Abstract
Acid-sensing ion channel 1 (ASIC1) is a H+-gated channel of the amiloride-sensitive epithelial Na+ channel (ENaC)/degenerin family. ASIC1 is expressed mostly in the central and peripheral nervous system neurons. ENaC and ASIC function is regulated by several serine proteases. The type II transmembrane serine protease matriptase activates the prototypical αβγENaC channel, but we found that matriptase is expressed in glioma cells and its expression is higher in glioma compared with normal astrocytes. Therefore, the goal of this study was to test the hypothesis that matriptase regulates ASIC1 function. Matriptase decreased the acid-activated ASIC1 current as measured by two-electrode voltage clamp in Xenopus oocytes and cleaved ASIC1 expressed in oocytes or CHO K1 cells. Inactive S805A matriptase had no effect on either the current or the cleavage of ASIC1. The effect of matriptase on ASIC1 was specific, because it did not affect the function of ASIC2 and no matriptase-specific ASIC2 fragments were detected in oocytes or in CHO cells. Three matriptase recognition sites were identified in ASIC1 (Arg-145, Lys-185, and Lys-384). Site-directed mutagenesis of these sites prevented matriptase cleavage of ASIC1. Our results show that matriptase is expressed in glioma cells and that matriptase specifically cleaves ASIC1 in heterologous expression systems.
Keywords: Acid-sensing Ion Channels (ASIC), Gel Electrophoresis, Serine Protease, Site-directed Mutagenesis, Xenopus, Matriptase, Two-electrode Voltage Clamp
Introduction
Acid-sensing ion channels (ASICs)2 are H+-gated members of the epithelial Na+ channel/degenerin (ENaC/Deg) family of amiloride-sensitive ion channels (1). Thus far, four ASIC genes have been cloned, ASIC1–ASIC4 (2). Human ASIC1 exists as two isoforms, ASIC1a and ASIC1b, with hASIC1a containing an extra 43 amino acids in the region just before the start of the second transmembrane domain (3). The recent crystallization of chicken ASIC1 has revealed that ASIC1 is likely to be a trimer. Each ASIC1 subunit consists of short intracellular N and C termini, two transmembrane domains, and a large extracellular loop, which in ASICs contains the pH-sensing region (4). ASICs are expressed mainly in the neurons of the central and peripheral nervous systems, where they are implicated in physiological functions such as learning and memory and pathologies such as neurodegenerative diseases and ischemia (1). ASIC1 is also expressed in glioblastoma multiforme (GBM) cells, highly invasive and proliferative primary brain tumors, where it plays an important role in cell migration and proliferation (5–8).
Several serine proteases modulate endogenous ENaC currents and ENaC/Deg subunits in in vitro expression systems (9). The large extracellular loops of ASICs and ENaCs contain many arginines and lysines that form putative sites for cleavage by serine proteases. For example, proteolytic cleavage of αENaC and γENaC subunits by furin convertases occurs in the trans-Golgi apparatus during ENaC maturation. These channels may be clipped further by secreted serine proteases (e.g. trypsin) or membrane-bound proteases (e.g. prostasin). Noncleaved αβγENaC channels also traffic to the plasma membrane, where they provide a pool of “near silent” channels, ready to be cleaved into active ones (9–13). The exact mechanism of how cleavage activates ENaC is incompletely understood, but one hypothesis is that cleavage removes inhibitory peptides from αENaC and γENaC (12). An alternative mechanism of ENaC activation by cleavage, proposed by Hu et al. (14), involves loss of the αENaC N terminus (including the first transmembrane domain) from the channel complex. Cleavage by proteases does not change the number of ENaC channels at the surface but rather increases channel open probability and is a mechanism for regulating ENaC activity (13). The sites for cleavage of ENaCs by many proteases such as prostasin, furin, and trypsin have been identified and are located in the N-terminal part of the extracellular loop (12).
ASIC1 homomers or ASIC1-containing heteromeric ASICs can also be modified by serine proteases such as trypsin, chymotrypsin, and proteinase K. Protease treatment disrupts the Psalmopoeus cambridgei venom block of heterologously expressed ASIC1 channels, decreases the peak acid-activated current, and shifts the pH50 of activation to a more acidic pH (15, 16). Because ASICs inactivate quickly upon exposure to a low pH, this shift could be important in extracellular acidosis, where a sustained decrease in extracellular pH could inactivate ASICs. The only known site of ASIC cleavage by proteases that has been identified is the trypsin cleavage site at Arg-145 (16).
Recently, the serine protease matriptase (also known as channel-activating protease 3 (CAP3)) has been identified as a modulator of ENaC activity in vitro. When matriptase is co-expressed with αβγENaC subunits in Xenopus laevis oocytes, it causes a 10-fold increase in INa (17, 18). However, this functional effect of matriptase on ENaC has not been correlated with the presence of ENaC proteolytic cleavage fragments.
Matriptase is an 80–90-kDa type II transmembrane protease that belongs to the S1 family of trypsin-like serine proteases. It contains an extracellular C-terminal catalytic domain and an intracellular N terminus. It was first described as a gelatinolytic activity in cultured breast cancer cells and was isolated in a complex with its cognate inhibitor, hepatocyte growth factor activator inhibitor-1 (HAI-1) from human milk (19). Matriptase is synthesized as an inactive, single-chain zymogen. Its activation is complex; it is first cleaved in the secretory pathway by an unknown protease at Gly-149, and later at Arg-614 in the serine protease domain, after it reaches the surface. This last cleavage converts matriptase into the active protease. Mutations in any of the catalytic triad (His-656, Asp-711, and Ser-805) amino acids render matriptase inactive by making it unable to undergo the activation site cleavage at Arg-614 (20, 21).
Matriptase is an epithelial protease and has important physiological functions. It is crucial for epidermal barrier formation and is involved in hair follicle growth and thymocyte development. In addition, matriptase has been implicated in many epithelial cancers. It is consistently expressed in human epithelial tumors of the head, neck, mesothelium, breast, ovary, cervix, prostate, lungs, and gastrointestinal tract, and its overexpression in keratinocytes results in spontaneous squamous cell carcinomas. In most tumors matriptase RNA and protein are up-regulated, and there is a positive correlation between matriptase expression and tumor grade (20, 22). Interestingly, matriptase inhibition in vitro or in xenografted tumors with siRNAs or antisense oligodeoxyribonucleotides decreases invasion (20, 22, 23). Additionally, overexpression of the serine protease inhibitor HAI-1 suppresses the in vitro invasion of glioblastoma cells (24). However, the target of HAI-1 in these assays is not known, because HAI-1 inhibits several other serine proteases in addition to matriptase.
The goal of this study was to test the hypothesis that matriptase can modulate the activity of ASIC1 channels through proteolytic cleavage. Matriptase cleaved ASIC1 when the two were co-expressed in X. laevis oocytes or CHO cells. Matriptase decreased ASIC1 function but had no effect on the function of ASIC2. The effects of matriptase on the function and proteolytic cleavage of ASIC1 could be prevented by mutagenesis of three matriptase recognition sites on the ASIC1 extracellular loop.
EXPERIMENTAL PROCEDURES
RNA Extraction and Reverse Transcription-PCR
Total RNAs were isolated from freshly excised human tissues (obtained from Birmingham Neurosurgery Brain Tissue Bank under Institutional Review Board approval) using TRIzol (Invitrogen) and following the manufacturer's instructions as described previously (5). Total RNAs from human primary cells or human cell lines were isolated with the RNeasy RNA extraction kit (Qiagen) as specified by the manufacturer. The integrity and quality of the isolated RNAs were checked with denaturing agarose-formaldehyde gel electrophoresis. The RT-PCR reaction was done using a One-Step RT-PCR kit (Qiagen) with 500 ng of RNA and each primer at 0.6 μm. The matriptase forward and reverse primer sequences were 5′-CACAAGGAGTCGGCTGTGAC-3′ (forward) and 5′-GAGGGTAGGTGCCACACAA-3′ (reverse). Standard RT-PCR conditions were used: at 50 °C, 1 cycle for 30 min; at 95 °C, 1 cycle for 15 min; at 94 °C 1 min, 54 °C 1 min, and 72 °C 1 min, for 35 cycles; at 72 °C, 1 cycle for 10 min. The RT-PCR product was visualized by electrophoresis in a 2% agarose gel. A negative control with no RNA in the RT-PCR reaction was included with each experiment to guard against contamination.
Cell Culture and Transfections
Primary non-tumor human astrocytes isolated from astrogliosis regions and primary human GBMs were obtained from the University of Alabama at Birmingham Neurosurgery Brain Tissue Bank (Institutional Review Board Approval X050415007). The cell lines used (U87MG, D54MG, SKMG, U251MG, and CHO K1) have been described previously (7, 8, 25). All cells were cultured in Dulbecco's modified Eagle's medium/F-12 (1:1) (Hyclone) with 10% fetal bovine serum (Hyclone) and were maintained in a 95% O2, 5% CO2 humidified incubator at 37 °C.
For transfections, CHO K1 cells were split into 6-well tissue culture dishes and transiently transfected with 2 μg of each plasmid DNA and 10 μl of Lipofectamine 2000 reagent (Invitrogen) per well following the manufacturer's protocol. Whole-cell lysates were obtained at 48 h post-transfection as described below.
Plasmids, Tagged Constructs, and Site-directed Mutagenesis
The plasmids used contained the DNA sequence of human ASIC1b (NM_001095), human ASIC2b (NM_001094), and human matriptase (NM_021978). N- or C-terminal fusions of eGFP on hASIC1b and hASIC2b were prepared as described previously (26). The channel sequences were modified with PCR by the addition of 5′-XhoI and 3′-BamHI restriction sites using Pfu polymerase (Stratagene). The constructs were then subcloned into the pEGFP-N1 or pEGFP-C1 vectors (Clontech). The DNA sequences coding for the eGFP fusion channel proteins were subcloned, using NheI and AflII for the C-terminal tags and NheI and BamHI for the N-terminal tag, into the pcDNA 3.1+ vectors (Invitrogen). The ASIC1-hemagglutinin (HA) construct with HA in the extracellular loop between Phe-147 and Lys-148 and the ASIC2-eYFP construct have been described previously (25, 26). For the ASIC2-HA construct, a new BstBI site was added in the extracellular loop of ASIC2 with PCR, and an HA tag was inserted into the BsTB1 site at Thr-239.
Site-directed mutagenesis to introduce point mutations in the matriptase sites on ASIC1 was carried out as described previously using the QuikChange II XL mutagenesis kit (Stratagene), sense and antisense primers, and ASIC1-EGFP-N1 in pcDNA3.1+ as template DNA (25). The primers contained the necessary base changes to mutate the desired arginines (Arg) or lysines (Lys) to alanines (Ala).
The catalytically inactive S805A-matriptase-pcDNA3.1+ was generated using the QuikChange II XL site-directed mutagenesis kit (Stratagene) and primers containing the appropriate base pairs (18). All of the generated mutations were confirmed by DNA sequencing (Heflin Genetics Center, University of Alabama at Birmingham).
Expression in Xenopus Oocytes
Oocytes were isolated surgically from anesthetized female X. laevis frogs and were collagenase-treated, sorted, and maintained as described previously (25). These procedures were in accordance with and approved by the Institutional Animal Care and Use Committee of the University of Alabama at Birmingham. cRNA preparations and injections into oocytes have also been described previously (25). cRNAs were transcribed using the high yield in vitro transcription kit and CAP analog (Ambion). Each oocyte was injected with 12 ng of channel cRNA in 50 nl of H2O with matriptase or with S805A-matriptase cRNAs as indicated in the experiments.
Electrophysiological Analysis
Two-electrode voltage clamp at a holding potential of −60 mV was performed at room temperature 1–4 days post-injection as described previously (25). Oocytes were placed in a recording chamber (500 μl) and impaled with two glass microelectrodes filled with 3 m KCl with resistances of 0.5 to 2 megohms. Whole-cell currents were recorded with a Geneclamp 500B amplifier with a steady-state restore switch modification (Axon Instruments). A SF-77B Perfusion Fast-Step (Warner Instruments) was used to rapidly exchange the solution bathing the oocyte from ND96, pH 7.4 (96 mm NaCl, 1 mm MgCl2, 2 mm CaCl2, 2 mm KCl, and 5 mm HEPES), to ND96, pH 4.0 (same as ND96, pH 7.4, but with 5 mm MES instead of HEPES). The oocyte was exposed to pH 7.4 for 13 s, to pH 4.0 for 13 s to maximally activate the channels, and again to pH 7.4 for 13 s to allow for recovery of the acid-activated channels. To obtain the pH activation curves, oocytes were exposed to sequentially lower activation pHs (pH 7.0, 6.5, 6.0, 5.5, 5.0, and 4.0). The peak current at each activation pH was normalized to the peak pH 4.0 current within each oocyte. The normalized values were fit to the Hill equation, I = Imax/[1+(10−pH50/10−pH)nH], where Imax is the maximal current (current at pH 4.0), pH50 is the pH at which half the maximal current is obtained, and nH is the Hill coefficient. pH50 values and Hill coefficients were obtained separately for each oocyte by means of a “brute force” algorithm. Predicted values for the normalized currents across the range of experimental pH values were obtained by means of the Hill equation, and the sum-squared error between predicted and observed currents for each combination was computed. The Hill coefficient and pH50 combination that minimized the sum-squared error was then selected.
Reagents
The recombinant human matriptase/ST14 catalytic domain was obtained from R&D Systems, Inc. and contained an N-terminal sequence corresponding to the beginning of the catalytic domain with Val-615. Purified and folded psalmotoxin (PcTX-1) peptide was obtained from Peptides International in the trifluoroacetate form and with formed disulfide bonds. The cell-permeable proteasome inhibitor MG132 was from Calbiochem.
Whole-cell Lysates and Immunoblot Analysis
Mammalian cell lines growing on tissue culture dishes were washed twice with ice-cold phosphate-buffered saline (PBS) and incubated in lysis buffer (150 mm NaCl, 5 mm EDTA, 50 mm Tris, pH 7.5, 1% Triton-X-100, and Complete® protease inhibitor mixture (Roche Applied Science)) at 4 °C for at least 30 min. The cells were scraped off the dishes; the lysate was transferred to a microcentrifuge tube and passed through a 21-gauge needle 5–7 times. After centrifugation at 12,000 rpm for 20 min at 4 °C, the supernatants were saved, and 30–50 μg of lysate was used for Western blot. All experiments were repeated at least three times.
Oocyte whole-cell lysates were obtained by lysing oocytes at 2–3 days post-injection in 20 μl of Triton homogenization buffer/oocyte as described previously (25). After protein assay, equal amounts of protein (50 μg) were used for Western blot. All experiments were repeated at least three times.
The lysates were boiled at 95 °C for 7 min in 1× Laemmli sample buffer (62.5 mm Tris HCl, pH 6.8, 25% glycerol, 2% SDS, 10% β-mercaptoethanol, and 0.01% bromophenol blue) and subjected to 8% (for matriptase blots only) or 12% SDS-PAGE with 4% stacking gels. The separated proteins were transferred to Immobilon-P transfer polyvinylidene difluoride (PVDF) membranes (Millipore). Membranes were blocked with 10% nonfat dry milk in Tris-buffered saline with Tween (TBS-T) (100 mm Tris, pH 7.5, 150 mm NaCl, and 0.1% Tween-20) for 30 min to 1 h at room temperature or overnight at 4 °C and probed with the appropriate antibodies in 3% milk in TBS-T overnight at 4 °C or 2 h at room temperature. The following antibodies were used: mouse anti-matriptase/ST14 monoclonal antibody (R&D Systems) at 1:500, mouse anti-GFP monoclonal antibody (Abgent) at 1:2000, and rat anti-HA antibody (Roche Applied Science) at 1:2000. The blots were washed with TBS-T and incubated in goat anti-mouse horseradish peroxidase (HRP)-conjugated antibody (Jackson ImmunoResearch Laboratories, 1:10,000) or goat anti-rat HRP-conjugated antibody (Thermo Scientific, 1:10,000), as appropriate, in 5% milk in TBS-T. The blots were developed in SuperSignal West Pico Substrate (Thermo Scientific) and exposed to x-ray film (Denville).
Data Analysis and Statistics
Data analysis was done with Clampfit 9 (Molecular Devices) or Excel. Data are presented as means ± S.D. or means ± S.E. as noted. Statistical significance was set at p < 0.05, and statistical tests were done with Excel or GraphPad Prism 5. Two-tailed t tests (unpaired or paired) or comparisons of multiple groups with post hoc tests were chosen as appropriate to determine the p values.
RESULTS
Detection of Matriptase RNA and Protein in Glioma Cells
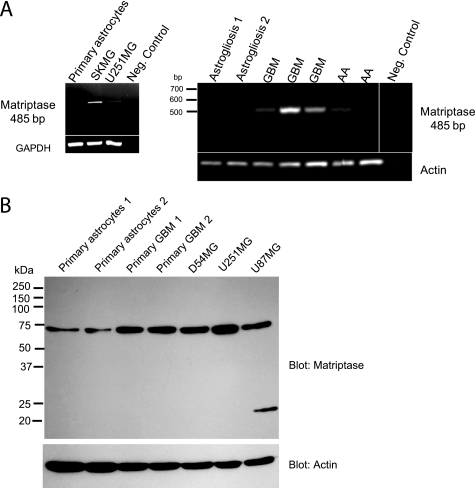
Matriptase expression has been reported in numerous epithelial tissues, both normal and cancerous (20). However, in studies in which matriptase localization was determined by Northern blot analysis of rodent tissues, matriptase RNA was not detected in the normal brain (17, 27). ENaC/Deg subunits are candidate substrates for matriptase, because matriptase increases the amiloride-sensitive current of αβγENaC when the two are co-expressed in Xenopus oocytes (18). Because we have previously shown that glioma cells express ENaC/Deg subunits and because matriptase is involved in malignant progression, we looked for matriptase expression in high grade glioma cells. RT-PCR results showed that matriptase mRNA was expressed in two glioma cell lines (U251 and SKMG), in three different freshly excised GBMs, and in two different freshly resected grade III anaplastic astrocytomas (AA); matriptase RNA was not detected either in primary human astrocytes (from astrogliosis regions) or in astrogliosis tissue (Fig. 1A). Matriptase protein expression in glioma was confirmed by Western blot of glioma cell lines and human primary GBM lysates. Matriptase was detected at ∼75 kDa. Even though we could not detect matriptase mRNA in the primary human normal astrocytes, we could detect matriptase protein there (Fig. 1B); therefore, we cannot rule out that very low levels of matriptase mRNA, undetected by standard RT-PCR, are expressed in normal astrocytes. Western blot data showed that matriptase protein expression was lower in normal astrocytes compared with the primary human GBMs or glioma cell lines (Fig. 1B).
FIGURE 1.
RT-PCR and Western blot detection of matriptase in glioma cell lines and fresh GBM tissues. A, agarose gels of RT-PCR products detect matriptase RNA in SKMG and U251MG cell lines, freshly resected GBM, and anaplastic astrocytoma (AA). A matriptase message is not detected in primary human astrocytes or fresh astrogliosis tissue. No RNA in the RT-PCR reaction is used as a negative control. B, Western blot of whole-cell lysates with a matriptase antibody shows that expression of matriptase in glioma cell lines (D54MG, U251MG, and U87MG) and two different human primary GBM is higher than its expression in two different primary human astrocytes. The blot was probed for actin as a loading control.
Matriptase Inhibits the Function of ASIC1 and Has No Effect on the Function of ASIC2 in Xenopus Oocytes
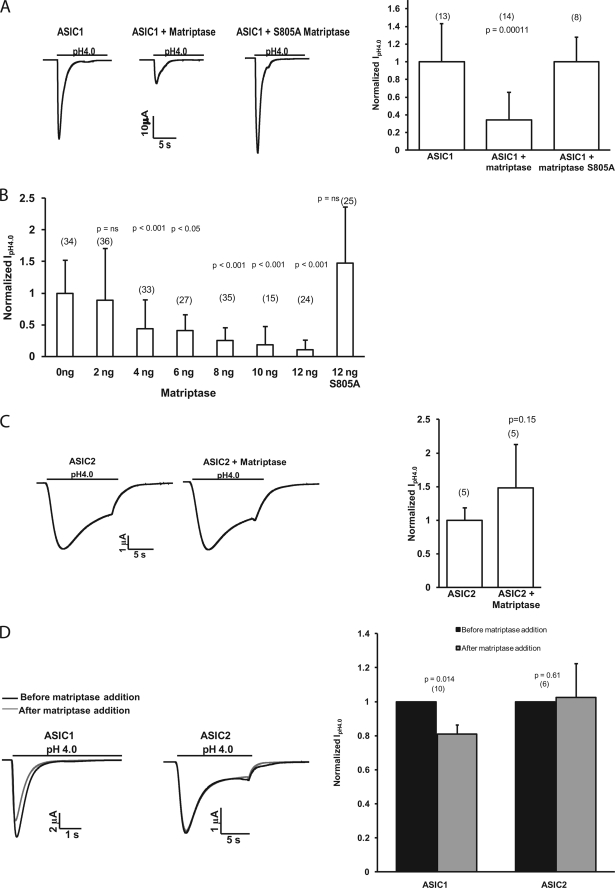
To test the hypothesis that matriptase regulates ASIC1, we assessed ASIC1 function by measuring the whole-cell peak pH 4.0-activated current of oocytes expressing ASIC1 ± matriptase. Unlike its effect on αβγENaC, matriptase decreased the peak pH 4.0 current of ASIC1. A similar effect of proteases on ASIC1 has been reported for trypsin (15). A catalytically inactive matriptase, with a mutation in one of the catalytic triad amino acids (Ser-805 to Ala), did not have an effect on the maximum ASIC1 current (Fig. 2A).
FIGURE 2.
Matriptase decreases the peak acid-activated current of ASIC1 but not that of ASIC2 in Xenopus oocytes. A, peak IpH4.0 were measured by two-electrode voltage clamp in oocytes injected with 12 ng of ASIC1 cRNA alone, ASIC1 with matriptase (8 or 12 ng), or ASIC1 with the catalytically inactive matriptase S805A (8 ng). Representative traces are shown above each graph. Graph values are means ± S.D. The numbers of oocytes measured are from two to three experiments and are shown in parentheses. The nonparametric Kruskal-Wallis test with Dunn's post hoc test was used to determine the p value. B, dose-response of effect of matriptase on the peak IpH4.0 of ASIC1. Oocytes were injected with 12 ng of ASIC1 and increasing amounts of matriptase cRNA. Values are means ± S.D. The numbers of oocytes measured are shown in parentheses and are from five experiments. The nonparametric Kruskal-Wallis test with Dunn's post hoc test was used to determine the p values. C, normalized IpH4.0 of oocytes injected with 12 ng of ASIC2 cRNA alone or ASIC2 with 12 ng of matriptase. Unlike the effect on ASIC1, matriptase does not have an effect on the peak IpH4.0 of ASIC2. Representative traces are shown. Values are means ± S.D., and the numbers of oocytes measured are shown in parentheses. The p value was determined with a two-tailed, unpaired t test. D, oocytes were injected with ASIC1 or ASIC2 cRNA. After the peak IpH4.0 current was measured, catalytically active purified matriptase was added to the bath for 5 min at 50 ng/ml, and the peak IpH4.0 was measured again. The graph shows normalized values: for each oocyte, the peak IpH4.0 measured “before” and “after” matriptase addition was normalized to the “before” IpH4.0. The peak IpH4.0 of ASIC2 did not decrease after the addition of active matriptase to the bath. Representative traces for ASIC1 and ASIC2 before and after matriptase are shown. The p values were obtained with a two-tailed paired Student's t test of the raw current values before and after matriptase. Values are means ± S.D. The numbers of oocytes measured are shown in parentheses.
The dose-response of effect of matriptase was obtained by measuring ASIC1 whole-cell currents at pH 4.0 in oocytes injected with 12 ng of ASIC1 cRNA and increasing amounts of matriptase cRNA (0–12 ng) as indicated in Fig. 2B. The injection of 2 ng of matriptase with ASIC1 did not have a statistically significant effect on the peak pH 4.0 current. However, ASIC1 current decreased significantly when 4 ng or higher amounts of matriptase cRNA were used. The inactive S805A matriptase did not have a statistically significant effect on ASIC1 even at the highest concentration used (Fig. 2B).
To determine whether the effect of matriptase on ASIC1 was specific, we injected oocytes with another human ASIC family member, ASIC2. Matriptase had no effect on the whole-cell peak current of ASIC2, suggesting some specificity of its action on ASIC1 (Fig. 2C).
When the purified recombinant matriptase catalytic domain was added to the recording bath (50 ng/ml for 5 min, the same time scale and concentration used in its activity assay by the manufacturer), the current of ASIC1 decreased, whereas that of ASIC2 did not change significantly (Fig. 2D). We used the purified matriptase catalytic domain because of the shorter time scale compared with co-injection with ASIC1 followed by a two-day expression. However, it was not possible to compare the amount of matriptase present in the bath and available to cleave the ASIC1 channels with the amount of matriptase protein that the oocyte could express in 2 days after injection. The addition of recombinant active matriptase to the bath reduced the ASIC1 current by 20%; this decrease was smaller than that observed when matriptase was co-injected in oocytes with ASIC1, but it was statistically significant. It has been shown previously that 2 μg/ml (about 1 μm) trypsin for 5 min decreases the peak pH 4.0 current of ASIC1 by only 10%, whereas 20 μg/ml trypsin decreases it by 30–40% (15). These trypsin concentrations are much higher than the 50 ng/ml (about 2 nm) used in our experiments for matriptase. Because of the limited amount of purified matriptase catalytic subunit, we were unable to use higher concentrations of matriptase in these experiments. The physical closeness of expressed matriptase versus the matriptase added to the ASIC1 channels, and the fact that matriptase could be more active in its native conformation compared with the purified protein, could also have contributed to the result obtained with the catalytically active purified matriptase. Although the purified matriptase decreased ASIC1 peak pH 4.0 current, it had no statistically significant effect on the peak current of ASIC2-injected oocytes (Fig. 2D).
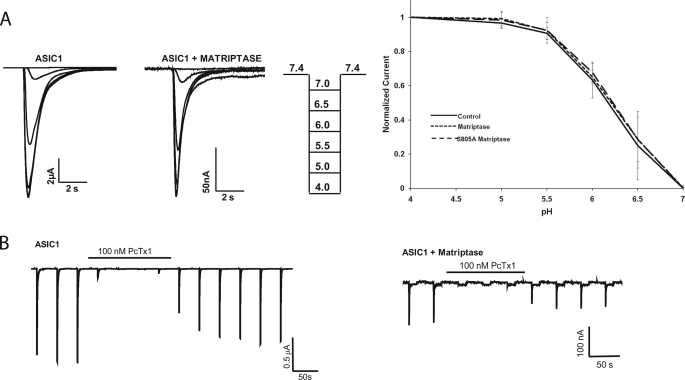
Matriptase Does Not Change the pH Activation Curve or PcTX-1 Block of ASIC1
Matriptase decreased the peak current of ASIC1, and for this decrease to occur, matriptase clearly required a functional catalytic domain (S805A has no effect on ASIC1), suggesting that matriptase cleaved the ASIC1 channels. Trypsin also cleaves ASIC1, and it affects the pH activation and channel inhibition by the venom of P. cambridgei (16). Trypsin shifts the pH activation curve of ASIC1 to a more acidic pH and disrupts the venom block of ASIC1 (16). Therefore, we hypothesized that matriptase would also affect these characteristics of the ASIC1 current. We obtained a pH activation curve and PcTX-1 sensitivity of ASIC1 + matriptase oocytes. For the pH activation curve, oocytes injected with ASIC1, ASIC1 + matriptase, or ASIC1 + matriptase S805A were subjected to two-electrode voltage clamp and first exposed to pH 7.4 followed by sequentially lower pH levels ranging from pH 7.0 to 4.0, returning to pH 7.4 after each activation. The peak current at each activation pH was normalized to the pH 4.0 current for each oocyte. The normalized values were then plotted against the activation pH to obtain the pH activation curves and were fitted to the Hill equation to obtain pH50 values and Hill coefficients. As shown in Fig. 3A, there is no difference in the pH activation curves in ASIC1, ASIC1 + matriptase, or ASIC1 + matriptase S805A. The pH50 values and Hill coefficients were also not statistically different (pH50 = 6.18 (S.D. = 0.18), nH = 1.77 (S.D. = 0.32), n = 10 for ASIC1; pH50 = 6.20 (S.D. = 0.08), nH = 1.64 (S.D. = 0.41), n = 10 for ASIC1 + matriptase; pH50 = 6.22 (S.D. = 0.13), nH = 1.71 (S.D. = 0.49), n = 7 for ASIC1 + S805A).
FIGURE 3.
Matriptase does not change the pH activation curve or inhibition by PcTX-1 of ASIC1. A, pH activation curve of currents measured in oocytes injected with 12 ng of ASIC1 alone, ASIC1 plus 8 or 12 ng of matriptase, or ASIC1 plus 12 ng of matriptase S805A. Oocytes were exposed to decreasing activation pH levels from 7 to 4.0. The peak current at each activation pH was normalized to the peak IpH4.0 for each oocyte. Representative pH activation curves and the pH protocol used are shown. The graphed values are means ± S.D. of 10 oocytes (ASIC1), 10 oocytes (ASIC1 + matriptase), and 7 oocytes (ASIC1 + matriptase S805A) from two or three different batches. B, peak IpH4.0 was measured in oocytes expressing ASIC1 alone or ASIC1 with 12 ng of matriptase. 100 nm PcTX-1 in the pH 7.4 solution abolished the IpH4.0 of control ASIC1-injected oocytes as well as of those co-injected with matriptase. Traces shown are representative of three oocytes for ASIC1 and three oocytes for ASIC1 and matriptase.
For the PcTX-1 block experiments, PcTX-1 was included in the pH 7.4 solution at 100 nm. PcTX-1 inhibited not only the ASIC1 current but also the current of ASIC1 + matriptase (Fig. 3B). This is different from trypsin, which removes the venom block of ASIC1 (15). Because the characteristics of the current (pH activation and PcTX-1 block) measured in ASIC1 + matriptase were not different from those of ASIC1 (but matriptase almost abolished the whole-cell peak current, and its catalytic site was required for this effect), it is likely that the current measured in ASIC1 + matriptase oocytes is from a population of noncleaved ASIC1 channels and that the cleaved channels are nonfunctional. The other possibility is that the cleaved channels have the same properties as the noncleaved ASIC1. However, the experiments below detecting the presence of ASIC1 cleavage products and determining the location of the possible sites where matriptase cleaves ASIC1 support the hypothesis that the current measured in ASIC1 + matriptase is due to a population of noncleaved channels.
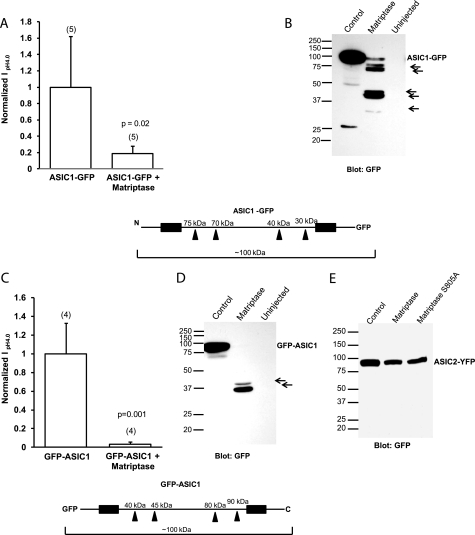
Matriptase-specific Fragments for ASIC1 but Not ASIC2 Are Observed in Xenopus Oocytes and in Transfected CHO K1 Cells
To determine whether ASIC1 channels are cleaved by matriptase, we obtained whole-cell lysates from oocytes expressing N- and C-terminal eGFP-tagged ASIC1 and matriptase. First, we measured whole-cell peak currents at pH 4.0 by two-electrode voltage clamp of ASIC1-GFP (C-terminal eGFP tag) + matriptase and GFP-ASIC1 (N-terminal eGFP tag) + matriptase to ensure that the tag did not affect the results. Matriptase decreased the current of ASIC1-GFP and that of GFP-ASIC1 similarly to the nontagged ASIC1 (Fig. 4, A and C). Following oocyte lysis and immunoblotting with a GFP antibody, in ASIC1-GFP + matriptase and GFP-ASIC1 + matriptase oocytes we observed several cleavage products that were not detected in the control lanes (Fig. 4, B and D). The full-length GFP-tagged ASIC1 was detected at ∼100 kDa, because the GFP tag adds 25 kDa to the 75-kDa ASIC1 protein (see Fig. 4 schematics). For the ASIC1-GFP constructs, five fragments were detected due to matriptase, and they are indicated by arrows in Fig. 4B.
FIGURE 4.
Detection of matriptase-specific cleavage fragments of GFP-tagged ASIC1 but not of tagged ASIC2 in Xenopus oocytes. A and C, normalized IpH4.0 of oocytes injected with C-terminal GFP-tagged ASIC1 (A) or with N-terminal GFP-tagged ASIC1 (C) with or without 12 ng of matriptase cRNA shows that matriptase significantly decreases the current of ASIC1-GFP (A) and GFP-ASIC1 (C). The values are means ± S.D. The numbers of oocytes measured are shown in parentheses; p values were determined with unpaired two-tailed t tests. B and D, Western blot of oocyte lysates with GFP antibody, which detects ASIC1 fragments in the matriptase lane for both the ASIC1-GFP (B) and GFP-ASIC1 (D) samples. The arrows point to GFP-tagged ASIC1 cleavage products. The schematics of ASIC1-GFP (below A and B) and GFP-ASIC1 (below C and D) show the locations of possible cleavage sites in each construct and the fragment sizes. The black boxes represent the two transmembrane domains. E, Western blot of oocyte whole-cell lysates from oocytes injected with ASIC2-YFP with or without 12 ng of matriptase cRNA or 12 ng of matriptase S805A cRNA. The GFP antibody does not detect any fragments for ASIC2 in the ASIC2-YFP plus matriptase samples, suggesting that matriptase does not cleave ASIC2.
The schematic below Fig. 4, A and B show the approximate positions of the possible cleavage sites that would result in the observed fragments. It also shows the approximate sizes of the fragments. Fragments that are not attached to GFP would not be detectable with the GFP antibody. The two largest fragments at 70–75 kDa could result from cleavage in the N-terminal part of the extracellular loop. The two fragments between 37 and 50 kDa could result from cleavage at a site near the C terminus of the extracellular loop, and the smallest fragment could be from cleavage at a site very near the second transmembrane domain. Alternatively, the smaller cleavage products could be due to further cleavage of the largest fragments by other endogenous oocyte serine proteases.
A schematic of GFP-ASIC1 is shown below Fig. 4, C and D. It illustrates the same possible cleavage sites as determined from the ASIC1-GFP construct, and the corresponding approximate fragment sizes that would be expected from cleavage at those sites. Two fragments at 35–40 kDa are detected for GFP-ASIC1. Their sizes correspond well to the two N-terminal sites, which would result in the two 70–75-kDa fragments for ASIC1-GFP (Fig. 4B). The schematic shows that larger fragments at 80–90 kDa are also expected for GFP-ASIC1, but no such fragments are detected in the blot in Fig. 4D. It is possible that the N-terminal 80–90 kDa fragments are cut further into smaller pieces by endogenous serine proteases.
Matriptase had no effect upon the function of ASIC2. A Western blot of lysates from oocytes expressing a GFP- or YFP-tagged ASIC2 and matriptase shows that there are no matriptase cleavage products of ASIC2-YFP and that the full-length ASIC2-YFP protein did not decrease as did the full-length ASIC1-GFP or GFP-ASIC1 (Fig. 4E). These results support the hypothesis that the functional effect of matriptase on the current of ASIC1 is accompanied by a decrease in full-length channels and the presence of cleavage products, whereas the lack of effect on ASIC2 is accompanied by a lack of ASIC2 cleavage products or substantial decrease in the full-length protein.
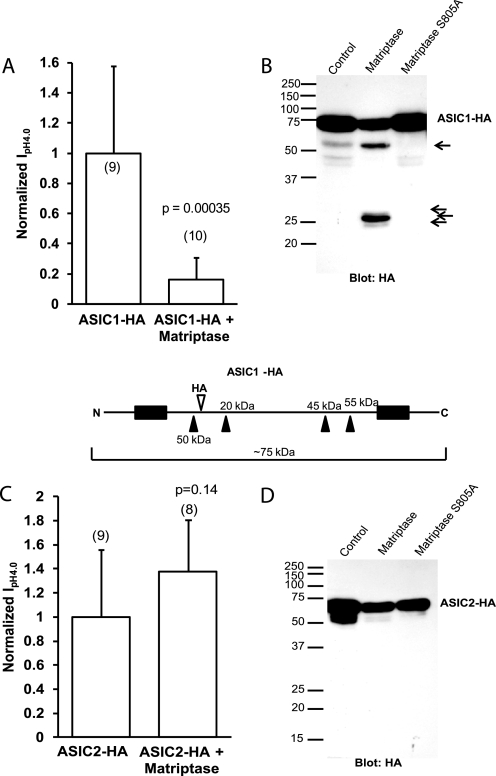
We also tested the effect of matriptase on an ASIC1 construct with an HA tag in the extracellular loop. The data with the eGFP-tagged ASIC1 and ASIC2 showed that matriptase cleaves ASIC1 but not ASIC2. ASIC1-HA and ASIC2-HA were used to further confirm these findings. Because the HA tag (at Phe-147 on ASIC1 and Thr-139 on ASIC2) is in a different position than the N or C terminus of the protein, we would detect different ASIC fragments than when using GFP-tagged ASICs. The ASIC1-HA and ASIC2-HA constructs did not differ from the untagged ASIC1 and ASIC2 in the way that they were functionally affected by matriptase. The current of ASIC1-HA + matriptase was decreased compared with control; matriptase had no effect on ASIC2-HA (Fig. 5, A and C). Western blots with an HA antibody detected full-length ASIC1-HA or ASIC2-HA at the expected size of about 75 kDa, and only ASIC1-HA was cleaved by matriptase.
FIGURE 5.
Detection of cleavage fragments of HA-tagged ASIC1 but not HA-tagged ASIC2 in Xenopus oocytes. A and C, normalized IpH4.0 of oocytes injected either with ASIC1-HA (A) or ASIC2-HA (C) with or without 8 or 12 ng of matriptase cRNA. Matriptase decreases the peak IpH4.0 of ASIC1-HA but not that of ASIC2-HA. Data are means ± S.D. The numbers of oocytes measured are shown in parentheses. The p values were determined with unpaired two-tailed t tests. B and D, consistent with the effect of matriptase on the function of the channels, Western blotting of whole oocyte lysates with an HA antibody detects fragments of ASIC1-HA (B) but not ASIC2-HA (D), suggesting that matriptase does not cleave ASIC2. The schematic of ASIC1-HA (below A and B) shows the locations of possible cleavage sites in the construct and the predicted fragment sizes. The black boxes represent the two transmembrane domains.
A schematic of the ASIC1-HA construct showing the relative position of the HA tag, the positions of the same possible cut sites that were deduced from Fig. 4B, and the approximate sizes of the fragments is shown below Fig. 5, A and B. Only the fragments that would contain the HA tag and would thus be detectable by an HA antibody were considered. The sizes (in kDa) at the top of the schematic are for fragments from the N terminus of ASIC1 to the indicated cleavage site. The size indicated below the schematic is for the fragment from the C terminus of ASIC1 to the first potential N-terminal cleavage site. This is ∼50 kDa, corresponding well to what is observed in the blot in Fig. 5B. As the schematic shows, there are a few fragments with sizes of 45–50 kDa; we could not tell the exact cleavage site that resulted in the 50-kDa fragment observed. It is possible that this fragment is from cleavage at the first N-terminal site and represents the C-terminal linked fragment, and the larger N-terminal linked fragments get cleaved further by proteases (especially as the largest N-terminal fragments for GFP-ASIC1 were not seen). The catalytically inactive matriptase S805A did not cleave ASIC1-HA, consistent with its lack of effect on the function of ASIC1. A few nonspecific smaller fragments were observed for ASIC1-HA and ASIC2-HA without matriptase (Fig. 5, B and D). These fragments could be degradation products and were not observed in noninjected oocytes (not shown). In agreement with the ASIC2 functional and Western blot data, no ASIC2-HA cleavage products specific to matriptase were detected, further supporting the hypothesis that matriptase does not cleave ASIC2.
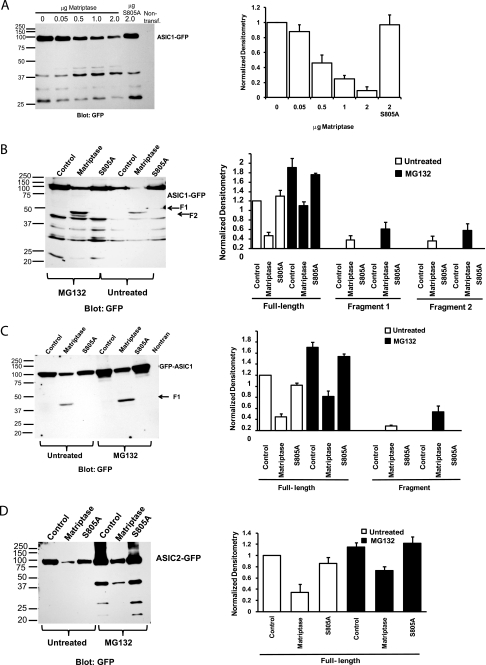
In the above experiments ASIC1 and matriptase were heterologously co-expressed in Xenopus oocytes. Because there may be differences between heterologous expression systems, we used mammalian CHO K1 cells to test the hypothesis that matriptase cleaves ASIC1 specifically, whereas it does not cleave ASIC2. In these experiments, ASIC1-GFP full-length protein was detected at 100 kDa in transfected CHO cells, and its intensity decreased with increasing amounts of matriptase DNA (Fig. 6A). The relative densitometry values for each band were normalized to the control ASIC1-GFP with no matriptase (Fig. 6A, first lane on the blot). The GFP antibody detected some nonspecific fragments that were present in all the samples transfected with ASIC1-GFP but were not present in nontransfected CHO cells. There were two faint fragments between 37 and 50 kDa specific to catalytically active matriptase. These two fragments were present only in the matriptase lanes and not in the control lane or the catalytically inactive matriptase S805A lane.
FIGURE 6.
Matriptase cleaves ASIC1-GFP and GFP-ASIC1, but not ASIC2-GFP, in transfected CHO cells. A, dose response of increasing amounts of matriptase DNA co-transfected in CHO cells with 2 μg of ASIC1-GFP DNA. Transfected CHO cells were lysed 2 days post-transfection, and whole-cell lysates were subjected to Western blot with a GFP antibody. The densitometry of the full-length ASIC1-GFP band shows a linear relationship between increasing amounts of matriptase DNA and decreasing amounts of full-length ASIC1-GFP and no effect of the catalytically inactive matriptase S805A. Data in the densitometry graph are means ± S.E. B, CHO cells transfected with 2 μg of ASIC1-GFP alone (control lanes) or plus 2 μg of matriptase or S805A were lysed after being left untreated or after treatment with the proteasome inhibitor MG132 (5 μg/ml, for 5 h or overnight). The whole-cell lysates were subjected to Western blot with GFP antibody. The amount of full-length ASIC1-GFP decreases when it is co-transfected with matriptase but not with the catalytically inactive matriptase S805A. Two small fragments are barely detected in the untreated samples of the matriptase lane. In the samples treated with MG132, the signal of the fragments in the matriptase lane (F1 and F2) is increased. There are no ASIC1-GFP fragments in ASIC1-GFP or ASIC1-GFP plus matriptase S805A in either the untreated or the MG132-treated samples. Data in the densitometry graph are means ± S.E. C, the same conditions apply as in B, with the exception that GFP-ASIC1 (ASIC1 with an N-terminal GFP tag) was used instead. One fragment of GFP-ASIC1 is detected only in the matriptase lanes, and its intensity increases with MG132 treatment. The densitometry data are means ± S.E. D, the same conditions apply as in B and C, with the exception that ASIC2-GFP was used instead and MG132 treatment was overnight. Even though matriptase decreases the full-length ASIC2-GFP, no ASIC2-GFP fragments appear in the matriptase lanes of untreated or MG-132-treated samples, suggesting that matriptase does not cleave ASIC2-GFP. The densitometry data are means ± S.E. of three separate experiments.
Because these fragments were difficult to detect, we hypothesized that after cleavage, they were being sent for degradation. We tested this hypothesis by treating the transfected CHO cells with the proteasome inhibitor MG132 (5 μg/ml for 5 h or overnight) at 48 h post-transfection with 2 μg of ASIC1-GFP alone, 2 μg of ASIC1-GFP + 2 μg of matriptase, or 2 μg of ASIC1-GFP + 2 μg matriptase S805A. As shown in Fig. 6B, in the samples treated with MG132, the two proteolytic fragments between 37 and 50 kDa could be clearly detected in the matriptase lane. Fig. 6B shows the normalized densitometry of the full-length bands and the two detected fragments. The nonspecific bands present in all of the lanes were ignored because they were not specific to matriptase. All of the bands were normalized to the signal of the full-length untreated ASIC1-GFP. The graph shows that the intensity of the full-length ASIC1-GFP protein decreases with matriptase, and two fragments, labeled F1 (fragment 1) and F2 (fragment 2), appear in the matriptase lanes (Fig. 6B). The intensities of all the bands, including the two fragments, increased when the cells were pretreated with the proteasome inhibitor. The inactive matriptase S805A did not decrease the signal of the full-length ASIC1-GFP.
The same experiment as shown in Fig. 6B was repeated for the N-terminal GFP-tagged ASIC1 (GFP-ASIC1). One fragment was observed for GFP-ASIC1 in the matriptase lanes (Fig. 6C). The nonspecific bands that were present in all of the lanes for the ASIC1-GFP construct were not detected with the GFP-ASIC1. MG132 pretreatment increased the intensities of all the bands, including the fragment (Fig. 6C densitometry). No fragments were observed either in the control or the matriptase S805A in the untreated and MG132-treated samples.
The effect of matriptase on a C-terminal GFP-tagged ASIC2 (ASIC2-GFP) in CHO cells was tested to determine if the effect of matriptase on ASIC1 was specific. Surprisingly, a substantial decrease in the full-length ASIC2-GFP was observed with matriptase, but no ASIC2-GFP cleavage fragments specific to matriptase could be detected even when cells were treated with MG132 (Fig. 6D). In the presence of MG132, a fragment appeared at 37 kDa. This fragment was present in all of the ASIC2-GFP lanes with MG132, including the control with ASIC2-GFP alone and the ASIC2-GFP with the inactive matriptase S805A. Because this fragment was not specific to matriptase (and its intensity actually decreased with matriptase compared with control or inactive matriptase) it was ignored and was not included in the densitometry. The absence of ASIC2-GFP fragments with matriptase suggests that matriptase does not cleave ASIC2. However, because we did detect a decrease in ASIC2-GFP protein with matriptase (in both untreated and MG132-treated as compared with the control/inactive matriptase) we cannot exclude the possibility that in CHO cells, matriptase could cleave ASIC2 and we were unable to observe ASIC2 fragments. Another possibility is that the decrease in ASIC2-GFP expression with matriptase is an indirect effect of matriptase on protein expression rather than cleavage of ASIC2 by matriptase. In all of these experiments, the catalytically inactive S805A matriptase was able to prevent the effect of matriptase on ASIC1 or ASIC2 (Fig. 6).
It is important to note some differences between expression in oocytes and mammalian CHO cells: several ASIC1 fragments were observed with the ASIC1-GFP construct in oocytes compared with only two fragments in CHO cells; two fragments were observed with the GFP-ASIC1 construct in oocytes compared with one fragment in CHO cells. Matriptase did not result in ASIC2 fragments either in oocytes or in CHO cells, and it did not dramatically decrease ASIC2 expression in oocytes; however, matriptase decreased ASIC2 expression in CHO cells. Despite these differences in the two expression systems, matriptase-specific fragments were detected for ASIC1, but not for ASIC2, in both oocytes and CHO cells. However, the sites of cleavage or downstream signaling events caused by matriptase may be different in the two expression systems.
Identification of Matriptase Cleavage Sites on ASIC1
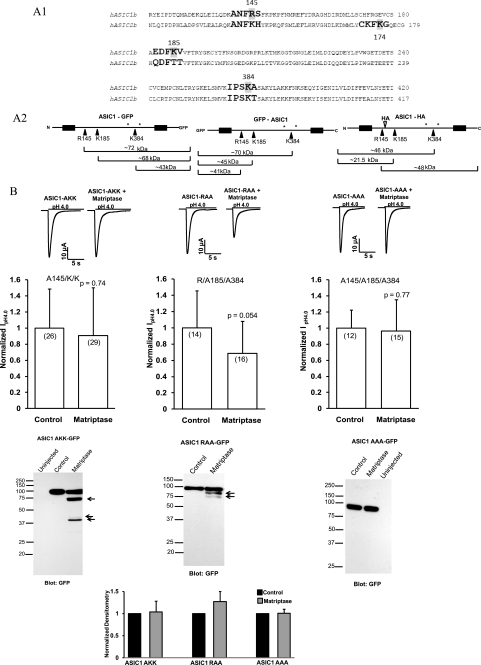
Because matriptase decreased the ASIC1 current and cleaved ASIC1 but had no effect on the function of ASIC2, we searched for matriptase cleavage sites on the ASIC1 and ASIC2 amino acid sequences. The preferred cleavage sequence of matriptase and the activation sites of known matriptase substrates were obtained from Uhland (20). The four preferred residues proximal to the cleavage site (P4, P3, P2, and P1) and also P1′, which is the position distal to the cleavage site, were entered in the FindPatterns program of Genetics Computer Group software. Arginine, lysine, and non-basic amino acids were entered for both P4 and P3; serine, alanine, phenylalanine, leucine, glycine, and arginine were entered for P2 and arginine/lysine for P1; finally, alanine, valine, isoleucine, serine, and glycine were entered for P1′. The search resulted in the identification of three extracellular cleavage sites on ASIC1 (Fig. 7A). Because the matriptase catalytic domain is extracellular and matriptase is activated after it reaches the cell surface (20), we ignored the intracellular N and C termini of ASIC1 and ASIC2. The software recognized one site (Lys-174) as a matriptase cleavage site on the extracellular loop of ASIC2. However, the electrophysiological and biochemical data showed no effect of matriptase on ASIC2 function, and no matriptase-specific ASIC2 fragments were detected, suggesting this site may not be accessible to cleavage.
FIGURE 7.
Identification and confirmation of the matriptase sites on ASIC1. A1, alignment of the human ASIC1b (NM_001095) and human ASIC2b (NM_001094) proteins showing the locations of the three matriptase cut sites on ASIC1 and the equivalent sites on ASIC2. All three locations are in the extracellular loop; the N and C termini of ASIC1 and ASIC2 were ignored because matriptase catalytic domain is extracellular. The matriptase cut sites were identified with software from Genetics Computer Group (University of Alabama at Birmingham) in which the information for positions P4, P3, P2, P1, and P1′, with P1 being the cut site, was entered manually (20). One matriptase site was identified by the software in the extracellular loop of ASIC2 at Lys-174, but because no ASIC2 fragments were detected, it may not be accessible to cleavage. A2, the schematics of ASIC1-GFP, GFP-ASIC1, and ASIC1-HA are shown, indicating the three matriptase sites in each protein and the sizes of the fragments expected from cleavage at these sites. The sizes take into account weight added by potential glycosylations. Only the fragments that are attached to tags, and therefore could be detected by Western blots, are shown. *, potential glycosylation sites (Asn-368 and Asn-395) (28) B, matriptase does not decrease the current of ASIC1 with Arg-145 mutated to Ala (R145A/K/K) or of the triple mutant R145A/K185A/K384A (AAA). Matriptase only slightly decreases the current of the R/K185A/K384A mutant. Representative traces from each experiment are shown. The normalized IpH4.0 are means ± S.D. The numbers of oocytes are shown in parentheses and are from two to three different experiments. C, oocytes were injected with 12 ng of ASIC1 AKK-GFP, ASIC1 RAA-GFP, or ASIC1 AAA-GFP with or without 12 ng of matriptase cRNA. The whole-cell lysates were subjected to 12% SDS-PAGE and Western blot with a GFP antibody. Some cleavage products were detected for the ASIC1 AKK and ASIC1 RAA. For the ASIC1 AAA, in which all three matriptase sites are mutated to alanines, no fragments were detected. The normalized densitometry of the full-length bands for ASIC1 AKK, ASIC1 RAA, and ASIC1 AAA, with and without matriptase, shows that despite the presence of some fragments, there is no dramatic decrease in the full-length protein (in contrast to the wild type ASIC1-GFP protein). The densitometry values are means ± S.E.
Alignment of the ASIC1 and ASIC2 amino acid sequences with ClustalW shows the locations of the three matriptase cleavage sites on ASIC1, the equivalent sites on ASIC2, and that the matriptase recognition sequences on ASIC1 are not conserved on ASIC2 (Fig. 7A1). A schematic of the ASIC1-GFP, GFP-ASIC1, and ASIC1-HA proteins indicating the three cleavage sites, the fragments, and their sizes is shown in Fig. 7A2. The size estimates take into account the weight added by potential glycosylations at Asn-368 and Asn-395 (28). For two glycosylations, ∼5 kDa was added to the calculated size of a peptide fragment (29).
We mutated the matriptase sites on the ASIC1-GFP construct with site-directed mutagenesis, measured whole-cell peak pH 4.0 currents of the mutants with or without matriptase, and detected the proteins with a GFP antibody. We chose the oocyte expression system instead of CHO cells for the mutagenesis experiments, because in oocytes we observed cleavage of ASIC1, but not ASIC2, without the substantial decrease in the expression of ASIC2 protein, which was caused by matriptase in CHO cells.
Mutation of the first matriptase site to Ala (R145A) (referred to as ASIC1 AKK) prevented the decrease in current observed when matriptase was co-injected with the wild type ASIC1 (Fig. 7B). ASIC1 AKK-GFP was not cleaved by matriptase to the same extent as wild type ASIC1; however, some fragments of ASIC1 AKK-GFP were detected (Fig. 7C). The fragments correspond to the same ones observed in wild type ASIC1-GFP, with the exception that the biggest cleavage product (the one observed right above 75 kDa, corresponding to cleavage at position 145) was missing (Fig. 7C). The densitometry of the 100-kDa band of ASIC1 AKK-GFP showed that this mutation prevented the decrease in full-length ASIC1-GFP, which was observed for the wild type ASIC1-GFP shown in Fig. 4B (Fig. 7C). Because the GFP tag is located in the C terminus of ASIC1, and the size of the fragment from Arg-145 to the C terminus of GFP would be ∼72 kDa, it was not surprising not to detect this fragment when Arg-145 was changed to the noncleavable Ala residue (Fig. 7, A1 and C). Arg-145 is the main trypsin cleavage site on ASIC1, although some cleavage of ASIC1 R145A by trypsin has also been observed (20).
Because the mutation of Arg-145 to Ala completely prevented the effect of matriptase on ASIC1, we hypothesized that this was the main site of ASIC1 cleavage by matriptase and that the other two sites (Lys-185 and Lys-384) were not as important. To test this hypothesis, we made an ASIC1-GFP construct with Lys-185 and Lys-384 mutated to Ala (ASIC1 RAA). If Arg-145 is the only site of matriptase cleavage, we would expect matriptase to decrease the current of the ASIC1 RAA-GFP construct similarly to the wild type ASIC1 and also to detect similar cleavage products on Western blots. However, although matriptase decreased the current of ASIC1 RAA-GFP by about 30%, this decrease was not statistically significant or comparable with the much larger decrease of the wild type ASIC1 current caused by matriptase (by about 70–90%) (Fig. 7C). Some cleavage of ASIC1 RAA-GFP was observed with cleavage products at ∼75 kDa, in agreement with the location of the Arg-145 site. Another fainter, smaller fragment was also observed and it could indicate further degradation of the fragment from the Arg-145 to the C terminus of the ASIC1-GFP or cleavage at another nonspecific site. The smaller cleavage products, which could result from cleavage at the other two sites (Fig. 7A2), are not present in the RAA construct, where Lys-185 and Lys-384 are changed to noncleavable amino acids (Fig. 7C). The densitometry shows that unlike with the wild type ASIC1-GFP, there is no decrease in the full-length ASIC1 RAA-GFP. These results suggest that Arg-145 is a matriptase cleavage site and that Lys-185 and Lys-384 are also important.
Mutation of all three matriptase cleavage sites to alanines (R145A, K185A, and K384A; referred to as ASIC1 AAA) prevented the effect of matriptase on ASIC1 current and also prevented ASIC1 protein cleavage (Fig. 7C). Compared with the effect of matriptase on the wild type ASIC1, the densitometry shows that there is no decrease in the full-length ASIC1 AAA-GFP protein. Some cleavage of ASIC1 AAA-GFP was observed with matriptase in overexposed films (not shown). Not unlike trypsin (16), matriptase might cleave at nonspecific sites (there are many additional Arg or Lys residues in the extracellular loop near the matriptase sites). Alternatively, the minimal cleavage observed for ASIC1 AAA-GFP could be the result of some other unknown endogenous proteases that get activated by matriptase in the oocyte expression system.
DISCUSSION
Matriptase is an epithelial cell surface serine protease that is expressed in a variety of carcinomas and has been implicated in the malignant progression of cancers (20). Although matriptase is expressed in many different epithelial tissues and organs, matriptase mRNA has not been detected in normal brain tissue (17, 27). In this study we show that matriptase is expressed in GBM cells. The data show that matriptase mRNA is expressed in glioma cell lines and in freshly excised tissues from GBM and grade III glioma (anaplastic astrocytoma). Matriptase RNA could not be detected in primary human normal astrocytes or in two different freshly excised astrogliosis tissues. Unlike the mRNA, matriptase protein is present in human primary normal astrocytes and in glioma cell lines, but its expression in normal astrocytes is much lower compared with glioma cells. It is possible that the levels of matriptase RNA in the normal astrocytes were too low to detect under our conditions.
The presence of matriptase protein suggests a role for this protease in glioma. Some of the physiological matriptase substrates (namely, urokinase plasminogen activator (uPA), hepatocyte growth factor/scatter factor (HGF/SF), and protease activated receptor-2 (PAR-2)) are associated with increased proliferation and invasiveness of glioma (30). In addition, overexpression of HAI-1, the cognate matriptase inhibitor, suppressed the in vitro invasiveness of the U251 glioblastoma cell line (24).
As a cell surface protease, matriptase is in an ideal position to cleave the extracellular loops of ion channels. In the Xenopus oocyte expression system, matriptase increased the activity of αβγENaC (17, 18). ASIC modulation by proteases has been less studied, but most proteases that have been tested have an effect on ASIC1 or ASIC1-containing heteromeric ASICs and no effect on ASIC2 or ASIC3 (15). The goal of this study was to test the hypothesis that matriptase can modulate ASIC1 function through proteolytic cleavage. We used heterologous expression systems and electrophysiological and biochemical approaches to determine the effect of matriptase on the function of ASIC1 and also to determine if/where in the extracellular loop of ASIC1 matriptase cleavage occurs.
Our data show that ASIC1 is a matriptase substrate in two heterologous expression systems, Xenopus oocytes and CHO cells, and that unlike its effect on ENaC, matriptase decreases the function of ASIC1. This effect requires the catalytic activity of matriptase and is associated with the presence of ASIC1 cleavage products. The putative matriptase cleavage sites on ASIC1 are Arg-145, Lys-185, and Lys-384. The crystal structure of chicken ASIC1 and alignments of ASICs from different species enabled us to locate these sites in the extracellular loop of ASIC1, which resembles a clenched hand and contains a palm (made of β-strands), knuckle, finger, thumb (all made of α-helices), and β-ball. The thumb, β-ball, and finger, along with the palm of an adjacent subunit, form the acidic pocket or proton-binding site of the channel (4). Arg-145 is located in the finger domain, Lys-185 in the β-ball, and Lys-384 between the palm and knuckle domains. Because all three sites are located at or close to the pH-sensing region (4), channels cleaved at these sites are likely not functional, suggesting that the small current measured in ASIC1 + matriptase is probably due to a few noncleaved channels. In agreement with this idea, the data show that the pH activation and PcTX-1 sensitivity of this current are not different from that of noncleaved ASIC1.
The first matriptase cleavage site, Arg-145, was previously characterized as a trypsin site (16). Similar to our results with matriptase, the application of extracellular trypsin decreased the peak acid-activated current of ASIC1 but had no effect on ASIC2 (16). The mutagenesis experiments strongly suggest that the effect of matriptase is direct, involving cleavage of ASIC1 in several locations in the extracellular loop. However, because some cleavage was observed in the ASIC1 AAA mutant, our results do not rule out the possibility that matriptase cleaves at adjacent arginines or lysines or that it activates another endogenous protease, which could cleave at other nearby sites. One such protease is prostasin. Matriptase co-localizes with and activates prostasin, which has been shown to activate αβγENaC (17, 18, 31). Therefore, the substrate recognition motif for prostasin, which is (R/K)(H/K/R)X(R/K) (where X is a basic or hydrophobic amino acid) (32), and the ASIC1 amino acid sequence were entered in the FindPatterns program in SeqWeb version 3.1.2, Genetics Computer Group (University of Alabama at Birmingham), to look for putative prostasin sites on ASIC1. However, the search identified no prostasin sites on ASIC1, suggesting that cleavage of ASIC1 by matriptase-activated prostasin is unlikely.
The ASIC1 cleavage fragments observed due to matriptase correlate well with what would be expected from cleavage at those three sites. Because many fragments were observed, it is possible that not all the channels are cleaved at all three sites. If that were the case, then the only size observed, for example, in the ASIC1-GFP construct, would be the smallest fragment still attached to GFP. This is the fragment from Lys-384 of ASIC1 to the C terminus of GFP with a calculated size of ∼40 kDa (Fig. 7A2). We do not know whether the ASIC1 fragments remain associated once cleaved by matriptase. Because cleaved fragments of αENaC and γENaC remain associated within the channel, it is possible that this is the case for ASIC1 as well (11).
The cleavage of ASIC1 by matriptase has implications for the pathologies in which ASIC1 is involved. ASIC1 is mainly a neuronal channel, and matriptase expression in neurons has not been reported; however, matriptase expressed by astrocytes could possibly modulate ASIC1 in neurons. Matriptase could modulate ASIC1 activity acting to decrease ASIC1 function through proteolytic cleavage.
Thus far the regulation of ASIC1 by proteases has been explored using heterologous expression systems (15, 16). We report matriptase expression in glioblastoma cells, where ASIC1 is also expressed (5–8). Although nothing is known about the proteolytic cleavage of ASIC1 in glioma, the expression of both matriptase and ASIC1 in the same cell type makes proteolytic cleavage of ASIC1 by matriptase in a nonheterologous system a realistic possibility.
Acknowledgments
The hASIC1b and hASIC2b constructs were a kind gift of Dr. David Corey. Matriptase DNA was a gift from Dr. C. Y. Lin (Lombardi Cancer Center, Georgetown University Medical Center). We thank Melissa McCarthy for technical assistance with cell culture, Ian Thornell and Xiaofeng Hu for technical assistance with Xenopus surgeries, Dr. Wanda Vila-Carriles for the ASIC2-HA DNA, and David Clark for assistance with fitting the pH activation data to the Hill equation. We also thank Drs. Kevin Kirk, Mark Bevensee, Niren Kapoor, and Yawar Qadri for helpful discussions and Brooke Goodman for some of the RNA isolations.
This work was supported, in whole or in part, by National Institutes of Health Grant DK 37206.
- ASIC
- acid-sensing ion channel
- hASIC
- human ASIC
- ENaC
- epithelial Na+ channel
- Deg
- degenerin
- GBM
- glioblastoma multiforme
- eGFP
- enhanced green fluorescent protein.
REFERENCES
- 1.Wemmie J. A., Price M. P., Welsh M. J. (2006) Trends Neurosci. 29, 578–586 [DOI] [PubMed] [Google Scholar]
- 2.Kellenberger S., Schild L. (2002) Physiol. Rev. 82, 735–767 [DOI] [PubMed] [Google Scholar]
- 3.García-Añoveros J., Derfler B., Neville-Golden J., Hyman B. T., Corey D. P. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 1459–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jasti J., Furukawa H., Gonzales E. B., Gouaux E. (2007) Nature 449, 316–323 [DOI] [PubMed] [Google Scholar]
- 5.Berdiev B. K., Xia J., McLean L. A., Markert J. M., Gillespie G. Y., Mapstone T. B., Naren A. P., Jovov B., Bubien J. K., Ji H. L., Fuller C. M., Kirk K. L., Benos D. J. (2003) J. Biol. Chem. 278, 15023–15034 [DOI] [PubMed] [Google Scholar]
- 6.Bubien J. K., Keeton D. A., Fuller C. M., Gillespie G. Y., Reddy A. T., Mapstone T. B., Benos D. J. (1999) Am. J. Physiol. Cell Physiol. 276, C1405–C1410 [DOI] [PubMed] [Google Scholar]
- 7.Kapoor N., Bartoszewski R., Qadri Y. J., Bebok Z., Bubien J. K., Fuller C. M., Benos D. J. (2009) J. Biol. Chem. 284, 24526–24541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vila-Carriles W. H., Kovacs G. G., Jovov B., Zhou Z. H., Pahwa A. K., Colby G., Esimai O., Gillespie G. Y., Mapstone T. B., Markert J. M., Fuller C. M., Bubien J. K., Benos D. J. (2006) J. Biol. Chem. 281, 19220–19232 [DOI] [PubMed] [Google Scholar]
- 9.Rossier B. C. (2004) Proc. Am. Thorac. Soc. 1, 4–9 [DOI] [PubMed] [Google Scholar]
- 10.Hughey R. P., Carattino M. D., Kleyman T. R. (2007) Curr. Opin. Nephrol. Hypertens. 16, 444–450 [DOI] [PubMed] [Google Scholar]
- 11.Hughey R. P., Mueller G. M., Bruns J. B., Kinlough C. L., Poland P. A., Harkleroad K. L., Carattino M. D., Kleyman T. R. (2003) J. Biol. Chem. 278, 37073–37082 [DOI] [PubMed] [Google Scholar]
- 12.Kleyman T. R., Carattino M. D., Hughey R. P. (2009) J. Biol. Chem. 284, 20447–20451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rossier B. C., Stutts M. J. (2009) Annu. Rev. Physiol. 71, 361–379 [DOI] [PubMed] [Google Scholar]
- 14.Hu J. C., Bengrine A., Lis A., Awayda M. S. (2009) J. Biol. Chem. 284, 36334–36345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poirot O., Vukicevic M., Boesch A., Kellenberger S. (2004) J. Biol. Chem. 279, 38448–38457 [DOI] [PubMed] [Google Scholar]
- 16.Vukicevic M., Weder G., Boillat A., Boesch A., Kellenberger S. (2006) J. Biol. Chem. 281, 714–722 [DOI] [PubMed] [Google Scholar]
- 17.Vuagniaux G., Vallet V., Jaeger N. F., Hummler E., Rossier B. C. (2002) J. Gen. Physiol. 120, 191–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andreasen D., Vuagniaux G., Fowler-Jaeger N., Hummler E., Rossier B. C. (2006) J. Am. Soc. Nephrol. 17, 968–976 [DOI] [PubMed] [Google Scholar]
- 19.Lin C. Y., Anders J., Johnson M., Dickson R. B. (1999) J. Biol. Chem. 274, 18237–18242 [DOI] [PubMed] [Google Scholar]
- 20.Uhland K. (2006) Cell. Mol. Life Sci. 63, 2968–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin C. Y., Tseng I. C., Chou F. P., Su S. F., Chen Y. W., Johnson M. D., Dickson R. B. (2008) Front. Biosci. 13, 621–635 [DOI] [PubMed] [Google Scholar]
- 22.List K., Bugge T. H., Szabo R. (2006) Mol. Med. 12, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.List K. (2009) Future Oncol. 5, 97–104 [DOI] [PubMed] [Google Scholar]
- 24.Miyata S., Fukushima T., Kohama K., Tanaka H., Takeshima H., Kataoka H. (2007) Hum. Cell 20, 100–106 [DOI] [PubMed] [Google Scholar]
- 25.Bashari E., Qadri Y. J., Zhou Z. H., Kapoor N., Anderson S. J., Meltzer R. H., Fuller C. M., Benos D. J. (2009) Am. J. Physiol. Cell Physiol. 296, C372–C384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meltzer R. H., Kapoor N., Qadri Y. J., Anderson S. J., Fuller C. M., Benos D. J. (2007) J. Biol. Chem. 282, 25548–25559 [DOI] [PubMed] [Google Scholar]
- 27.Kim M. G., Chen C., Lyu M. S., Cho E. G., Park D., Kozak C., Schwartz R. H. (1999) Immunogenetics 49, 420–428 [DOI] [PubMed] [Google Scholar]
- 28.Kadurin I., Golubovic A., Leisle L., Schindelin H., Gründer S. (2008) Biochem. J. 412, 469–475 [DOI] [PubMed] [Google Scholar]
- 29.Saugstad J. A., Roberts J. A., Dong J., Zeitouni S., Evans R. J. (2004) J. Biol. Chem. 279, 55514–55519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gessler F., Voss V., Dutzmann S., Seifert V., Gerlach R., Kogel D.Neuroscience 165, 1312–1322 [DOI] [PubMed] [Google Scholar]
- 31.List K., Hobson J. P., Molinolo A., Bugge T. H. (2007) J. Cell Physiol. 213, 237–245 [DOI] [PubMed] [Google Scholar]
- 32.Shipway A., Danahay H., Williams J. A., Tully D. C., Backes B. J., Harris J. L. (2004) Biochem. Biophys. Res. Commun. 324, 953–963 [DOI] [PubMed] [Google Scholar]