonists and by γORF4, a Dominant Negative PPARγ Isoform
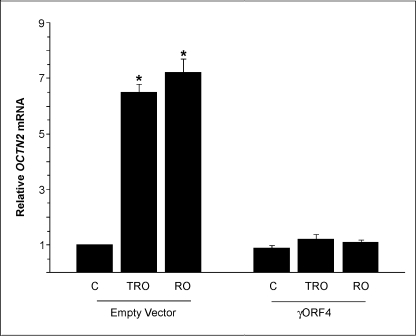
To examine whether the TZD-induced expression of OCTN2 was mediated by PPARγ activation, cells were pretreated with a selective PPARγ antagonist GW9662 (5 μm for 1 h) before incubation with troglitazone. qRT-PCR demonstrated that GW9662 blocked the troglitazone-induced increase of OCTN2 mRNA (Fig. 2). Similarly, another PPARγ antagonist, BADGE (10 μm), also reduced the induction of OCTN2 expression by troglitazone, whereas the treatment with the selective PPARβ antagonist GSK0660 (1 μm) did not affect TDZ-induced OCTN2 expression (Fig. 2). The same results were obtained using rosiglitazone instead of troglitazone. Activation of PPARγ results in a cascade of reactions, including its nuclear translocation and binding to specific DNA sequences at target gene level (30). Therefore, nuclear proteins were extracted to examine the activation of PPARγ by PPAR transfactor assay and to verify whether the decreased expression of OCTN2 was related to a decreased PPARγ nuclear translocation. Cell exposure to TDZ (troglitazone or rosiglitazone) for 4 h resulted in a significant increase in PPARγ activity, whereas GW9662 or BADGE pretreatment inhibited PPARγ activation (Fig. 2c). To further confirm the role of endogenous PPARγ on the expression of OCTN2 mRNA, the cells were transfected with expression vectors containing γORF4 cDNA and then treated or not with TDZs. γORF4 is a PPARγ isoform, generated by PPARγ alternative splicing, that has impaired transcriptional potential and dominant negative activity. We have previously demonstrated by analysis of the cytosol and nuclear PPARγ level that γORF4 transfection significantly inhibited the PPARγ DNA binding capacity and, thus, the amount of nuclear PPARγ (31). The γORF4 capacity to inhibit PPARγ activation also remained significant in γORF4-transfected cells after TDZs treatment. In γORF4-transfected colonocytes, OCTN2 expression analysis extended and confirmed the finding that the functional inactivation of the endogenous PPARγ by γORF4 significantly decreased OCTN2 expression (Fig. 3).
FIGURE 3.
Inhibition of PPARγ activity by γORF4, a dominant negative PPARγ isoform, decreases OCTN2 expression. PPARα−/− MCE cells transfected with empty or γORF4 expression vector (250 ng) were treated with troglitazone (TRO; 50 μm) or rosiglitazone (RO; 5 μm) for 24 h. Relative mRNA levels of OCTN2 were determined by using qRT-PCR. Results are the mean ± S.E. of five experiments. *, p < 0.01 versus control (C).
Carnitine Uptake
We examined the [3H]carnitine uptake in MCE and PPARα−/− MCE cell lines before and after TDZs treatment. As expected, TDZs-treated cells, independent of PPARα expression, accumulated a larger amount of carnitine than untreated cells (41.5 ± 1.7 versus 25.8 ± 1.3 pmol/30 min/mg of protein, respectively; p < 0.05). The carnitine uptake was inhibited by unlabeled carnitine, acetylcarnitine, and tetraethylammonium or by substitution of sodium, present in the transport medium, with N-methylglucamine.
Identification of a Putative PPARγ Binding Site in the OCTN2 Gene
Sequence analysis of the human OCTN2 gene revealed no typical binding site for PPARs. As previous characterization of PPREs has shown that a wide range of PPREs that diverge significantly from the canonical binding motif are also found to be active in various contexts (32), we applied an improved in silico screening approach based on the response element classification schema described in M. Heinäniemi et al. (33). This method integrates both experimental and in silico-derived evidence and has been demonstrated to have better sensitivity for PPAR binding site detection compared with traditional approaches.
The analysis was carried out on a chromosomal region containing the TSS of the human OCTN2 gene plus 10,000 nucleotides upstream and downstream from the TSS. These parameters were the same used by Heinäniemi et al. (33) for their study. This analysis led to the identification of 11 potential PPREs, with different degrees of predicted binding specificity (Table 1). In particular one motif, localized in the first intron at 2885 nucleotides below the TSS, was predicted to be highly specific for PPARγ. This site is also integrated in a genomic region of ∼350 nucleotides, highly conserved in 31 eutherian mammals, as reported by the ENSEMBL web site, thus reinforcing the idea of a potential functional role of this sequence. Notably, on the basis of this prediction, this motif can also bind PPARα with good efficiency and, to a lesser extent, PPARβ/δ.3 Based on this insight, it was decided to functionally characterize the identified sequences.
Characterization of the Predicted PPRE
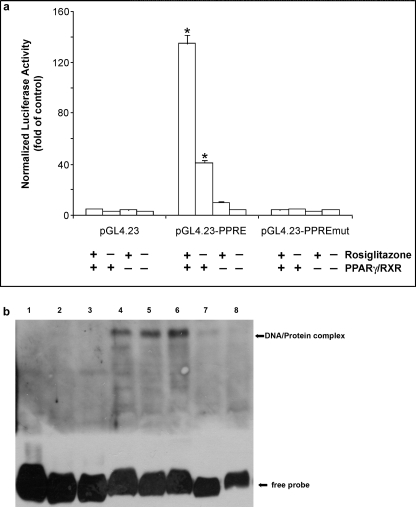
Next, we evaluated whether the putative PPRE identified were able to mediate the transactivation of OCTN2 by PPARγ. Thus, we generated different OCTN2 reporter gene constructs, each containing the selected PPRE, and transiently transfected these OCTN2 reporter constructs into PPARα−/− MCE cell line with co-transfection of either pCMX-human PPARγ and pCMX-human RXRα or empty vector (pCMX). Only in cells transiently transfected with the reporter construct PPRE located in the first OCTN2 intron at 2885 position (Table 1), the luciferase activity increased ∼40-fold by co-expression of human PPARγ/RXRα and about 135-fold by co-expression of human PPARγ/RXRα and subsequent stimulation with rosiglitazone when compared with cells cultured without rosiglitazone and without co-expression of human PPARγ/RXRα (p < 0.01; Fig. 4a). Because the luciferase activity remained at basal level without any increase in response to rosiglitazone and/or co-expression of human PPARγ/RXRα in cells transiently transfected with the other reporter constructs, these data were not depicted in Fig. 4. To further evaluate whether the putative PPRE identified in the first intron of mouse OCTN2 is functional, we carried out gel shift assays (EMSA) using in vitro translated PPARγ and RXRα and DIG-labeled oligonucleotide corresponding to the selected PPRE as well as its mutant counterpart. DNA-protein complex formation between the oligonucleotide corresponding to the PPRE and in vitro translated PPARγ/RXRα heterodimer was obtained (Fig. 4b). No DNA-protein complex formation was observed between the construct PPRE harboring a mutation in PPRE and in vitro translated PPARγ/RXRα heterodimer (Fig. 4b). These results indicated that the DNA sequence identified within the OCTN2 gene was a functional responsive element and that PPARγ was able to activate OCTN2 expression through this PPRE.
FIGURE 4.
Functional analysis of the putative PPRE of human OCTN2 intron. a, shown is the effect of exogenous human PPARγ/RXR and PPARγ ligand rosiglitazone on transcriptional activity of OCTN2 intron reporter constructs. PPARα−/− MCE cells were transiently transfected with wild-type or mutated OCTN2 intron reporter constructs, and a renilla luciferase expression vector for normalization. Cells were also co-transfected with or without (empty vector) expression vectors for human PPARγ and RXR. After transfection, cells were stimulated or not with 5 μm rosiglitazone for 24 h and lysed. Afterward, luciferase activities were determined (see “Materials and Methods”). Results represent the means ± S.E. for one of three independent experiments each performed in triplicate. *, p < 0.01 versus control. b, binding of in vitro translated human PPARγ/RXR to the putative PPRE of human OCTN2 intron is shown. EMSA was performed using in vitro translated human PPARγ/RXR and DIG-labeled oligonucleotide corresponding to either wild-type or mutated putative PPRE. Cold competition assays were done by adding molar excess of the unlabeled specific (unlabeled OCTN2-PPRE) or nonspecific probe (corresponding to random oligonucleotides of OCTN2 intron 1) to the binding reactions. DIG-labeled specific probe corresponding to rat-ACO-PPRE oligonucleotides was used as positive control. Lane 1, DIG-labeled OCTN2-PPRE w/o PPARγ/RXR; lane 2, PPARγ/RXR+NS probe; lane 3, DIG-labeled mutated OCTN2-PPRE+PPARγ/RXR; lane 4, DIG-labeled ACO-PPRE+PPARγ/RXR; lane 5, DIG-labeled OCTN2-PPRE+PPARγ/RXR; lane 6, as in lane 5 but with 100-fold excess of NS probe; lane 7, DIG-labeled OCTN2-PPRE+PPARγ/RXR+10-fold excess of unlabeled OCTN2-PPRE; lane 8, as in lane 3 but with a 50-fold excess of unlabeled OCTN2-PPRE.
Effects of PPARγ Agonist and Antagonist on the Expression of OCTN2 Gene in Vivo
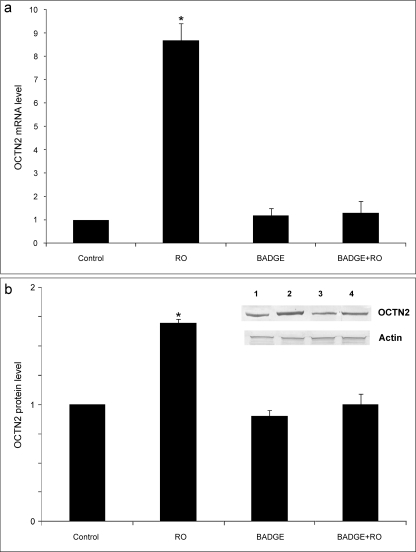
To determine whether OCTN2 is a target gene of PPARγ in vivo, PPARα−/− mice were treated with or without rosiglitazone (20 mg/kg/day) for 3 days, and the colon OCTN2 expression, at both mRNA and protein levels was measured (Fig. 5). The qRT-PCR (Fig. 5a) showed that the OCTN2 mRNA level was significantly increased in the colon epithelium of rosiglitazone-treated mice compared with the control group independently of PPARα expression. Similarly, the protein level of OCTN2 was also increased by rosiglitazone (Fig. 5b). To examine whether a different PPARγ agonist elicited the same effect in vivo, mice were also treated with troglitazone. The results demonstrated that troglitazone at a higher dose (200 mg/kg/day) elicited a similar induction of OCTN2 gene expression in the colon (data not shown). Importantly, treatment with the PPARγ antagonist BADGE (10 mg/kg/day) significantly decreased the TZD-induced OCTN2 expression, suggesting that PPARγ activation is required for the OCTN2 induction by TZD in vivo (Fig. 5). The same results were obtained using wild-type mice instead of PPARα−/− mice.
FIGURE 5.
TDZ increases in vivo the expression of OCTN2 in mouse colon epithelium. a, to assess OCTN2 mRNA expression levels, qRT-PCR was carried out with the use of total RNA of colon epithelium from the PPARα−/− mice (control) and those given rosiglitazone (RO), BADGE, or both for 3 days. There were three animals in each group. b, protein samples were isolated from the colon epithelium of the same animals and subjected to Western blotting using antibodies against OCTN2 or β-Actin (lane 1, control; lane 2, rosiglitazone; lane 3; BADGE; lane 4, RO+BADGE). The results of immunoblots were scanned, quantified using image analysis, and expressed as -fold increase compared with the control. Data represent the means±S.E. of three independent experiments. *, p < 0.01 versus control.
Induction of Colonic Expression of OCTN2 Gene by Adenovirus-mediated Overexpression of Constitutively Active Mutant of PPARγ
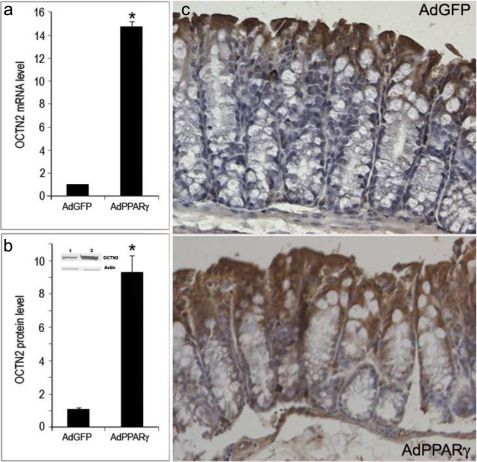
To assess the effect of PPARγ signaling in vivo without using a ligand that could influence additional pathways, a constitutively active mutant of PPARγ was created by fusing the activation domain of the herpes simplex virus Vp16 protein to the γ1 isoform of PPARγ. This mutant can transactivate a PPAR reporter to a similar magnitude as that observed with the natural receptor in the presence of ligand. To further investigate whether PPARγ activation is sufficient to induce the OCTN2 expression in vivo, the constitutively active PPARγ was adenovirally infected (AdPPARγ) into the colon of PPARα knock-out mice. As shown by qRT-PCR and Western blotting, AdPPARγ but not AdGFP (control) infection achieved the overexpression of PPARγ in colonocytes, which subsequently resulted in a marked increase in OCTN2 both at mRNA and protein levels (Fig. 6, a and b). The immunohistochemical studies also demonstrated that infection of AdPPARγ clearly induced OCTN2 expression (Fig. 6c). There was no staining when the primary antibody was omitted (not shown). Therefore, the results suggest that PPARγ activation is sufficient to induce the OCTN2 expression in situ. Interestingly the methodology we used to infuse viruses avoided the expression of PPARγ in other part of gastrointestinal tract or in extraintestinal tissues.
FIGURE 6.
Adenovirus-mediated overexpression of PPARγ increases the expression of OCTN2 in mouse colon epithelium. Mice were intra-colon injected with Ad-GFP or Ad-PPARγ. Total RNA and protein were extracted 48 h later and subjected to qRT-PCR for the detection of OCTN2 mRNA (a) or Western blotting for OCTN2 (b, lane 1, Ad-GFP; lane 2, Ad-PPARγ). The results were expressed as -fold increase compared with the control. *, p < 0.01 versus control. c, immunohistochemical staining shows that overexpression of PPARγ resulted in an increased expression of OCTN2 protein in colon epithelium (original magnification ×200).
Plasma Carnitine Concentration in PPARα−/− Mice Treated with TDZ or Overexpressing Colon PPARγ
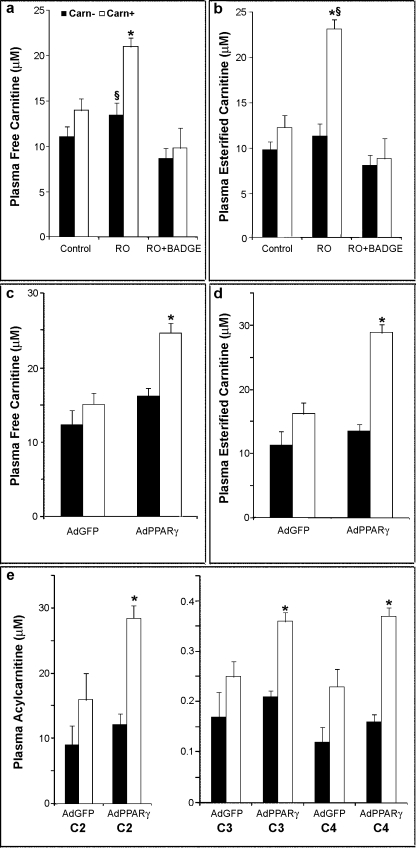
To elucidate whether the OCTN2 expression in colon epithelium maintained its physiological role of carnitine carrier, we analyzed carnitine and carnitine derivatives in the plasma of rosiglitazone- or rosiglitazone- and BADGE-treated mice and in AdPPARγ- or AdGFP-infected mice before and after the supplementation of carnitine in drinking water. Carnitine supplementation did not affect blood glucose, non-esterified fatty acids, ketones, or insulin in all the groups of mice. The supplementation regimen increased plasma levels of free carnitine in all animal groups; however, the increase was significant only in the rosiglitazone-treated mice and in AdPPARγ mice (Fig. 7, a and c). An increase in plasma total acylcarnitine concentration was observed in all groups, but it was significant only in rosiglitazone-treated mice and in AdPPARγ mice (Fig. 7, b and d). Co-treatment of BADGE counteracted the rosiglitazone-induced increase of plasma-free carnitine and plasma-esterified carnitine (Fig. 7, a and b). It is interesting to note that the increase of plasma-esterified carnitine was mainly due to short-chain acylcarnitines (namely acetyl-, propionyl-, and butyryl-carnitine), which showed a significant increase in AdPPARγ mice if compared with AdGFP controls (Fig. 7e).
FIGURE 7.
Plasma carnitine and acylcarnitine profiles after carnitine supplementation. Carnitine (Carn, a and c), and total esterified carnitines (b and d) were measured in plasma from untreated (Control) and treated (rosiglitazone (RO), RO+BADGE, AdGFP or AdPPARγ) PPARα−/− mice after consuming carnitine-supplemented or vehicle-treated drinking water for 1 week. Short chain acylcarnitines (e) were measured in AdGFP or AdPPARγ treated PPARα−/− mice. Data are presented as the mean ± S.E. from six animals per group. *, p < 0.01 versus control. §, p < 0.01 versus RO+BADGE. C2, acetylcarnitine; C3, propionylcarnitine; C4, butyrylcarnitine.
DISCUSSION
In this study a new transcriptional mechanism that controls the OCTN2 expression in the large intestine is found by demonstrating that OCTN2 is a target gene of PPARγ in cultured cell lines as well as in mouse colon in vivo. This finding is supported by both the loss-of-function and the gain-of-function approaches using the agonist and antagonist of PPARγ and suitable animal models. TZD induction of OCTN2 was significantly decreased by specific PPARγ antagonists, BADGE and GW9662, both in vitro and in vivo or the inhibition of endogenous PPARγ activity by γORF4. This effect was not attenuated by knockdown of endogenous PPARα or by PPARβ inhibitor. On the contrary, PPARγ agonists and the ligand-independent constitutive PPARγ activation resulted in the induction of OCTN2. PPARγ and the other members of this nuclear receptor family are believed to share a common mechanism of action. It appears to function as obligate heterodimers with RXRs and to bind as PPAR/RXR dimers to characteristic DNA sequences, the so-called PPREs, within the target genes region. Although in silico analysis of the promoter region of rat OCTN2 revealed several putative PPRE upstream of the transcription start site (17), a recent study employing reporter gene and gel shift assays concluded that the contribution of this proximal PPRE to the OCTN2 promoter activity is low and that a more functional PPRE might be located in other regulatory regions of the OCTN2 gene (36). By using bioinformatic analysis we have identified several putative PPRE with high similarity to the conserved consensus PPRE within the OCTN2 gene region. We have demonstrated that one of these putative PPRE contained in the first intron of OCTN2 human gene is recognized by PPARγ and may mediate its inductive effect. Moreover, the reporter assay showed that this noncanonical PPRE is functionally activated by TZD, and the activation is abolished by the PPARγ antagonist GW0742.
The fact that OCTN2 gene is transcriptionally induced by PPARγ via functional PPRE located in an intronic region of the OCTN2 gene is not so uncommon as regulatory elements are frequently observed in intronic regions (37, 38). Noncanonical motifs for PPARγ have been identified in many PPARγ target genes that apparently did not show any consensus PPRE within their regulatory region (17). More importantly, our analysis of OCTN2 gene has demonstrated that this noncanonical intronic PPRE is highly conserved in different species (i.e. rat and mouse) and can bind with good efficiency also PPARα and, to a lesser extent, PPARβ/δ.3 This is indicative of the importance of PPAR for transcription regulation of OCTN2 gene in different tissues and organs and probably represents an evolutionarily conserved mechanism with relevant physiological significance. It has been reported that juvenile visceral steatosis mice, characterized by loss-of-function/mutation in the OCTN2 carnitine transporter, spontaneously developed intestinal villous atrophy, breakdown, and inflammation with intense lymphocytic and macrophage infiltration, leading to ulcer formation and gut perforation (40). All these histopathological findings were associated with an increased apoptosis of gut epithelial cells. We have previously demonstrated that gene expression of OCTN2 was reduced in rats with experimental colitis. The down-regulation of the OCTN2 gene in the inflamed colon could exacerbate the epithelial damage through the impairment of mitochondrial β-oxidation of fatty acids, a metabolic pathway regulated by a carnitine-dependent mechanism (21). When activated by TZD, PPARγ reduced mucosal damage and inflammation in animal models of colon inflammation (41, 42). Furthermore, PPARγ natural and synthetic ligands were equally effective in the treatment of chronic and acute colitis (42), and the beneficial effects were reflected in reduced mortality and a lower intensity of lesion in experimental animals (43). An association between decreased PPARγ expression and ulcerative colitis has been demonstrated (43), whereas short-term therapy with rosiglitazone resulted in clinical remission for patients with active ulcerative colitis (44). Given our results that OCTN2 is a colon target of PPARγ, it is suggested that the up-regulation of OCTN2 expression by PPARγ may contribute to the protective effects of TZDs in inflammatory bowel disease. These findings may have important pathophysiological significance because they link the TZD efficacy not only to a direct effect of PPARγ activation on local immune response but also to an indirect effect secondary to an improved colonocyte metabolism. Intriguingly, the positive feedback loop between PPARγ activation and OCTN2 expression could be strongly regulated by the enteric microbioma. PPARγ expression in colon epithelial cells is associated with intestinal microorganisms. LPS and Saccharomyces boulardii increase PPARγ expression in colon epithelial cells (HT-29 and Caco-2) (45). Extremely low levels of PPARγ have been reported in mouse colon that has nonfunctional Toll-like receptor 4 (TLR4) (45). Thus, LPS recognition of TLR4 likely is involved in the enhancement of PPARγ expression by microorganisms. These results indicate the pivotal role of bacteria in the regulation of PPARγ expression in colon epithelial cells. This may account for the PPARγ expression pattern observed in the colon compared with other parts of the digestive tract. Fujiya et al. (22) revealed that a B. subtilis quorum-sensing signal molecule, the competence- and sporulation-stimulating factor (CSF), is internalized via OCTN2 into the colonocytes where it induces the heat shock protein Hsp27, which protects intestinal cells against oxidant-mediated tissue damage and loss of barrier function. Our results may supply the missing piece to complete the molecular connections between bacteria, quorum-sensing signal molecules, and human host in the colon ecological niche. Thus, colon PPARγ provides the host with the ability to respond or adapt to changes in the microbiome, inducing OCTN2 expression and protecting intestinal epithelial cells via competence- and sporulation-stimulating factor (CSF) and carnitine uptake; in this regard, PPARγ could potentially mediate some actions of probiotic microorganisms. Because PPARγ is an important therapeutic target for the treatment of ulcerative colitis, the functional role of OCTN2 regulation by PPARγ warrants further investigation in colon inflammation. Finally, our data demonstrate that OCTN2 expressed in the large intestine can contribute to intestinal carnitine absorption and systemic carnitine homeostasis. Previous studies investigating the intestinal absorption of carnitine after single oral doses have shown that carnitine is preferentially adsorbed in the small intestine with the oral bioavailability reported as ∼18 and 5% of 2 and 6 g, respectively (34, 39). However, limitations in the study design must be considered during interpretation of the published data. For example, these studies did not investigate the absorption of oral carnitine from small and large intestine when carnitine was administered as a repeated dose regimen. In addition, because carnitine is rapidly taken up from the intestinal lumen, but it is only slowly released into the circulation after an extensive intracellular acylation, it is important to analyze after oral carnitine administration not only the blood level of carnitine and its derivatives but also the amount of carnitine stored in the enterocytes (39). The effects of TZDs and fenofibrate on intestinal carnitine absorption in patients using these drugs have not been evaluated. In addition, there are no studies about the effects of TZDs and fenofibrate on absorption of oral carnitine in normal volunteers in whom the confounding effects of insulin sensitization and associated changes in metabolic parameters are minimized. We might also predict that carnitine absorption through colon epithelium would achieve greater importance when OCTN2 in the small intestine is compromised (35) and also during therapeutic supplementation with oral doses of carnitine as in this case the absolute bioavailability is very low (34). In conclusion, we demonstrated that PPARγ rather than PPARα modulates OCTN2 gene expression in the colon with important implications in colonocyte welfare and carnitine homeostasis.
G. Paolella, personal observation.
- OCTN
- organic cation transporters
- PPARα
- peroxisome proliferator-activated receptor-α
- BADGE
- bisphenol A diglycidyl ether
- PPRE
- PPAR response elements
- RXR
- retinoid X receptor
- DIG
- digoxigenin
- Ad
- adenoviruse
- TZD
- thiazolidinediones
- qRT
- quantitative RT
- TSS
- transcription start site.
REFERENCES
- 1.Rebouche C. J., Seim H. (1998) Annu. Rev. Nutr. 18, 39–61 [DOI] [PubMed] [Google Scholar]
- 2.Rebouche C. J. (2004) Ann. N.Y. Acad. Sci. 1033, 30–41 [DOI] [PubMed] [Google Scholar]
- 3.Lohninger A., Pittner G., Pittner F. (2005) Chem. Month. 136, 1255–1266 [Google Scholar]
- 4.Steiber A., Kerner J., Hoppel C. L. (2004) Mol. Aspects Med. 25, 455–473 [DOI] [PubMed] [Google Scholar]
- 5.Lahjouji K., Mitchell G. A., Qureshi I. A. (2001) Mol. Genet. Metab. 73, 287–297 [DOI] [PubMed] [Google Scholar]
- 6.Tein I. (2003) J. Inherit. Metab. Dis. 26, 147–169 [DOI] [PubMed] [Google Scholar]
- 7.Koepsell H., Lips K., Volk C. (2007) Pharm. Res. 24, 1227–1251 [DOI] [PubMed] [Google Scholar]
- 8.Wu X., George R. L., Huang W., Wang H., Conway S. J., Leibach F. H., Ganapathy V. (2000) Biochim. Biophys. Acta 1466, 315–327 [DOI] [PubMed] [Google Scholar]
- 9.Tamai I., Ohashi R., Nezu J., Yabuuchi H., Oku A., Shimane M., Sai Y., Tsuji A. (1998) J. Biol. Chem. 273, 20378–20382 [DOI] [PubMed] [Google Scholar]
- 10.Alnouti Y., Petrick J. S., Klaassen C. D. (2006) Drug Metab. Dispos. 34, 477–482 [DOI] [PubMed] [Google Scholar]
- 11.Ohashi R., Tamai I., Yabuuchi H., Nezu J. I., Oku A., Sai Y., Shimane M., Tsuji A. (1999) J. Pharmacol. Exp. Ther. 291, 778–784 [PubMed] [Google Scholar]
- 12.Ohashi R., Tamai I., Nezu Ji. J., Nikaido H., Hashimoto N., Oku A., Sai Y., Shimane M., Tsuji A. (2001) Mol. Pharmacol. 59, 358–366 [DOI] [PubMed] [Google Scholar]
- 13.Wu X., Huang W., Prasad P. D., Seth P., Rajan D. P., Leibach F. H., Chen J., Conway S. J., Ganapathy V. (1999) J. Pharmacol. Exp. Ther. 290, 1482–1492 [PubMed] [Google Scholar]
- 14.Slitt A. L., Cherrington N. J., Hartley D. P., Leazer T. M., Klaassen C. D. (2002) Drug Metab. Dispos. 30, 212–219 [DOI] [PubMed] [Google Scholar]
- 15.Xuan W., Lamhonwah A. M., Librach C., Jarvi K., Tein I. (2003) Biochem. Biophys. Res. Commun. 306, 121–128 [DOI] [PubMed] [Google Scholar]
- 16.Nezu J., Tamai I., Oku A., Ohashi R., Yabuuchi H., Hashimoto N., Nikaido H., Sai Y., Koizumi A., Shoji Y., Takada G., Matsuishi T., Yoshino M., Kato H., Ohura T., Tsujimoto G., Hayakawa J., Shimane M., Tsuji A. (1999) Nat. Genet. 21, 91–94 [DOI] [PubMed] [Google Scholar]
- 17.Ringseis R., Pösel S., Hirche F., Eder K. (2007) Pharmacol. Res. 56, 175–183 [DOI] [PubMed] [Google Scholar]
- 18.Koch A., König B., Stangl G. I., Eder K. (2008) Exp. Biol. Med. 233, 356–365 [DOI] [PubMed] [Google Scholar]
- 19.Braissant O., Foufelle F., Scotto C., Dauça M., Wahli W. (1996) Endocrinology 137, 354–366 [DOI] [PubMed] [Google Scholar]
- 20.Meier Y., Eloranta J. J., Darimont J., Ismair M. G., Hiller C., Fried M., Kullak-Ublick G. A., Vavricka S. R. (2007) Drug Metab. Dispos. 35, 590–594 [DOI] [PubMed] [Google Scholar]
- 21.D'Argenio G., Calvani M., Casamassimi A., Petillo O., Margarucci S., Rienzo M., Peluso I., Calvani R., Ciccodicola A., Caporaso N., Peluso G. (2006) FASEB J. 20, 2544–2546 [DOI] [PubMed] [Google Scholar]
- 22.Fujiya M., Musch M. W., Nakagawa Y., Hu S., Alverdy J., Kohgo Y., Schneewind O., Jabri B., Chang E. B. (2007) Cell. Host Microbe 1, 299–308 [DOI] [PubMed] [Google Scholar]
- 23.Auboeuf D., Rieusset J., Fajas L., Vallier P., Frering V., Riou J. P., Staels B., Auwerx J., Laville M., Vidal H. (1997) Diabetes 46, 1319–1327 [DOI] [PubMed] [Google Scholar]
- 24.Kaiser G. C., Polk D. B. (1997) Gastroenterology 112, 1231–1240 [DOI] [PubMed] [Google Scholar]
- 25.Corredor J., Yan F., Shen C. C., Tong W., John S. K., Wilson G., Whitehead R., Polk D. B. (2003) Am. J. Physiol. Cell. Physiol. 284, C953–C961 [DOI] [PubMed] [Google Scholar]
- 26.Whitehead R. H., VanEeden P. E., Noble M. D., Ataliotis P., Jat P. S. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 587–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sahi J., Goldstein J. L., Layden T. J., Rao M. C. (1994) Am. J. Physiol. Gastrointest. Liver Physiol. 266, 846–855 [Google Scholar]
- 28.Laplante M., Festuccia W. T., Soucy G., Gélinas Y., Lalonde J., Berger J. P., Deshaies Y. (2006) Diabetes 55, 2771–2778 [DOI] [PubMed] [Google Scholar]
- 29.Diehl K. H., Hull R., Morton D., Pfister R., Rabemampianina Y., Smith D., Vidal J. M., van de Vorstenbosch C. (2001) J. Appl. Toxicol. 21, 15–23 [DOI] [PubMed] [Google Scholar]
- 30.Rousseaux C., Lefebvre B., Dubuquoy L., Lefebvre P., Romano O., Auwerx J., Metzger D., Wahli W., Desvergne B., Naccari G. C., Chavatte P., Farce A., Bulois P., Cortot A., Colombel J. F., Desreumaux P. (2005) J. Exp. Med. 201, 1205–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabatino L., Casamassimi A., Peluso G., Barone M. V., Capaccio D., Migliore C., Bonelli P., Pedicini A., Febbraro A., Ciccodicola A., Colantuoni V. (2005) J. Biol. Chem. 280, 26517–26525 [DOI] [PubMed] [Google Scholar]
- 32.Degenhardt T., Matilainen M., Herzig K. H., Dunlop T. W., Carlberg C. (2006) J. Biol. Chem. 281, 39607–39619 [DOI] [PubMed] [Google Scholar]
- 33.Heinäniemi M., Uski J. O., Degenhardt T., Carlberg C. (2007) Genome Biol. 8, R147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans A. M., Fornasini G. (2003) Clin Pharmacokinet. 42, 941–967 [DOI] [PubMed] [Google Scholar]
- 35.Ciacci C., Peluso G., Iannoni E., Siniscalchi M., Iovino P., Rispo A., Tortora R., Bucci C., Zingone F., Margarucci S., Calvani M. (2007) Dig. Liver Dis. 39, 922–928 [DOI] [PubMed] [Google Scholar]
- 36.Maeda T., Wakasawa T., Funabashi M., Fukushi A., Fujita M., Motojima K., Tamai I. (2008) Biol. Pharm. Bull. 31, 1230–1236 [DOI] [PubMed] [Google Scholar]
- 37.Mandard S., Stienstra R., Escher P., Tan N. S., Kim I., Gonzalez F. J., Wahli W., Desvergne B., Müller M., Kersten S. (2007) Cell. Mol. Life Sci. 64, 1145–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schupp M., Lefterova M. I., Janke J., Leitner K., Cristancho A. G., Mullican S. E., Qatanani M., Szwergold N., Steger D. J., Curtin J. C., Kim R. J., Suh M. J., Suh M., Albert M. R., Engeli S., Gudas L. J., Lazar M. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 1105–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hamilton J. W., Li B. U., Shug A. L., Olsen W. A. (1986) Gastroenterology 91, 10–16 [DOI] [PubMed] [Google Scholar]
- 40.Shekhawat P. S., Srinivas S. R., Matern D., Bennett M. J., Boriack R., George V., Xu H., Prasad P. D., Roon P., Ganapathy V. (2007) Mol. Genet. Metab. 92, 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dubuquoy L., Rousseaux C., Thuru X., Peyrin-Biroulet L., Romano O., Chavatte P., Chamaillard M., Desreumaux P. (2006) Gut 55, 1341–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Desreumaux P., Dubuquoy L., Nutten S., Peuchmaur M., Englaro W., Schoonjans K., Derijard B., Desvergne B., Wahli W., Chambon P., Leibowitz M. D., Colombel J. F., Auwerx J. (2001) J. Exp. Med. 193, 827–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dubuquoy L., Jansson E. A., Deeb S., Rakotobe S., Karoui M., Colombel J. F., Auwerx J., Pettersson S., Desreumaux P. (2003) Gastroenterology 124, 1265–1276 [DOI] [PubMed] [Google Scholar]
- 44.Lewis J. D., Lichtenstein G. R., Deren J. J., Sands B. E., Hanauer S. B., Katz J. A., Lashner B., Present D. H., Chuai S., Ellenberg J. H., Nessel L., Wu G. D. (2008) Gastroenterology 134, 688–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang K., Wan Y. J. Y. (2008) Exp. Biol. Med. 233, 496–506 [DOI] [PMC free article] [PubMed] [Google Scholar]