Abstract
Patients carrying mutations within the amyloid-β (Aβ) sequence develop severe early-onset cerebral amyloid angiopathy with some of the related variants manifesting primarily with hemorrhagic phenotypes. Matrix metalloproteases (MMPs) are typically associated with blood brain barrier disruption and hemorrhagic transformations after ischemic stroke. However, their contribution to cerebral amyloid angiopathy-related hemorrhage remains unclear. Human brain endothelial cells challenged with Aβ synthetic homologues containing mutations known to be associated in vivo with hemorrhagic manifestations (AβE22Q and AβL34V) showed enhanced production and activation of MMP-2, evaluated via Multiplex MMP antibody arrays, gel zymography, and Western blot, which in turn proteolytically cleaved in situ the Aβ peptides. Immunoprecipitation followed by mass spectrometry analysis highlighted the generation of specific C-terminal proteolytic fragments, in particular the accumulation of Aβ-(1–16), a result validated in vitro with recombinant MMP-2 and quantitatively evaluated using deuterium-labeled internal standards. Silencing MMP-2 gene expression resulted in reduced Aβ degradation and enhanced apoptosis. Secretion and activation of MMP-2 as well as susceptibility of the Aβ peptides to MMP-2 degradation were dependent on the peptide conformation, with fibrillar elements of AβE22Q exhibiting negligible effects. Our results indicate that MMP-2 release and activation differentially degrades Aβ species, delaying their toxicity for endothelial cells. However, taking into consideration MMP ability to degrade basement membrane components, these protective effects might also undesirably compromise blood brain barrier integrity and precipitate a hemorrhagic phenotype.
Keywords: Alzheimer Disease, Amyloid, Brain, Endothelium, Protease, Amyloid-β, Familial Aβ Variant, Cerebral Amyloid Angiopathy, Matrix Metalloproteases
Introduction
Cerebral amyloid angiopathy (CAA)3, the deposition of amyloid in vessel walls of the central nervous system, compromises leptomeningeal and cortical vessels affecting medium and small size arteries and arterioles as well as capillary endothelium. The most common form of CAA is associated with amyloid-β (Aβ) deposition, particularly in elderly individuals and in patients with Alzheimer disease (AD) (1, 2). In these cases the deposited protein, Aβ, is originated by sequential processing of the amyloid precursor protein primarily generating peptides constituted by 40- and 42-amino acid residue-long peptides, Aβ40 and Aβ42, respectively. For reasons not completely understood, Aβ42 is one of the main components of parenchymal amyloid plaques, whereas Aβ40 is the predominant species present in vascular deposits (2–4). The fibrillar and compact Aβ deposition in the brain vessel walls has been related to abnormalities in the vasculature including microaneurysms and fibrinoid necrosis (5), which can subsequently compromise the integrity of the blood brain barrier (BBB). Indeed, one of the most relevant pathological consequences of CAA is the development of intracerebral hemorrhages (ICH) usually affecting cortical and subcortical areas. In turn, ICH results in elevated mortality rates and a higher risk of suffering subsequent strokes (3, 6, 7). CAA translates also in other common clinical manifestations involving white matter changes associated with cortical microbleeds and progressive dementia (8, 9). Although it is clear that the relation between CAA and dementia is complex and probably encompasses a range of different pathogenic mechanisms, to date vascular deposits and not parenchymal plaques are more sensitive predictors of dementia in AD (10, 11).
CAA is mainly a sporadic disorder concomitant with the deposition of wild-type (WT) Aβ40; nevertheless, a relatively small group of cases consist of autosomal dominant hereditary forms. Patients carrying mutations within the Aβ precursor protein concentrated in the amino acid cluster comprising Aβ residues 21–23 primarily develop severe CAA while presenting with dissimilar clinical manifestations (12–14). One of the most aggressive clinical phenotypes described among the intra-Aβ mutations is associated with the replacement of a Glu for Gln at residue 22 (AβE22Q) in a disorder known as hereditary cerebral hemorrhage with amyloidosis Dutch type (HCHWA-D) (15–17). HCHWA-D is characterized clinically by recurrent hemorrhages and neuropathologically by extensive Aβ deposition in cerebral vessels and diffuse amyloid plaques in parenchyma in the absence of neurofibrillar pathology (18, 19). Parenchymal mature plaques characteristic of AD are rare in this kindred. A new intra-Aβ mutation manifesting with comparable, albeit less aggressive features, has been found in Piedmont, Italy. The mutation, resulting from the substitution of Leu by Val at position 34 (AβL34V), is also associated with severe CAA and fatal ICH, with amyloid deposition selectively affecting the cerebral vessel walls in the absence of parenchymal plaques (20).
In vitro studies have demonstrated an exacerbated toxic potential of Aβ mutants for different cell types. In particular AβE22Q, one of the most studied genetic variants, exhibits an enhanced capability to induce cellular damage in neurons (21), cerebrovascular smooth muscle (22, 23), and endothelial cells (24–26). This toxicity has often been related to the stronger fibrillogenic properties of AβE22Q exhibiting accelerated in vitro formation of amyloid aggregates with shorter lag-phase and faster kinetics (27, 28). Much less is known about the cellular effects elicited by AβL34V, although recent evidence indicates that, like AβE22Q, this variant triggers cell death mechanisms in cerebral endothelial cells (ECs) affecting the mitochondrial apoptotic pathway (29). In both genetic variants, the induction of apoptosis precedes fibril formation and correlates with the presence of intermediate-size oligomeric assemblies. Both mutants, as well as the WT-Aβ40 peptide, activate analogous cell death pathways, albeit at different timeframes and with different intensity largely correlating with the severity of the respective clinical phenotypes (29).
Matrix metalloproteinases (MMPs) are a family of zinc-dependent endopeptidases responsible for the degradation and remodeling of extracellular matrices. A role for overexpression of MMPs in CAA-hemorrhagic complications has been suggested by the known association of the proteases with BBB disruption in many vasculopathies and with hemorrhagic transformations after ischemic stroke (30–32). In vitro, the members of the MMP family with gelatinase activity, MMP-2 and MMP-9, exhibit the ability to degrade Aβ peptides (33–36) and have, therefore, been implicated as central players in Aβ catabolism contributing to the protein homeostasis and the maintenance of its endogenous brain levels. Studies have mainly focused on MMP-9 based on the ability of the protein to degrade fibrillar structures and compact plaques in ex vivo experiments (34). Found at low basal levels in pyramidal neurons of normal human hippocampus, MMP-9 expression is elevated in AD cases in which it localizes in neuritic plaques, vascular walls, and neurofibrillary tangles. Much less is known about MMP-2 with a few controversial reports indicating either elevated activity in AD hippocampal homogenates or no difference with respect to normal control individuals in AD frontal cortices (for review, see Ref. 37). Herein, we investigated the MMP brain-endothelial cell response after challenge with WT and Aβ variant peptides, evaluating their impact in Aβ degradation and fibrillization homeostasis.
EXPERIMENTAL PROCEDURES
Aβ Peptides
Synthetic homologues of WT Aβ40, the C-terminal-truncated fragment Aβ-(1–16), and the Aβ40 genetic variants containing the E22Q and L34V substitutions as well as the standards for quantitative mass spectrometry (MS) Aβ40 and the C-terminal-truncated fragment Aβ-(1–16) labeled with deuterated (2H) phenylalanine residues (Aβ-(1–16) deuterated at position 4, mass = 1963.0 Da, mass difference with unlabeled Aβ-(1–16) +8 Da; Aβ40 labeled at positions 4, 19, and 20, mass = 4353.9 Da, mass difference with unlabeled Aβ40 +24 Da) were synthesized using N-tert-butyloxycarbonyl chemistry by James I. Elliott at Yale University. All synthetic homologues were purified by reverse phase-high performance liquid chromatography, molecular masses were corroborated by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) MS, and concentrations were assessed by amino acid analysis as previously described (26, 29).
Peptides were pretreated with 0.05% trifluoroacetic acid for 10 min at room temperature (RT) with intermittent vigorous vortexing. After lyophilization, peptides were reconstituted in distilled water to a 1 mm concentration (stock solution) and immediately dissolved in either phosphate-buffered saline (PBS) or culture medium before their use in electron microscopy (EM) analysis for the assessment of their structural conformations or in the cell culture experiments, respectively, as described below. AβE22Q preparations enriched in fibrillar components were obtained by incubating the 1 mm stock solution for 6 days at 37 °C. The quality and purity of the resulting fibrillar elements were analyzed by fluorescence evaluation of Thioflavin-T binding and EM before their use.
Cell Cultures and Preparation of Short Hairpin RNA-MMP-2 Cells
Human cerebral microvascular endothelial cells (hCMEC/D3) were provided by B. Weksler (38), Weill Medical College, Cornell University, New York, NY. Cells were grown in EGM-2 medium (Lonza, Allendale, NJ) supplemented with VEGF, insulin-like growth factor, bovine FGF, hydrocortisone, ascorbate, and 2.5% fetal bovine serum (FBS) as previously described (29). For MMP-2 RNA silencing, cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Four different MMP-2 shRNAs as well as scrambled shRNA plasmids (SureSilencingTM shRNA plasmid for human MMP-2; SABiosciences, Frederick, MD) were prepared in accordance with the manufacturer's instructions and tested for their capacity to knock down MMP-2 expression. At least two different clones of shMMP-2 (from the four different shRNA sequences targeting different MMP-2 regions) and shControl cells were used to confirm the MMP-2 gene silencing specificity. Stable transfected cell lines of both shMMP-2 and scrambled shRNA controls were selected by their resistance to G418 exposure (800 μg/ml; Fisher) and subcultured with complete medium containing 200 μg/ml G418. In all cases gene silencing was verified by quantitative RT-PCR (qRT-PCR), Western blot analysis, gelatin zymography, and ELISA.
Cell Culture Treatment with the Different Aβ Homologues
Aβ-(1–40) peptides, either WT or containing the E22Q or L34V substitutions, as well as Aβ-(1–16) were pretreated, solubilized, and reconstituted in EGM-2, 1% FBS at a 50 μm final concentration as indicated above. Cells, either D3, shMMP-2, or shControl, were challenged with the different Aβ peptides for 1, 2, or 3 days. At the respective time points apoptosis induction was assessed by Cell Death ELISA, MMP secretion was evaluated by multiplex MMP array, and MMP-2 activation was determined by zymography together with Western blot analysis. Degradation of the different Aβ peptides in the culture supernatants was evaluated by Western and dot-blot analysis in conjunction with MALDI-TOF MS as described below.
Cell Death ELISA
Apoptosis induced by the pertinent Aβ treatments was evaluated by quantitation of DNA fragmentation using Cell Death ELISA (Roche Applied Science) in accordance with the manufacture's specifications, as we previously described (29). After amyloid incubation, culture plates were centrifuged in a Beckman J-6B centrifuge (10 min; 1000 rpm) to collect floaters, cells were lysed, and quantitation of DNA-histone complexes (nucleosomes) was assessed by absorbance at 405 nm. Results are expressed as -fold of change compared with no-peptide controls.
Multiplex MMP Array
MMP secretion in Aβ-challenged ECs was evaluated in culture supernatants (SN) after a 3-day peptide treatment using SearchLight® human MMP Arrays (Pierce). The respective SNs were incubated with the antibody arrays followed by immunoreaction with biotin-conjugated detection antibodies and streptavidin DyLight 649 red fluorescent conjugate, in accordance to the manufacturer's specifications. Chemiluminescence signals were captured with a cooled charged-coupled device (CCD), images were analyzed by ArrayVision Version 8.0 software (Imaging Research, Ontario, Canada), and MMP levels were calculated based on standard curves values. Results are illustrated in -fold change with respect to untreated controls and represent MMP levels obtained in five independent experiments in which each sample was assayed in duplicate.
Gelatin Zymography
Secretion and activation of gelatinases was assessed by substrate-specific zymography. Equal volumes of the respective culture SNs were diluted with sample buffer (80 mm Tris-HCl, pH 6.8, containing 4% SDS, 10% glycerol, and 0.01% bromphenol blue) and loaded onto 10% SDS-PAGE gels co-polymerized with 0.1% gelatin (Invitrogen) without boiling or the addition of reducing agents. After electrophoresis, proteases were re-natured by removing SDS with unbuffered 2.5% Triton X-100 for 1 h, and the gelatinolytic activity was developed by incubation with 50 mm Tris-HCl buffer, pH 7.5, containing 10 mm CaCl2 for 48 h at 37 °C. Enzymatically digested bands were visualized after staining with 0.1% Amido Black for 1 h and de-staining for 30 min with 30% (v/v) methanol/10% (v/v) acetic acid. Images were acquired with the Gel Logic 440 Imaging System (Eastman Kodak Co.), and band intensity was normalized to the untreated control samples for each experimental set with the aid of 1-D Image Analysis software (Kodak).
MMP-2 Quantitation by ELISA
MMP-2 levels in cell SNs were determined using human MMP-2 Quantikine ELISA (R&D Systems, Inc., Minneapolis, MN), a quantitative sandwich enzyme-linked inmunoabsorbent assay employing a polyclonal anti-MMP-2 as the capture antibody. Briefly, 50 μl of each of the cell culture supernatants were loaded in duplicate onto the microtiter plate and analyzed following the manufacturer's instructions. Final color was quantitated at 450 nm on a Spectracount microplate reader (Packard Instrument Co., Meriden, CT), and MMP-2 concentration was calculated by interpolation with a standard curve generated with recombinant enzyme, which is included in the kit.
Quantitative Real-time PCR Analysis
MMP-2 mRNA relative expression levels were evaluated by qRT-PCR. Total RNA was extracted from Aβ-treated and untreated control cells with RNeasy mini kit (Qiagen, Valencia, CA) in accordance with the manufacturer's instructions. Quality control monitoring and quantitation of isolated RNA were performed using the RNA 6000 Nano Chip kit in an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Pertinent cDNAs were synthesized with the High Capacity cDNA Archive kit (Applied Biosystems, Foster city, CA), and mRNA levels were quantitated by real-time PCR using TaqMan fluorogenic probes. qRT-PCR reactions were run in triplicate and analyzed using Applied Biosystems SDS 7500 system software (Applied Biosystems). Results were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression levels to account for variability in the initial sample concentration. Data were expressed as relative quantitation units, calculating the -fold change expression values with respect to the average expression of untreated samples.
In Vitro Aβ Proteolytic Digestion
Demonstration of the ability of D3-secreted MMP-2 to proteolytically cleave Aβ peptides was assessed by in vitro experiments. For this purpose culture supernatants of confluent cells that had been maintained for 2 days in culture in the absence of Aβ peptides and in media without FBS supplement to assure both, the lack of exogenous MMP-2 and the maximum production of the endogenous enzyme were evaluated for total protein content by BCA assay (Pierce). For digestion experiments, Aβ peptides (2.5 μl of 0.1 mm solution) were incubated for 2 and 6 days at 37 °C with the equivalent of 1 mg of total protein of each cell supernatant, either D3, MMP-2 shRNA, or shRNA controls, in a 50-μl final volume. Digestion was stopped by freezing at −80 °C, and samples were maintained under these conditions until use.
As controls, 125 pmol of the different Aβ peptides were proteolytically cleaved by incubation with recombinant human MMP-2 (R&D Systems; 1- and 5-h time points; 37 °C) at an enzyme:peptide ratio of 1: 25. Before the enzymatic assay, rhMMP-2 was activated by a 37 °C incubation with 1 mm para-aminophenylmercuric acetate (Sigma) in 50 mm Tris, pH 7.5, containing 10 mm CaCl2, 150 mm NaCl, and 0.05% Brij-35. MMP-2 activation reaction was stopped by the addition of EDTA, pH 8.0, and the resulting activated enzyme samples were stored at −80 °C until use.
In all cases degradation of the different Aβ peptides, either by rhMMP-2 or by the enzymes released to the culture supernatants, was evaluated by Western- and dot-blot analysis, a decrease in aggregated/fibrillar components estimated by thioflavin-T binding assays, and generation of specific Aβ degradation products assessed by immunoprecipitation (IP) followed by MS analysis, as described below.
Western Blot Analysis
Detection of Aβ peptides by WB was performed as previously described (39). Briefly, 5 μl of 1:10 dilutions of the respective cell culture supernatants or 2.5 μl of rhMMP-2 digestion samples were separated on 16.5% Tris-Tricine SDS-PAGE under reducing conditions and transferred onto 0.45-μm pore size polyvinylidene difluoride membranes (Millipore Corp., Billerica, MA) for 45 min at 400 mA using N-cyclohexyl-3-aminopropanesulfonic acid (CAPS) buffer, pH 11. Membranes were subsequently blocked for 1 h with 5% nonfat milk in PBS containing 0.1% Tween 20 (PBS-T), and probed with a combination of monoclonal antibodies 4G8 and 6E10 (Covance, Princeton, NJ; each diluted 1:3000 in PBS-T) either for 1 h at RT or overnight at 4 °C. After thorough washes with PBS-T, membranes were immunoreacted with HRP-conjugated anti-mouse IgG (BioSource-Invitrogen; 1:3000) for 1 h at RT and further developed by enhanced chemiluminescence using Supersignal West Pico Luminol/Enhancer and Stable Peroxide solutions (Thermo Scientific).
For MMP-2 detection, 30 μg of the respective cell lysates were separated by SDS-PAGE and electrotransferred onto nitrocellulose using the same conditions as above. Membranes were blocked for 1 h with 10% nonfat milk in PBS-T and incubated overnight at 4 °C with rabbit polyclonal anti-MMP-2 antibodies (Chemicon, Millipore; 1:2000). Membranes were subsequently reacted with HRP-conjugated anti-rabbit IgG (Dako, Glostrup, Denmark; 1:2000) for 1 h at RT and developed with ECL® detection reagents (Millipore). To corroborate similar protein loads among the different samples, the membranes were stripped for 15 min with Restore Western blot Stripping Buffer (Pierce) and re-probed with mouse monoclonal anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH, 1/5000; Imgenex, San Diego, CA). In all cases images from exposed films were analyzed with ImageJ software (rsbweb.nih.gov).
Dot-blot Analysis
Proteolytic cleavage of the different Aβ peptides by rhMMP-2 was assessed by dot-blot analysis. Briefly, 500 ng of the in vitro digested samples were loaded on a nitrocellulose membrane assembled into a Bio-Dot Microfiltration Apparatus (Bio-Rad) and allowed to diffuse passively for about 30 min before vacuum application. The membrane was then blocked in situ for 1 h with 1% nonfat milk in Tris-buffered saline, pH 7.4, containing 0.1% Tween 20 (TBST) followed by vacuum application and two subsequent washes with TBST. After removal from the dot-blot apparatus and further blocking with 1% milk in TBST (1 h, RT), the nitrocellulose membrane was further incubated with a combination of monoclonal antibodies 4G8 and 6E10 (each at 1:3000 dilution in 0.1% TBST; 1 h at RT) followed by HRP-conjugated sheep anti-mouse IgG (BioSource-Invitrogen; 1:3000, 1 h at RT). Immunoreactivity was assessed by ECL as for the WB above, and proteolytic cleavage was estimated by a decrease in dot intensity.
Immunoprecipitation/Mass Spectrometry
Identification of the Aβ degradation products in the culture supernatants of ECs challenged with the different Aβ peptides as well as in the in vitro digestion assays employing rhMMP-2 was assessed via IP followed by MS analysis. Aβ molecules were immunoprecipitated with paramagnetic beads (Dynabeads M-280, Invitrogen) coated with a combination of 4G8 and 6E10 antibodies, as previously described (39–41). Recombinant MMP-2 in vitro digestion samples and cell culture supernatants were separately incubated overnight at 4 °C with the anti-Aβ-coated beads resuspended in 500 μl of PBS containing 0.025% Tween 20. After subsequent washes with PBS-T and 50 mm NH4HCO3, the IP material was eluted with 0.5% formic acid, dried down in a speed vacuum without heat, and further reconstituted in 5 μl of 0.1% formic acid in 50% acetonitrile. One-fifth of the reconstituted sample (1 μl) was combined with an equal volume of α-4-hydroxycinnamic acid (AHCA; Agilent Technologies) matrix (15 g/liter), and 1 μl of the resulting mixture was spotted onto a Bruker Daltonics MTP 384 massive target T aluminum plate pre-seeded with 0.5 μl of AHCA (1 g/liter). All samples were spotted in duplicate and analyzed at the NYU Protein Analysis Facility using a Bruker Daltonics Autoflex MALDI-TOF mass spectrometer (Bremen, Germany) in linear mode with standard instrument settings.
For quantitative MS assessment of the rhMMP-2-generated Aβ-(1–16) as well as the remaining uncleaved Aβ40, known amounts of Aβ-(1–16) and Aβ40 deuterated standards were spiked into the digestion mixture before performing IP and MS analysis essentially as above. In all cases MS spectra were processed and analyzed by FlexAnalysis, and quantitation was performed by comparison with peak intensities of the pertinent labeled standards. Both Aβ-(1–16)-generated and uncleaved remaining peptides were expressed as a percentage with respect to the starting quantity of the respective Aβ peptides incubated with rhMMP-2.
Assessment of the Different Aβ Peptide Conformations
Evaluation of the respective peptide structures and the presence of fibrillar components was performed by EM and thioflavin-T binding assays as described below.
Electron Microscopy
Peptide samples prepared as described above were placed onto carbon-coated 400 mesh copper/rhodium grids (Ted Pella, Inc., Redding, CA) and stained with 1% uranyl acetate in distilled water (Polysciences, Inc., Warrington, PA). Stained grids were examined in a Philips CM-12 transmission electron microscope and photographed with a Gatan (4,000 × 4,000 pixels) digital camera at the Image Core Facility of the Skirball Institute of Biomedical Medicine, NYU School of Medicine, as previously described (42).
Thioflavin-T Binding Assay
The presence of fibrillar components in culture supernatants of ECs challenged with the different Aβ peptides as well as in the in vitro digestion samples of WT and AβE22Q peptides was assessed by thioflavin-T binding assays. Briefly, 24-μl aliquots of the respective culture SNs were added to 10 μl of freshly prepared thioflavin-T (Sigma; 0.1 mm) and 50 mm Tris buffer, pH 8.5, to a final volume of 200 μl, as previously described (43). For in vitro Aβ digestion samples, in which the respective peptides were incubated with the conditioned media in the absence of cells, the sample volume was increased to 50 μl while maintaining constant the amount of thioflavin-T. In all cases fluorescence was recorded after 300 s in a LS-50B luminescence spectrometer (PerkinElmer Life Sciences) with excitation and emission wavelengths of 435 nm (slit width = 10 nm) and 490 nm (slit width = 10 nm), respectively, as described (25, 42). Each sample was analyzed in duplicate.
Statistics
Statistical analyses were performed by one-way analysis of variance with Newman-Keuls Multiple Comparison post hoc test using Graph-Pad Prism 4.0 (GraphPad Software, La Jolla, CA). Values of p < 0.05 were considered significant.
RESULTS
Pre-fibrillar AβE22Q and L34V Variants Induce the Preferential Release and Activation of MMP-2 in Human Brain Microvascular Endothelial Cells
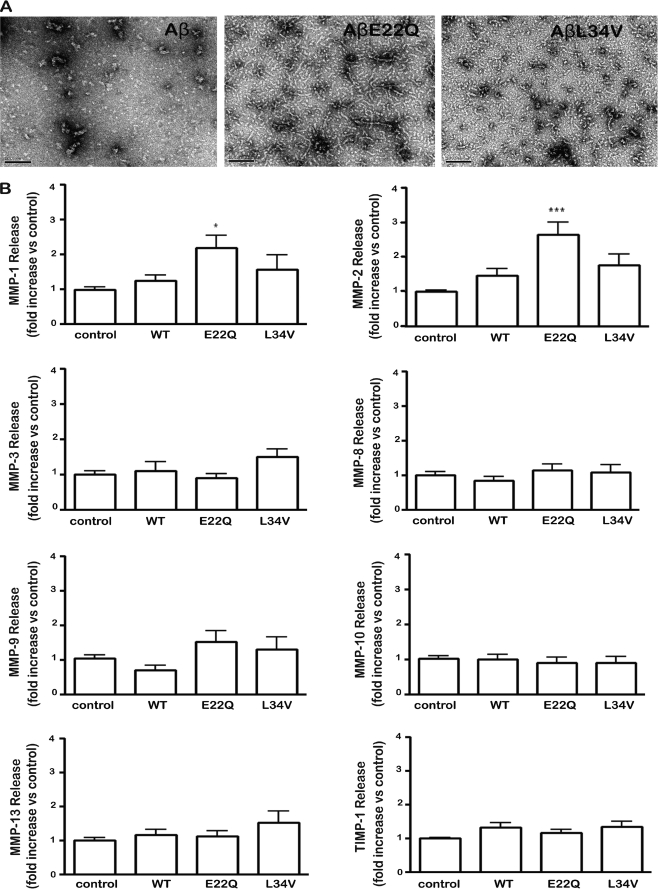
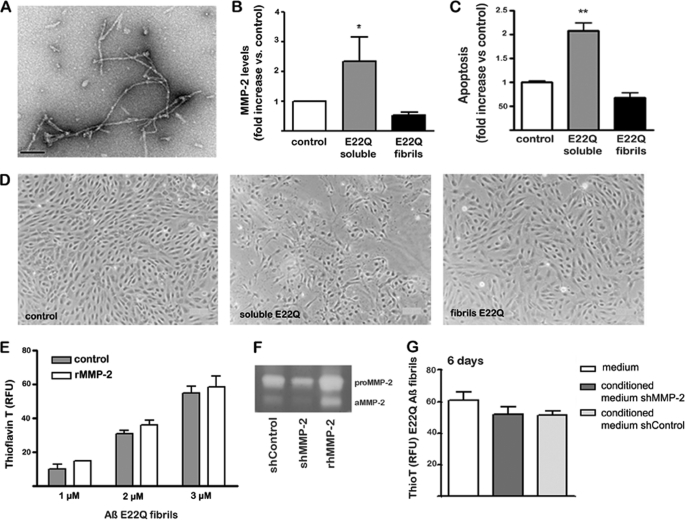
The structural conformation of the different Aβ peptides used to challenge the EC cultures was analyzed by EM. As illustrated in Fig. 1A, WT-Aβ40 mainly exhibited small globular structures, which typically precede the formation of protofibrils (44). On the contrary, both genetic variants showed larger oligomeric components and string-like structures. These elements together with abundant protofibrillar components were profuse and of larger size in the case of the aggressive E22Q variant, which is associated in vivo with very early onset clinical phenotypes.
FIGURE 1.
Evaluation of Aβ-induced MMP secretion by cerebral microvascular endothelial cells. Aβ40, AβE22Q, and AβL34V peptides, solubilized as described under “Experimental Procedures,” were incubated with microvascular EC cultures at 50 μm concentration and MMP secretion, evaluated by SearchLight® human MMP Array. A, shown is an electron microscopic assessment of the conformational structure of the different Aβ peptides used to challenge the EC cultures. B, MMP-1, MMP-2, MMP-3, MMP-8, MMP-9, MMP-10, MMP-13, and TIMP-1 levels were analyzed after a 3-day peptide challenge. Results are expressed as -fold change with respect to untreated control values. In all cases, bars represent the mean ± S.E. of at least five different experiments; statistically significant differences are expressed as p < 0.05 (*) and p < 0.001 (***).
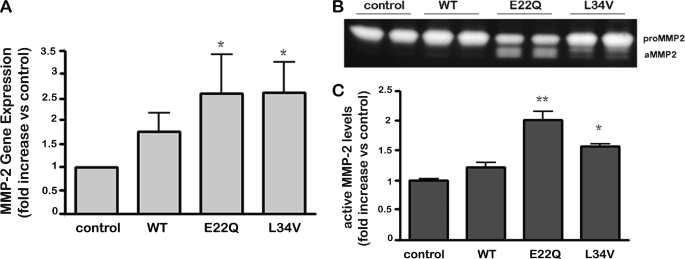
Human cerebral microvascular endothelial cells (hCMEC/D3) were challenged for 3 days with WT, E22Q, and L34V peptides followed by evaluation of metalloproteases content in the culture supernatants using the Multiplex Human MMP Protein Array (SearchLight®), which allows the simultaneous detection of eight proteins of the MMP family: the gelatinases MMP-2 and MMP-9, the collagenases MMP-1, MMP-8, and MMP-13, and the stromelysins MMP-3 and MMP-10 together with the MMP endogenous inhibitor, TIMP-1. The assay showed high reproducibility with a mean intra-assay coefficient of variation of <15% for all proteins in the array. Under the conditions tested, the only significant differences between Aβ-treated samples and no-peptide control cell supernatants were detected at the level of MMP-2 secretion followed by smaller changes in MMP-1 (Fig. 1B). MMP-2 secretion was particularly enhanced in the case of AβE22Q with a mean a 2.65 ± 0.36-fold increase with respect to untreated controls, whereas the other two peptides had less enhancing effect (AβL34V, 1.63 ± 0.28-fold increase); WT Aβ, 1.5 ± 0.18-fold increase) as illustrated in Fig. 1B. Consistent with the enhanced secretion of MMP-2 observed with the multiplex arrays, both Aβ mutants also showed a statistically significant increase in MMP-2 gene expression after a 2-day peptide challenge as evaluated by qRT- PCR (Fig. 2A), whereas the increase in MMP-2 RNA expression induced by WT-Aβ under identical conditions did not reach statistical significance.
FIGURE 2.
Evaluation of Aβ-induced MMP-2 secretion and activation in cerebral microvascular endothelial cells. A, MMP-2 gene expression after a 2-day Aβ treatment was evaluated by quantitative RT-PCR; results illustrate -fold increase in MMP-2 mRNA with respect to untreated control cells. B, a gel zymography shows MMP-2 release into cell supernatants after a 3-day treatment with the different Aβ peptides; the higher molecular mass band corresponds to pro-MMP-2, and the lower molecular mass band to the active form of the enzyme (aMMP-2). Zymography illustrates results of duplicate experiments for each of the peptides as well as the untreated controls. C, densitometric quantitation analysis of the active MMP-2 band in the gelatin zymograph is shown. Bars represent the mean ± S.E. of at least five different experiments; statistically significant differences are expressed as p < 0.05 (*) and p < 0.01 (**).
Release and activation of gelatinases after Aβ treatment was further analyzed by zymography. Both AβE22Q and AβL34V mutants induced a statistically significant release and activation of MMP-2, whereas MMP-9 was not affected by Aβ treatment under our experimental conditions (data not shown), confirming the MMP array results. As shown in Fig. 2B, MMP-2 activation was especially pronounced in AβE22Q. Densitometric analysis of the active MMP-2 zymogram band (Fig. 2C) showed a highly significant 2-fold increase in activation for AβE22Q followed by a 1.5-fold change in the case of AβL34V. In contrast, treatment with WT-Aβ yielded a negligible difference in MMP-2 activation with untreated control cells.
MMP-2 Proteolytically Cleaves WT-Aβ40, the AβL34V Variant, and Less Efficiently, AβE22Q
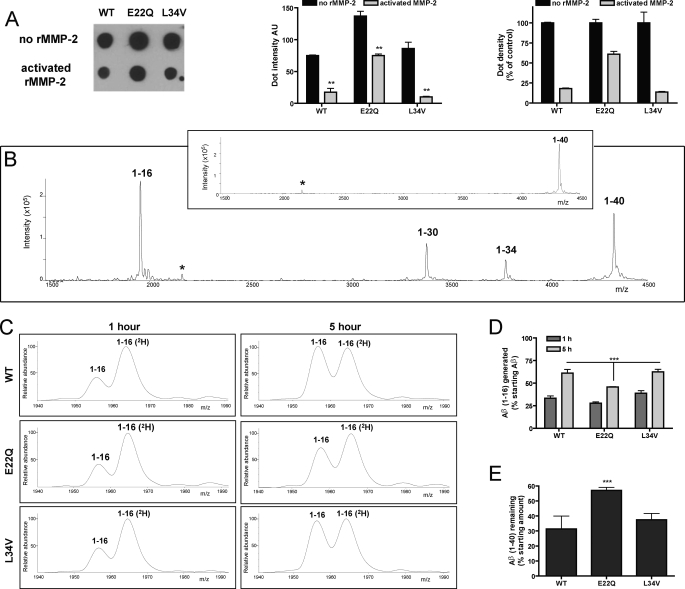
To evaluate whether activated MMP-2 had the capability to enzymatically cleave the different Aβ peptides used in the cell culture treatments, a set of in vitro digestion experiments was designed. The WT, E22Q, and L34V peptides were incubated with rhMMP-2 for either 1 or 5 h, and Aβ proteolysis evaluated by a combination of dot-blot analysis and identification of Aβ cleavage fragments by IP followed by MS. As indicated in Fig. 3A, dot-blot studies revealed that para-aminophenylmercuric acetate-activated rhMMP-2 was able to digest the three Aβ peptides. Densitometric quantitation of dot-blot immunoreactivity showed a statistically significant decrease in dot intensity after enzymatic cleavage for all peptides tested (Fig. 3A, central panel). Normalization of the densitometry data indicated that AβE22Q was notoriously less degraded by the enzyme (after 5 h of incubation, ∼65% of the peptide remained at the sample spot; Fig. 3A, right panel).
FIGURE 3.
Proteolytic digestion of Aβ-variant peptides by human recombinant MMP-2. Aβ40 peptides (WT, E22Q, and L34V) were incubated in the presence or absence of activated human recombinant MMP-2 (1 and 5 h; 37 °C), as described under “Experimental Procedures.” Enzymatic digestion and generation of Aβ proteolytic fragments were evaluated by a combination of dot-blot analysis and IP followed by MALDI-TOF MS. A, shown is a dot-blot assay (probed versus 4G8 anti-Aβ antibodies) of the different Aβ peptides incubated for 5 h in the presence of activated rhMMP-2; the top row illustrates control peptides incubated under identical conditions in the absence of enzyme. The central panel illustrates the dot-blot densitometric quantitation; bars represent mean ± S.E. of three independent experiments. AU, absorbance units. ** indicates statistically significant differences with p < 0.01. The right panel illustrates the normalized densitometry data signals expressed as percentage of signal intensity compared with the immunoreactivity of respective peptides incubated in the absence of recombinant MMP-2 which were considered as 100% intensity. B, a representative qualitative MALDI-TOF MS spectrum of WT Aβ-(1–40) after in vitro digestion with rhMMP-2 for 5 h is shown. The profile illustrates the characteristic peaks of the proteolytically generated fragments 1–16, 1–30, and 1–34 as well as uncleaved remaining Aβ-(1–40). The inset shows the MS profile of controls in which Aβ40 was incubated under identical conditions in the absence of rhMMP-2. * indicates the double-charged peak ((M+H)/2) for Aβ40. C, zoomed-in quantitative MALDI-TOF MS spectra of the 1–16 region of rhMMP-2-digested Aβ peptides at 1- and 5-h digestion time points (left and right panels, respectively) obtained using isotopically labeled Aβ-(1–16) as the internal standard. All Aβ-(1–16) (2H) standard peaks were normalized to the same height to allow direct peak intensity comparison with proteolytically generated Aβ-(1–16) fragments; in all spectra standard peaks represent 625 fmol of Aβ-(1–16) (2H). In all cases 6250 fmol of labeled Aβ-(1–16) standard were added before the immunoprecipitation step (and, therefore, subjected to IP and MS analysis simultaneously with the unknown samples), and 10% of the IP material was analyzed in each spectrum. D, shown is the Aβ-(1–16) fragment generated, expressed in percentage with respect to the initial amount of the respective Aβ peptides incubated with rhMMP-2 for 1 and 5 h. E, shown are the remaining uncleaved Aβ peptides after a 5-h digestion calculated based on the use of 2500 fmol of Aβ-(1–40) (2H) as the internal standard in the MS analysis; results are expressed in percentage with respect to the initial amount of Aβ incubated with the recombinant enzyme. In D and E each bar represents data from duplicate matrix spot quantifications. *** indicates statistically significant differences with p < 0.01 in three independent experiments.
IP followed by MALDI-TOF MS studies revealed the generation of the same three Aβ degradation products after incubation of WT, E22Q, and L34V peptides with rhMMP-2. A representative qualitative mass spectrum of WT-Aβ40 after 5 h of in vitro degradation, shown in Fig. 3B, illustrates the presence of the newly generated fragments Aβ-(1–16), Aβ-(1–30), and Aβ-(1–34) together with the remaining un-cleaved Aβ40. In the absence of rhMMP-2, Aβ40 remained intact under identical experimental conditions (inset). For quantitative MS assessment, we focused on the major fragment generated after 5 h of incubation, Aβ-(1–16), previously shown to have a relevant presence in human CSF (45, 46). The addition of Aβ40- and Aβ-(1–16)-deuterated homologues provided the necessary internal standards for quantitative evaluation, as described under “Experimental Procedures.” Fig. 3C illustrates the zoomed-in MS spectra of the 1940–1990 m/z area showing a time-dependent increase in peak intensity, from the 1-h (left panels) to the 5-h digestion values (right panels), of endogenously generated Aβ-(1–16) (expected average mass, 1955.0 Da) in comparison with that of 6.25 pmol of the isotopically labeled internal standard (expected average mass, 1963.0 Da; mass difference between labeled and unlabeled peptides, +8 Da). Representation of the percentage of Aβ-(1–16) generated with respect to the starting amount of the respective Aβ peptides (Fig. 3D) illustrates the significant increase of the fragment resulting from extending the enzymatic reaction from 1 to 5 h. The data clearly demonstrate that Aβ-(1–16) is a major product of MMP-2 proteolysis, representing from ∼45% to 60% of all the degradation products at the end of the 5-h digestion depending on the Aβ peptide. It is also evident that E22Q is the least susceptible of the three Aβ molecules studied herein to MMP-2 digestion yielding the lowest levels of Aβ-(1–16). Consistent with these data, the levels of undigested intact molecules were higher in E22Q in comparison with L34V and WT peptides, as illustrated in Fig. 3E for the 5-h digestion, corroborating the dot-blot studies (Fig. 3A).
Aβ-(1–16) was also a major element in the MS profile of culture supernatants of Aβ-challenged ECs, and consistent with the rhMMP-2 in vitro digestion data, the fragment was of lower intensity in the case of E22Q than in WT and L34V (not shown). Interestingly, also present in these culture supernatants were other Aβ degradation products (Aβ1–18 and Aβ1–28; not shown), consistent with insulin-degrading enzyme and neprilysin activity, both known expressed enzymes in ECs (47–49).
MMP-2 Silencing Enhances the Apoptotic Response Induced by AβE22Q and AβL34V
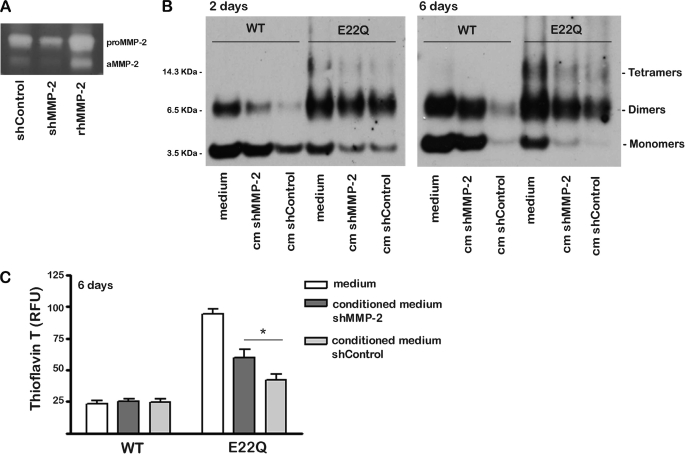
The participation of MMP-2 up-regulation and activation in apoptosis induction was evaluated via the use of shMMP-2 cells, which as described under “Experimental Procedures,” were prepared through stable transfection with plasmids containing MMP-2 shRNA sequences. All clones selected for the current studies displayed a growth rate equivalent to non-transfected cells; supplemental Fig. 1 exemplifies the growth curves of two shMMP-2 cell lines and one control line generated by transfection with scramble RNA sequences (shControl) in comparison with non-transfected cells. Quantitative RT-PCR demonstrated that shMMP-2 cells, in contrast to shControls, expressed only minimal levels of MMP-2 RNA, <80% of the RNA levels of non-transfected cells (Fig. 4A). Corroborating the specificity of the gene silencing, WB analysis indicated that MMP-2 protein expression levels in the pertinent shMMP-2 cell lysates were also highly decreased compared with shControls (Fig. 4B), whereas gel zymography (Fig. 4C) of culture supernatants demonstrated a concomitant reduction in the metalloprotease secretion. This reduction in the secreted enzyme levels was further quantitated by ELISA, which as illustrated in Fig. 4D, revealed that MMP-2 concentration in shMMP-2-conditioned medium was decreased to a mean of 3 ng/ml, whereas shControls exhibited mean values of 20 ng/ml.
FIGURE 4.
Generation of short hairpin RNA-MMP-2 (shMMP-2) cerebral endothelial cells. A, shown is relative quantitation of MMP-2 gene expression by qRT-PCR; the graph illustrates -fold increase expression of MMP-2 RNA in shMMP-2 and shControl cells compared with non-transfected cells. B, shown is a MMP-2 Western blot analysis of shMMP-2 cell lysates compared with the corresponding control cells transfected with scrambled shRNAs. Immunoreactivity of GAPDH, used as loading control, is indicated in the bottom panel. C, shown is gel zymography of shMMP-2 and shControl conditioned media. The figure also illustrates enzymatically digested bands corresponding to rhMMP-2 used as standard in the zymogram. aMMP-2, active MMP-2. D, total MMP-2 levels, measured by ELISA, in cell supernatants from shMMP-2 and shControl cells are shown. cm, conditioned media. In B and E, bars illustrate the mean ± S.E. of at least three different experiments.
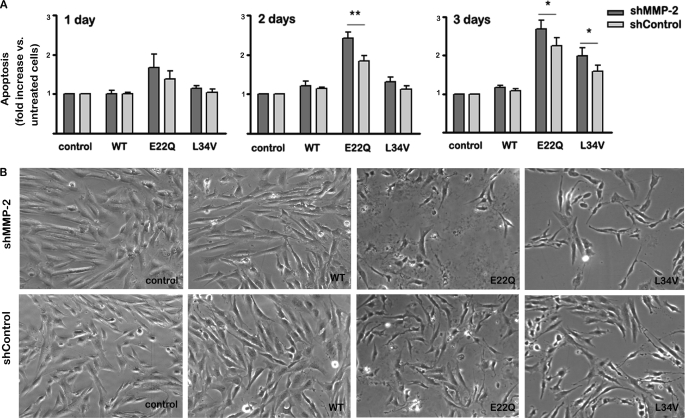
Quantitation of nucleosome formation by Cell Death ELISA indicated that AβE22Q elicited a rapid apoptotic response with enhanced levels observed after only 1 day of peptide challenge (Fig. 5), in agreement with our previous reports (29). In the case of AβL34V, consistent with its lower rate of formation of oligomeric/protofibrillar assemblies, there was a delay in apoptosis induction. Nucleosome formation started at day 2 but only reached significant levels, a 1.5-fold increase with respect to untreated control cells, after a 3-day treatment. In contrast, Aβ-(1–16), the main proteolytic fragment generated by MMP-2 cleavage of WT and variant Aβ peptides, failed to induce apoptosis even after a 3-day challenge (supplemental Fig. 2A). Confirming its lack of toxicity, EC 3-day treatment with Aβ-(1–16) also failed to elicit a differential LDH release compared with untreated control cells (supplemental Fig. 2B).
FIGURE 5.
Effect of MMP-2 silencing on Aβ-induced apoptosis in microvascular ECs. Cells were challenged with Aβ-(1–40) peptides (WT, E22Q, and L34V) for 1, 2, and 3 days, and induction of apoptosis was assessed by quantitation of nucleosome formation by Cell Death ELISA together with morphological evaluation by contrast phase microscopy, as described under “Experimental Procedures.” A, apoptosis rate in shMMP-2 and shControl cells is shown. Nucleosome generation values are expressed as -fold increase with respect to untreated control cells. Bars indicate the mean ± S.E. of at least five independent experiments; statistically significant differences with p < 0.01 are indicated by ** and with p < 0.05 by *. B, shown are phase contrast images (400× magnifications) of shMMP-2 and shControl cells after a 3-day treatment with the respective Aβ peptides.
Notably, MMP-2 silencing enhanced the apoptotic response elicited by both E22Q and L34V variant peptides (Fig. 5A). At the 2- and 3-day time points a statistically significant difference between the nucleosome levels generated in shMMP-2 cells compared with shControls could be observed. Contrast phase microscopy confirmed the Cell Death ELISA data (Fig. 5B). Whereas challenge with WT-Aβ40 did not induce noticeable morphological changes in the time frame of the experiments, treatment with both Aβ mutants resulted in fewer cells remaining attached to the culture dishes as well as in abnormal cell morphology with the appearance of characteristic features of apoptotic cells including cell shrinking and formation of apoptotic bodies (bottom panels). Corroborating the ELISA findings, MMP-2 silencing accentuated the signature morphological apoptotic changes (top panels).
MMP-2 Silencing Restricts Aβ Degradation and Increases Fibrillization
WB analysis and thioflavin-T binding assays were used to determine whether the secretion and activation of MMP-2 induced by treatment with the different amyloid peptides resulted in turn in their proteolytic degradation. As illustrated by the WB in Fig. 6A, MMP-2 silencing resulted in higher levels of monomeric and dimeric forms of WT-Aβ40, AβE22Q, and AβL34V in comparison with the peptides present in supernatants of shControl cells. In the case of the oligomerization-prone AβE22Q, exhibiting tetrameric species under the conditions tested, MMP-2 silencing also translated in elevated levels of these assemblies. Although all visible species appear to be sensitive to proteolysis, an enhanced susceptibility of the monomeric assemblies was evident. The effect of MMP-2 silencing on larger oligomeric and fibrillar components was evaluated by fluorescence quantitation of thioflavin-T binding in culture supernatants after a 3-day challenge with the amyloid peptides. Consistent with its demonstrated proclivity for fibrillogenesis (29), only AβE22Q showed elevated thioflavin-T fluorescence values at this time point. MMP-2 silencing translated in statistically significant ∼2-fold higher levels of thioflavin-T fluorescence in comparison to shControl supernatants (Fig. 6B). Confirming the thioflavin-T results, EM imaging revealed that conditioned media from AβE22Q-treated shMMP-2 cells exhibited higher levels of fibrillar structures (Fig. 6C). Furthermore, the fibrillar elements also showed different morphological features being longer than those in shControl supernatants, many of them over the typical 200-nm length of protofibrillar assemblies (43).
FIGURE 6.
Proteolytic degradation of Aβ peptides in EC-conditioned media. shMMP-2 and shControl cells were incubated with Aβ-(1–40) peptides (WT, E22Q, and L34V) for 3 days, and subsequent Aβ degradation resulting from enzymatic release and activation was evaluated by Western blot analysis and a decrease in thioflavin-T binding as described under “Experimental Procedures.” A, shown is a Western blot analysis of the respective Aβ peptides in conditioned media from shMMP-2 and shControl cells probed with a combination of 4G8 and 6E10 anti-Aβ antibodies. Electrophoretic mobility of the molecular weight standards is indicated on the left of the panel. B, thioflavin-T fluorescence assay of the respective conditioned media is shown; results are expressed in fluorescence units (RFU) at 300 s and represent the mean ± S.E. of at least three different experiments. C, shown are electron microscopy images of shMMP-2- and shControl-conditioned media after a 3-day challenge with AβE22Q. Fibrillar elements in shMMP-2 cell supernatants are more abundant and longer than those in shControl, typically 200–600 nm compared with <200 nm. The figure shows representative images for each condition; the bar indicates 100 nm. The graph on the right panel indicates quantitation of AβE22Q fibrils in the EM analysis; results represent the mean ± S.E. of number of fibrils present per field (n = 30 high magnification fields (×110,000)) in at least three different experiments. * indicates statistically significant differences with p < 0.05.
To confirm the cell culture experiments, WT and E22Q Aβ peptides were incubated in vitro with conditioned media from shMMP-2 and shControl cells that had not been previously challenged with the amyloid peptides and, therefore, contained only the respective endogenous MMP-2 levels. Aβ degradation after incubation with the respective cell supernatants was evaluated by WB and thioflavin-T binding, as described under “Experimental Procedures.” Fig. 7A illustrates the levels of pro- and active MMP-2 in each of the culture supernatants as evaluated by gel zymography. WB revealed that incubation with conditioned medium from control cells sharply reduced monomeric and dimeric assemblies of WT-Aβ40, in comparison with the peptide incubated with medium that had not been in contact with cells for the same period of time (Fig. 7B). This effect increased with time of incubation, and at 6 days both monomeric and dimeric components were practically absent in shControl medium. AβE22Q was also susceptible to proteolytic degradation by this conditioned medium. However, in this case peptide degradation was more evident after a 6-day incubation, further indicating the lower susceptibility of AβE22Q to enzymatic cleavage. Confirming the role of MMP-2 in Aβ degradation, silencing of the protease in shMMP-2 cells translated in decreased degradation of both Aβ peptides. Evaluation of fibrillar/protofibrillar components by thioflavin-T fluorescence revealed the highest signals upon incubation of AβE22Q with media in the absence of cells and, therefore, with minimum levels of MMP-2. Conditioned medium from shControl cells, consistent with the higher content of MMP-2 than in supernatants of shMMP-2 cells (illustrated in Fig. 7A), caused a significant decrease in thioflavin-T fluorescence, indicating the enzyme was capable of additionally degrading higher order oligomeric/fibrillar assemblies (Fig. 7C).
FIGURE 7.
In vitro digestion of Aβ peptides by conditioned media of shMMP-2 and shControl cells. Aβ peptides (WT and E22Q) were incubated in vitro for 2 and 6 days at 37 °C with conditioned media from amyloid-unchallenged shMMP-2 and shControl cells. Aβ degradation was evaluated by Western blot analysis and thioflavin-T binding, as described under “Experimental Procedures.” As controls, the respective Aβ peptides were incubated under identical conditions with culture media that had not been preincubated with the corresponding cell lines. A, gel zymography of conditioned media from amyloid unchallenged shMMP-2 and shControl cells grown to confluence to maximize endogenous MMP-2 secretion levels is shown. The electrophoretic bands corresponding to rhMMP-2, both the pro-form and the activated enzyme, are shown for comparison. aMMP-2, active MMP-2. B, shown is Western blot analysis of Aβ peptides after incubation with the respective conditioned media, probed with a combination of 4G8 and 6E10 anti-Aβ. Lanes labeled medium represent media that had not been in contact with the respective cell lines; cm indicates conditioned medium. Electrophoretic mobility of the molecular weight standards is indicated on the left of the panel. C, shown is a thioflavin-T fluorescence assay of the respective conditioned media. RFU, relative fluorescence units. (Bars represent the mean ± S.E. of three independent experiments. * indicates statistically significant differences with p < 0.05.
AβE22Q Fibrillar Assemblies Do Not Induce MMP-2 Release and Are Not Susceptible to Proteolysis
Fibril-enriched AβE22Q preparations generated after a 6-day peptide incubation, as described under “Experimental Procedures,” exhibited, as expected, abundant elongated fibrillar components under EM examination (Fig. 8A). These AβE22Q assemblies failed to induce secretion of MMP-2 by EC cultures, as evaluated with MMP antibody arrays. In contrast to soluble AβE22Q assemblies that elicited a >2-fold increase in MMP-2 secretion with respect to untreated control cells, fibril-enriched preparations had no effect (Fig. 8B). This lack of MMP-2 release correlated with the inability of fibril-enriched AβE22Q species to induce apoptosis. Cell Death ELISA indicated no difference in nucleosome formation with respect to untreated controls, in contrast to the 2-fold increase induced by AβE22Q soluble oligomeric/protofibrillar structures (Fig. 8C). The absence of morphological changes after EC challenge with AβE22Q fibrillar components, in contrast to cells exposed to AβE22Q soluble oligomeric assemblies in which shrunk cells and abundant apoptotic bodies were observed (Fig. 8D), corroborated the Cell Death ELISA data. To confirm the cell culture experiments, fibril-enriched AβE22Q was incubated in vitro with rhMMP-2, and the presence of fibrillar elements was evaluated by thioflavin-T binding. As illustrated in Fig. 8E, the fluorescence levels remained unchanged after 24 h of incubation with the enzyme in comparison with control samples in which the peptide was incubated under identical conditions in the absence of MMP-2. Further corroborating these results, incubation of shControl cell-conditioned media, exhibiting active MMP-2 by gelatin zymography analysis (Fig. 8F), failed to degrade AβE22Q fibrillar components, therefore, yielding unchanged thioflavin-T levels (Fig. 8G). These fluorescence levels remained practically constant even after a 6-day incubation.
FIGURE 8.
AβE22Q fibrillar assemblies fail to elicit MMP-2 release and activation or an apoptotic response in microvascular endothelial cell cultures. EC cultures were challenged with AβE22Q soluble and fibril-enriched assemblies for 3 days followed by evaluation of MMP-2 secretion, assessment of Aβ fibrillar components in culture supernatants by thioflavin-T-binding assays, and estimation of apoptosis induction by morphological studies and Cell Death ELISA. A, electron microscopy images illustrating AβE22Q fibrillar assemblies were prepared as described under “Experimental Procedures”; the bar represents 100 nm. Note: please refer to Fig. 1A for EM visualization of AβE22Q-soluble assemblies. B, evaluation of MMP-2 secretion by SearchLight® human MMP antibody arrays is shown. Results are expressed as -fold increase with respect to control cells incubated in the absence of Aβ peptides. Bars represent mean ± S.E. of triplicate experiments; * indicates statistically significance differences with p < 0.05. C, evaluation of apoptosis induction by nucleosome quantitation via Cell Death ELISA is shown. Apoptosis rate is represented in -fold change with respect to untreated control cells. Bars indicate the mean ± S.E. of three independent experiments; ** highlights statistically significant differences with p < 0.01. D, contrast phase microscopy images at 100× magnification exemplify changes in cell density and morphology after Aβ treatment. E, shown is a thioflavin-T fluorescence assay of AβE22Q fibrils after 24 h in vitro incubation with rhMMP-2; as controls, the fibrillar assemblies were incubated under identical conditions in the absence of the enzyme. Bars represent the mean ± S.E. of at least three independent experiments. F, gel zymography of conditioned media from untreated shMMP-2 and shControl cells is shown; the electrophoretic bands corresponding to rhMMP-2, both the pro-form and the activated enzyme, are also shown as control. aMMP-2, active MMP-2. G, shown is a thioflavin-T fluorescence assay of AβE22Q fibrils after in vitro incubation for 6 days with conditioned media from shMMP-2 and shControl cells. As negative controls, AβE22Q fibrils were incubated with plain media that had not been in contact with cells. Results are expressed as the mean values ± S.E. of at least three independent experiments. RFU, relative fluorescence units.
DISCUSSION
The cross-talk among various cell types, endothelial, astrocytes, pericytes, and neurons, constitutes a dynamic entity, the neurovascular unit, that regulates CNS development, modulates cerebral blood flow, and influences the permeability properties of the blood-brain barrier (50). Recent epidemiologic, clinical and pathologic studies emphasized the importance of the microvascular system as well as the consequences of its compromise by amyloid deposition in the functional integrity of the neurovascular unit, revealing a strong association between cognitive decline, microvessel abnormalities, and cerebrovascular dysfunction (51–55). Puzzlingly, vascular amyloid load, albeit affecting the morphology and functionality of the vessels, does not directly correlate with the presence of hemorrhagic transformations. This is clearly exemplified by the existence of certain non-Aβ forms of cerebral amyloidosis, such as the chromosome-13 dementias, associated with striking vascular deposition in practical absence of hemorrhagic strokes. Reinforcing the notion of differential mechanisms leading to BBB leakage and hemorrhage, specific vasculotropic Aβ variants predominantly associate with hemorrhagic manifestations, whereas others, although correlating with severe deposition in the vasculature, do not translate in hemorrhagic episodes (14). In this context, the data presented herein demonstrate that challenging cerebral endothelial cells with AβE22Q and AβL34V, both Aβ genetic variants associated with ICH, results in the exacerbated production and activation of MMP-2, an endopeptidase with the ability to specifically cleave type IV and V collagen, as well as laminin and proteoglycans, all major structural components of basement membranes (56, 57). In fact, direct intracerebral injection of MMP-2 has been shown to induce opening of the BBB causing intracerebral hemorrhage (58–60), a deleterious effect reduced by administering the MMP-2 natural inhibitor TIMP-2 (61). The action of MMP-2, which was differentially secreted and activated in our in vitro model in comparison with cells challenged with WT-Aβ, could explain at least in part the severe ICH observed in vivo in Dutch and Piedmont kindreds. Our findings extend previous reports indicating MMP-2 activation in smooth muscle cells after treatment with AβE22Q (62) and suggest the existence of more general mechanisms associated with CAA variants correlating with hemorrhagic complications.
Supporting a role for MMPs in the pathogenesis of spontaneous intracerebral hemorrhage in patients with CAA, various groups have demonstrated an association of MMPs to CAA-related pathology in different experimental models. MMP-9 overexpression was evident in CAA vessels exhibiting microhemorrhages in aged amyloid precursor protein APPsw transgenic mice, in contrast with wild-type and young transgenic animals (63, 64). Additionally, and more relevant to the current data, MMP-2 activation was demonstrated as a pivotal component of the tight junction protein disruption occurring concomitantly to BBB permeability alterations in a rat model of ischemia reperfusion (65). This model, in which a predominant increase in MMP-2 over the gelatinase MMP-9 could be clearly demonstrated during the early phase of the BBB disruption, provided direct evidence implicating the proteolytic degradation of the tight junction proteins occludin and claudin in the BBB opening leading to increased permeability. These changes could be prevented through pharmacologic modulation by a broad-spectrum MMP inhibitor, a strategy also able to decrease the frequency of hemorrhage in rat and rabbit stroke animal models (66, 67). Interestingly, MMP pharmacologic inhibition was also successful in another experimental model in which it significantly reduced the concomitant oxidative stress associated with vascular deposits in amyloid precursor protein transgenic lines (68), suggesting a multifactorial effect for MMPs in vessel pathology.
The role of MMPs in the induction of cell toxicity remains controversial, perhaps reflecting the kaleidoscope of functions exhibited by these enzymes. In addition to causing a direct loss of vessel wall integrity, as described above, MMP secretion is known to affect specific ECM-integrin interactions resulting in loss of cell survival signals and induction of apoptotic vascular cell death (69, 70). On the other hand, MMPs appear to counterbalance Aβ-mediated neurotoxicity occurring through activation of death receptor pathways (71), in particular the FAS receptor. In this sense, MMP pharmacologic inhibition was shown to act synergistically with Aβ to dramatically increase neurotoxicity, an effect that correlated with the modulation of membrane-bound FAS ligand levels. Consistent with these studies, our results add another element by which MMP inhibition may contribute to cell death and apoptosis. In the in vitro paradigms presented herein, silencing of MMP-2 also contributed to the enhancement of the Aβ pro-apoptotic role through a decrease of in situ peptide degradation. Assessment of Aβ degradation products in culture supernatants of amyloid-challenged cells revealed that the peptides were actively cleaved in situ with production of different Aβ-cleavage fragments and concomitant reduction of apoptosis induction. Quantitative MS analysis indicated that Aβ-(1–16) was a prominent fragment representing more than 50% of all Aβ degradation products. Other enzymes likely present in the EC-conditioned media have also been reported to cleave Aβ at peptide bond 16–17, including endothelin converting enzyme, plasmin, and to a lower extent MMP-9 (33, 72, 73). Nevertheless, our in vitro studies using recombinant MMP-2 confirm the protease activity at this peptide bond and suggest that EC-released MMP-2 contributes to the generation of Aβ-(1–16). Notably, this Aβ degradation fragment is not only the major MMP-2 generated cleavage product of Aβ40 and Aβ42 peptides purified from AD brain (35), but it is also a constituent of human CSF (45). In this biological fluid, Aβ-(1–16) appears to constitute a robust biomarker and the most important C-terminally truncated Aβ peptide for discriminating AD from non-demented controls (45). In fact, cerebrospinal fluid Aβ-(1–16) likely reflects Aβ brain clearance mechanisms as the peptide exhibits poor aggregation propensity and lacks neurotoxicity (74). Our data demonstrating the additional lack of toxicity of Aβ-(1–16) for ECs provides further support for the notion that proteolytic degradation of Aβ may play a key role in the regulation of the detrimental role of amyloid for disease pathogenesis.
Degradation of Aβ peptides in culture supernatants as well as in vitro digestion assays using recombinant MMP-2 indicated that variant Aβ molecules were susceptible to proteolytic cleavage at the same peptide bonds as WT Aβ40, generating the same degradation fragments. However, the efficiency of proteolysis differed among the different peptides, with AβE22Q being the most resistant of the ones studied, in agreement with previous findings with other proteases including insulin-degrading enzyme and neprilysin, as reported by our and other groups (75–77). This resistance to proteolysis with the potential for reducing brain clearance in vivo and thereby exacerbating the detrimental effect of amyloid most likely relates to the enhanced aggregation propensity of AβE22Q, as illustrated in our data by its highest thioflavin-T fluorescence and tendency to form protofibrillar/fibrillar assemblies revealed by EM. In this sense, our data indicate that, although soluble Aβ monomeric and oligomeric components are susceptible to MMP-2 action, fibril-enriched preparations of AβE22Q were practically unaffected by MMP-2. Notably, these fibril-enriched samples were also incapable of inducing MMP-2 secretion and activation or eliciting an apoptotic cellular response. The concept of oligomeric assemblies as the active species is not new (78). Originally supporting a pivotal role for intermediate assemblies in neurotoxicity, it was further extended to cover the potent effects of these species in inducing neuronal synaptic disruption (for review, see Refs. 78 and 79). Our data further extend the relevance of intermediate oligomeric/protofibrillar forms to other elements of disease pathogenesis, the enhanced secretion and activation of proteases with potential to disrupt BBB permeability. In this context as well as in cell toxicity, fibrillar elements also fail to induce a relevant response. Furthermore, based on the data presented herein, it is conceivable that fibrillar components may even exert a protective cellular effect. In agreement with this notion, it has been demonstrated that promoting Aβ42 fibrillization proves beneficial at inhibiting neurotoxicity, likely through a decrease in the formation of more toxic oligomeric assemblies, a concept postulated to have potential therapeutic implications (80, 81).
In summary, our data highlight the dichotomy of EC responses to Aβ challenge. At one end, MMP-2 secretion translates in Aβ in situ degradation with a concomitant delay in apoptosis induction. On the other end, the enhanced MMP-2 secretion and activation observed, particularly in association with Aβ variants correlating with clinical phenotypes of ICH, together with the known capacity of MMPs to degrade extracellular matrices, basement membrane components, and tight junction proteins suggest that CAA-Aβ might undesirably compromise BBB integrity and precipitate a hemorrhagic phenotype. Overall, our results emphasize the complexity of the cellular mechanisms elicited by Aβ. Further studies will elucidate the preponderance of the individual responses under specific disease conditions or at different stages of disease progression, events crucial for the rational design of future therapeutic strategies.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants NS051715, AG30539, and NS050276. The work was also supported by Spanish Ministry of Health Grant PI070737, the Instituto de Salud Carlos III, the Alzheimer's Association, and the American Heart Association.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- CAA
- cerebral amyloid angiopathy
- Aβ
- amyloid-β
- AD
- Alzheimer disease
- BBB
- blood brain barrier
- ICH
- intracerebral hemorrhage
- MMP
- matrix metalloproteinases
- rhMMP
- recombinant human MMP-2
- qRT
- quantitative RT
- SN
- supernatant
- IP
- immunoprecipitation
- WB
- Western blot
- EC
- endothelial cell
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- CAPS
- 3-(cyclohexylamino)propanesulfonic acid
- WT
- wild type.
REFERENCES
- 1.Yamada M. (2000) Neuropathology 20, 8–22 [DOI] [PubMed] [Google Scholar]
- 2.Kalaria R. N. (1992) Brain Metab. Rev. 4, 226–260 [PubMed] [Google Scholar]
- 3.Castaño E. M., Prelli F., Soto C., Beavis R., Matsubara E., Shoji M., Frangione B. (1996) J. Biol. Chem. 271, 32185–32191 [DOI] [PubMed] [Google Scholar]
- 4.Rensink A. A., de Waal R. M., Kremer B., Verbeek M. M. (2003) Brain Res. Rev. 43, 207–223 [DOI] [PubMed] [Google Scholar]
- 5.Vonsattel J. P., Myers R. H., Hedley-Whyte E. T., Ropper A. H., Bird E. D., Richardson E. P., Jr. (1991) Ann. Neurol. 30, 637–649 [DOI] [PubMed] [Google Scholar]
- 6.Mandybur T. I. (1986) J. Neuropathol. Exp. Neurol. 45, 79–90 [PubMed] [Google Scholar]
- 7.O'Donnell H. C., Rosand J., Knudsen K. A., Furie K. L., Segal A. Z., Chiu R. I., Ikeda D., Greenberg S. M. (2000) N. Engl. J. Med. 342, 240–245 [DOI] [PubMed] [Google Scholar]
- 8.Maia L. F., Vasconcelos C., Seixas S., Magalhães R., Correia M. (2006) Cerebrovasc. Dis. 22, 155–161 [DOI] [PubMed] [Google Scholar]
- 9.Greenberg S. M., Gurol M. E., Rosand J., Smith E. E. (2004) Stroke 35, 2616–2619 [DOI] [PubMed] [Google Scholar]
- 10.Brayne C., Matthews F. E., Xuereb J. H., Broome J. C., McKenzie J., Rossi M., Ince P. G., McKeith I. G., Lowe J., Esiri M. M., Morris J. H. (2001) Lancet 357, 169–17511213093 [Google Scholar]
- 11.Rostagno A., Ghiso J. (2008) Neurodegener. Dis. 5, 173–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar-Singh S. (2009) Int. J. Mol. Sci. 10, 1872–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Revesz T., Holton J. L., Lashley T., Plant G., Frangione B., Rostagno A., Ghiso J. (2009) Acta Neuropathol. 118, 115–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rostagno A., Holton J. L., Lashley T., Revesz T., Ghiso J. (2010) Cell. Mol. Life Sci. 67, 581–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levy E., Carman M. D., Fernandez-Madrid I. J., Power M. D., Lieberburg I., van Duinen S. G., Bots G. T., Luyendijk W., Frangione B. (1990) Science 248, 1124–1126 [DOI] [PubMed] [Google Scholar]
- 16.Van Broeckhoven C., Haan J., Bakker E., Hardy J. A., Van Hul W., Wehnert A., Vegter-Van der Vlis M., Roos R. A. (1990) Science 248, 1120–1122 [DOI] [PubMed] [Google Scholar]
- 17.Bornebroek M., Haan J., Van Duinen S. G., Maat-Schieman M. L., Van Buchem M. A., Bakker E., Van Broeckhoven C., Roos R. A. (1997) Ann. Neurol. 41, 695–698 [DOI] [PubMed] [Google Scholar]
- 18.Maat-Schieman M., Roos R., van Duinen S. (2005) Neuropathology 25, 288–297 [DOI] [PubMed] [Google Scholar]
- 19.van Duinen S. G., Castaño E. M., Prelli F., Bots G. T., Luyendijk W., Frangione B. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 5991–5994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Obici L., Demarchi A., de Rosa G., Bellotti V., Marciano S., Donadei S., Arbustini E., Palladini G., Diegoli M., Genovese E., Ferrari G., Coverlizza S., Merlini G. (2005) Ann. Neurol. 58, 639–644 [DOI] [PubMed] [Google Scholar]
- 21.Murakami K., Irie K., Morimoto A., Ohigashi H., Shindo M., Nagao M., Shimizu T., Shirasawa T. (2003) J. Biol. Chem. 278, 46179–46187 [DOI] [PubMed] [Google Scholar]
- 22.Davis J., Van Nostrand W. E. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 2996–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muñoz F. J., Opazo C., Gil-Gómez G., Tapia G., Fernández V., Valverde M. A., Inestrosa N. C. (2002) J. Neurosci. 22, 3081–3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eisenhauer P. B., Johnson R. J., Wells J. M., Davies T. A., Fine R. E. (2000) J. Neurosci. Res. 60, 804–810 [DOI] [PubMed] [Google Scholar]
- 25.Viana R. J., Nunes A. F., Castro R. E., Ramalho R. M., Meyerson J., Fossati S., Ghiso J., Rostagno A., Rodrigues C. M. (2009) Cell. Mol. Life Sci. 66, 1094–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miravalle L., Tokuda T., Chiarle R., Giaccone G., Bugiani O., Tagliavini F., Frangione B., Ghiso J. (2000) J. Biol. Chem. 275, 27110–27116 [DOI] [PubMed] [Google Scholar]
- 27.Wisniewski T., Ghiso J., Frangione B. (1991) Biochem. Biophys. Res. Commun. 179, 1247–1254 [DOI] [PubMed] [Google Scholar]
- 28.Van Nostrand W. E., Melchor J. P., Ruffini L. (1998) J. Neurochem. 70, 216–223 [DOI] [PubMed] [Google Scholar]
- 29.Fossati S., Cam J., Meyerson J., Mezhericher E., Romero I. A., Couraud P. O., Weksler B. B., Ghiso J., Rostagno A. (2010) FASEB J. 24, 229–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montaner J., Molina C. A., Monasterio J., Abilleira S., Arenillas J. F., Ribó M., Quintana M., Alvarez-Sabín J. (2003) Circulation 107, 598–603 [DOI] [PubMed] [Google Scholar]
- 31.Alvarez-Sabín J., Delgado P., Abilleira S., Molina C. A., Arenillas J., Ribó M., Santamarina E., Quintana M., Monasterio J., Montaner J. (2004) Stroke 35, 1316–1322 [DOI] [PubMed] [Google Scholar]
- 32.Abilleira S., Montaner J., Molina C. A., Monasterio J., Castillo J., Alvarez-Sabín J. (2003) J. Neurosurg. 99, 65–70 [DOI] [PubMed] [Google Scholar]
- 33.Backstrom J. R., Lim G. P., Cullen M. J., Tökés Z. A. (1996) J. Neurosci. 16, 7910–7919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan P., Hu X., Song H., Yin K., Bateman R. J., Cirrito J. R., Xiao Q., Hsu F. F., Turk J. W., Xu J., Hsu C. Y., Holtzman D. M., Lee J. M. (2006) J. Biol. Chem. 281, 24566–24574 [DOI] [PubMed] [Google Scholar]
- 35.Roher A. E., Kasunic T. C., Woods A. S., Cotter R. J., Ball M. J., Fridman R. (1994) Biochem. Biophys. Res. Commun. 205, 1755–1761 [DOI] [PubMed] [Google Scholar]
- 36.Crouch P. J., Tew D. J., Du T., Nguyen D. N., Caragounis A., Filiz G., Blake R. E., Trounce I. A., Soon C. P., Laughton K., Perez K. A., Li Q. X., Cherny R. A., Masters C. L., Barnham K. J., White A. R. (2009) J. Neurochem. 108, 1198–1207 [DOI] [PubMed] [Google Scholar]
- 37.Miners J. S., Baig S., Palmer J., Palmer L. E., Kehoe P. G., Love S. (2008) Brain Pathol. 18, 240–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weksler B. B., Subileau E. A., Perrière N., Charneau P., Holloway K., Leveque M., Tricoire-Leignel H., Nicotra A., Bourdoulous S., Turowski P., Male D. K., Roux F., Greenwood J., Romero I. A., Couraud P. O. (2005) FASEB J. 19, 1872–1874 [DOI] [PubMed] [Google Scholar]
- 39.Rostagno A., Ghiso J. (2009) Curr. Protoc. Cell Biol. Chapter 3, 3.33.31–33.33.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomidokoro Y., Lashley T., Rostagno A., Neubert T. A., Bojsen-Møller M., Braendgaard H., Plant G., Holton J., Frangione B., Révész T., Ghiso J. (2005) J. Biol. Chem. 280, 36883–36894 [DOI] [PubMed] [Google Scholar]
- 41.Tomidokoro Y., Rostagno A., Neubert T. A., Lu Y., Rebeck G. W., Frangione B., Greenberg S. M., Ghiso J. (2010) Am. J. Pathol. 176, 1841–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Solito R., Corti F., Fossati S., Mezhericher E., Donnini S., Ghiso J., Giachetti A., Rostagno A., Ziche M. (2009) Exp. Cell Res. 315, 385–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walsh D. M., Hartley D. M., Kusumoto Y., Fezoui Y., Condron M. M., Lomakin A., Benedek G. B., Selkoe D. J., Teplow D. B. (1999) J. Biol. Chem. 274, 25945–25952 [DOI] [PubMed] [Google Scholar]
- 44.Harper J. D., Wong S. S., Lieber C. M., Lansbury P. T., Jr. (1999) Biochemistry 38, 8972–8980 [DOI] [PubMed] [Google Scholar]
- 45.Portelius E., Zetterberg H., Andreasson U., Brinkmalm G., Andreasen N., Wallin A., Westman-Brinkmalm A., Blennow K. (2006) Neurosci. Lett. 409, 215–219 [DOI] [PubMed] [Google Scholar]
- 46.Gelfanova V., Higgs R. E., Dean R. A., Holtzman D. M., Farlow M. R., Siemers E. R., Boodhoo A., Qian Y. W., He X., Jin Z., Fisher D. L., Cox K. L., Hale J. E. (2007) Brief Funct. Genomic Proteomic 6, 149–158 [DOI] [PubMed] [Google Scholar]
- 47.Morelli L., Llovera R., Ibendahl S., Castaño E. M. (2002) Neurochem. Res. 27, 1387–1399 [DOI] [PubMed] [Google Scholar]
- 48.Chen K. L., Wang S. S., Yang Y. Y., Yuan R. Y., Chen R. M., Hu C. J. (2009) Biochem. Biophys. Res. Comm. 378, 57–61 [DOI] [PubMed] [Google Scholar]
- 49.Graf K., Koehne P., Gräfe M., Zhang M., Auch-Schwelk W., Fleck E. (1995) Hypertension 26, 230–235 [DOI] [PubMed] [Google Scholar]
- 50.McCarty J. H. (2005) Assay Drug Dev. Technol. 3, 89–95 [DOI] [PubMed] [Google Scholar]
- 51.Farkas E., Luiten P. G. M. (2001) Prog. Neurobiol. 64, 575–611 [DOI] [PubMed] [Google Scholar]
- 52.Bailey T. L., Rivara C. B., Rocher A. B., Hof P. R. (2004) Neurol. Res. 26, 573–578 [DOI] [PubMed] [Google Scholar]
- 53.de la Torre J. C. (2004) Neurol. Res. 26, 517–524 [DOI] [PubMed] [Google Scholar]
- 54.Gorelick P. B. (2004) Stroke 35, 2620–2622 [DOI] [PubMed] [Google Scholar]
- 55.Petty M. A., Wettstein J. G. (2001) Brain Res. Rev. 36, 23–34 [DOI] [PubMed] [Google Scholar]
- 56.Rosenberg G. A. (2009) Lancet Neurol. 8, 205–216 [DOI] [PubMed] [Google Scholar]
- 57.Yong V. W., Power C., Forsyth P., Edwards D. R. (2001) Nat. Rev. Neurosci 2, 502–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosenberg G. A., Mun-Bryce S., Wesley M., Kornfeld M. (1990) Stroke 21, 801–807 [DOI] [PubMed] [Google Scholar]
- 59.Rosenberg G. A., Estrada E., Kelley R. O., Kornfeld M. (1993) Neurosci. Lett. 160, 117–119 [DOI] [PubMed] [Google Scholar]
- 60.Rosenberg G. A., Estrada E. Y., Dencoff J. E., Stetler-Stevenson W. G. (1995) Brain Res. 703, 151–155 [DOI] [PubMed] [Google Scholar]
- 61.Rosenberg G. A., Kornfeld M., Estrada E., Kelley R. O., Liotta L. A., Stetler-Stevenson W. G. (1992) Brain Res. 576, 203–207 [DOI] [PubMed] [Google Scholar]
- 62.Jung S. S., Zhang W., Van Nostrand W. E. (2003) J. Neurochem. 85, 1208–1215 [DOI] [PubMed] [Google Scholar]
- 63.Lee J. M., Yin K. J., Hsin I., Chen S., Fryer J. D., Holtzman D. M., Hsu C. Y., Xu J. (2003) Ann. Neurol. 54, 379–382 [DOI] [PubMed] [Google Scholar]
- 64.Lee J. M., Yin K., Hsin I., Chen S., Fryer J. D., Holtzman D. M., Hsu C. Y., Xu J. (2005) J. Neurol. Sci. 229–230, 249–254 [DOI] [PubMed] [Google Scholar]
- 65.Yang Y., Estrada E. Y., Thompson J. F., Liu W., Rosenberg G. A. (2007) J. Cereb. Blood Flow Metab. 27, 697–709 [DOI] [PubMed] [Google Scholar]
- 66.Lapchak P. A., Chapman D. F., Zivin J. A. (2000) Stroke 31, 3034–3040 [DOI] [PubMed] [Google Scholar]
- 67.Sumii T., Lo E. H. (2002) Stroke 33, 831–836 [DOI] [PubMed] [Google Scholar]
- 68.Garcia-Alloza M., Prada C., Lattarulo C., Fine S., Borrelli L. A., Betensky R., Greenberg S. M., Frosch M. P., Bacskai B. J. (2009) J. Neurochem. 109, 1636–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Du R., Petritsch C., Lu K., Liu P., Haller A., Ganss R., Song H., Vandenberg S., Bergers G. (2008) Neuro Oncol. 10, 254–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Frisch S. M., Vuori K., Ruoslahti E., Chan-Hui P. Y. (1996) J. Cell Biol. 134, 793–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ethell D. W., Kinloch R., Green D. R. (2002) Curr. Biol. 12, 1595–1600 [DOI] [PubMed] [Google Scholar]
- 72.Eckman E. A., Reed D. K., Eckman C. B. (2001) J. Biol. Chem. 276, 24540–24548 [DOI] [PubMed] [Google Scholar]
- 73.Tucker H. M., Kihiko M., Caldwell J. N., Wright S., Kawarabayashi T., Price D., Walker D., Scheff S., McGillis J. P., Rydel R. E., Estus S. (2000) J. Neurosci. 20, 3937–3946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liao M. Q., Tzeng Y. J., Chang L. Y., Huang H. B., Lin T. H., Chyan C. L., Chen Y. C. (2007) FEBS Lett. 581, 1161–1165 [DOI] [PubMed] [Google Scholar]
- 75.Morelli L., Llovera R., Gonzalez S. A., Affranchino J. L., Prelli F., Frangione B., Ghiso J., Castano E. M. (2003) J. Biol. Chem. 278, 23221–23226 [DOI] [PubMed] [Google Scholar]
- 76.Tsubuki S., Takaki Y., Saido T. C. (2003) Lancet 361, 1957–1958 [DOI] [PubMed] [Google Scholar]
- 77.Betts V., Leissring M. A., Dolios G., Wang R., Selkoe D. J., Walsh D. M. (2008) Neurobiol. Dis. 31, 442–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Walsh D. M., Selkoe D. J. (2007) J. Neurochem. 101, 1172–1184 [DOI] [PubMed] [Google Scholar]
- 79.Caughey B., Lansbury P. T. J. (2003) Annu. Rev. Neurosci. 26, 267–298 [DOI] [PubMed] [Google Scholar]
- 80.Feng Y., Yang S. G., Du X. T., Zhang X., Sun X. X., Zhao M., Sun G. Y., Liu R. T. (2009) Biochem. Biophys. Res. Commun. 390, 1250–1254 [DOI] [PubMed] [Google Scholar]
- 81.Necula M., Breydo L., Milton S., Kayed R., van der Veer W. E., Tone P., Glabe C. G. (2007) Biochemistry 46, 8850–8860 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.