Abstract
Art v 1, the major pollen allergen of the composite plant mugwort (Artemisia vulgaris) has been identified recently as a thionin-like protein with a bulky arabinogalactan-protein moiety. A close relative of mugwort, ragweed (Ambrosia artemisiifolia) is an important allergen source in North America, and, since 1990, ragweed has become a growing health concern in Europe as well. Weed pollen-sensitized patients demonstrated IgE reactivity to a ragweed pollen protein of apparently 29–31 kDa. This reaction could be inhibited by the mugwort allergen Art v 1. The purified ragweed pollen protein consisted of a 57-amino acid-long defensin-like domain with high homology to Art v 1 and a C-terminal proline-rich domain. This part contained hydroxyproline-linked arabinogalactan chains with one galactose and 5 to 20 and more α-arabinofuranosyl residues with some β-arabinoses in terminal positions as revealed by high field NMR. The ragweed protein contained only small amounts of the single hydroxyproline-linked β-arabinosyl residues, which form an important IgE binding determinant in Art v 1. cDNA clones for this protein were obtained from ragweed flowers. Immunological characterization revealed that the recombinant ragweed protein reacted with >30% of the weed pollen allergic patients. Therefore, this protein from ragweed pollen constitutes a novel important ragweed allergen and has been designated Amb a 4.
Keywords: Carbohydrate Structure, Defensins, Glycoprotein, Immunology, Mass Spectrometry (MS), Allergen, Arabinogalactan, Hydroxyproline, Weed Pollen
Introduction
The pollen of common ragweed (Ambrosia artemisiifolia) is a major cause of hay fever and associated asthma in Northern America. During the past few decades, ragweed has started to spread in many parts of central Europe, where it has become a serious health problem in the sensitized population. Several initiatives have formed to prevent further spread in e.g. France, Austria or southern Germany. The Compositae (or Asteraceae) family is one of the largest families of flowering plants, but only a few are important allergenic sources. These include Ambrosia (ragweed), Artemisia (mugwort), Helianthus (sunflower), and Parthenium (feverfew). It was further demonstrated that sera of mugwort allergic patients show considerable cross-reactivity with ragweed pollen extracts (1, 2). IgE-binding to mugwort allergens in immunoblots was inhibited effectively by ragweed pollen extract (1), which indicates close homology of the essential allergens in ragweed and mugwort pollen. So far, six groups of allergens have been identified in ragweed pollen. Most patients were classified as ragweed allergic if they reacted with the pectate lyases of the Amb a 1/2 group (3, 4). The homologous pectate lyase Art v 6 in mugwort has been reported to play only a minor role in allergic disease. Amb a 6 (lipid transfer protein), Amb a 8 (profilin), Amb a 9 and Amb a 10 (both calcium-binding proteins) are small proteins belonging to the group of well known cross-reactive pan-allergens (1, 4–8). Amb a 7 and the fragment Amb a 3 are plastocyanins and are described only as minor ragweed allergens (9).
In mugwort pollen, the major allergen is Art v 1, a protein with a globular domain homologous to thionins (or defensins) and a hydroxyproline-rich extensively glycosylated moiety (10, 11). Although the molecular weight of Art v 1 is 13–16 kDa, it is migrating between 24–28 kDa in SDS-PAGE, which results from this rigid and glycosylated C-terminal domain (11). SDS-PAGE and IgE immunoblot experiments using ragweed pollen extract revealed the presence of a glycoprotein similar in size and migrating with the same diffuse double band pattern as Art v 1 (1, 2). Nevertheless, no homologue to Art v 1 in ragweed pollen has been described until now.
In this work, we describe the molecular cloning and structural characterization of the ragweed homologue of Art v 1. Furthermore, an initial assessment of the IgE binding capacity of this new ragweed allergen, designated as Amb a 4, with sera from ragweed and mugwort sensitized patients was performed.
EXPERIMENTAL PROCEDURES
Purification of Amb a 4
5 g pollen of A. artemisiifolia (Allergon AB) were extracted for 15 min at room temperature with 120 ml of water. The extract was centrifuged, filtered, and mixed with 0.1 volumes of 0.2 m sodium phosphate buffer of pH 7.2. A CM-Sepharose column (1 × 18 cm, GE Healthcare) was equilibrated with 20 mm sodium phosphate, pH 7.2, and the extract was applied. Elution was performed with a 100-ml gradient from 0.02 to 0.3 m sodium phosphate.
Fractions were analyzed by SDS-PAGE for the occurrence of a 30-kDa protein, which was shown to cross-react with a rabbit anti-Art v 1 serum available from a previous Art v 1 study (10). In later purifications, Amb a 4 was identified via its N-terminal peptide KLCEKPSVTWSGK by tryptic digestion of 1% of each fraction and subsequent reversed phase-HPLC-ESI-MS as described (12). Pooled fractions were subjected to size exclusion chromatography (Sephacryl S100 HR, GE Healthcare).
Characterization of the Carbohydrate Moiety
Monosaccharide composition was determined after hydrolysis with 4 m trifluoroacetic acid at 100 °C for 3 h by HPLC of 1-phenyl-3-methyl-5-pyrazolone derivatives (13, 14) as well as of 2-aminobenzoic acid derivatives (15). From the same Amb a 4 preparation, amino acids were determined by HPLC of o-phthalaldehyde derivatives (16). This allowed a correlation of sugar and protein content.
For characterization of the polysaccharide moiety, the protein backbone was hydrolyzed with 0.22 m BaOH2 at 100 °C for 6 h (11, 17). The alkaline hydrolyzate was neutralized with acetic acid and fractionated over Sephadex G15 using 1% acetic acid as the eluent. The void volume fraction was subjected to MALDI-TOF-MS on an Ultraflex TOF/TOF MS (Bruker) using 2,5-dihydroxybenzoic acid as matrix. Additionally, the samples were measured by off-line ESI-Q-TOF on a Q-TOF Ultima Global (Waters-Micromass) for MS/MS experiments. Alternatively, hydroxyproline (Hyp)-polysaccharide was subjected to reversed phase-LC-ESI-MS on an Aquastar C18 column (0.32 × 5 mm, Thermo Fisher Scientific) using 0.1% formic acid in water and a gradient from 5 to 25% acetonitrile at a flow rate of 5 μl/min. The spray voltage was set to 3400 V, the RF lens value to 60 and the MS profile to 250/850/1450.
Short arabinan chains linked to Hyp were detected by liquid-chromatography with mass spectrometric detection after alkaline hydrolysis of the protein (11, 17) and dansylation (18). In this case, alkaline hydrolysis was followed by neutralization with sulfuric acid in the presence of sodium acetate buffer. After vacuum drying and dansylation of the Hyp-(Ara)n chains, the samples were subjected to porous graphitic carbon chromatography with MS detections using a 100 mm ammonia formate puffer of pH 9.0 and a 20-min acetonitrile gradient (14–38%). Mass spectrometric detection was done in positive ion mode on the Q-TOF instrument.
A rabbit antiserum recognizing patches of β-arabinosyl residues was available from a previous Art v 1 study and used as described previously (11). Aspergillus niger α-arabinofuranosidase was purchased from Megazyme (Wicklow, Ireland).
800 MHz NMR analysis of Amb a 4
NMR spectra were recorded at 25 °C in 5-mm tubes in a Cryo probe on a Bruker Avance 800 instrument at 799.3 MHz for proton and 200.98 MHz for carbon, using acetone as reference for proton (2.225 ppm) and 1,4-dioxane for carbon (67.4 ppm). Bruker standard programs cosydfphpr, noesyphpr (mixing time, 100 ms), mlevphpr (spinlock time, 80 ms), HSQC, HSQCTOCSY (spinlock time, 80 ms), HSQCNOESY (mixing time, 200 ms), HMBC, and H2BC were used with digital resolution in F2 dimension <2 Hz/pt (19). Processed spectra were assigned using the computer program PRONTO (20).
Molecular Cloning of cDNA and Expression of Amb a 4
RNA from A. artemisiifolia young blossoms were extracted using SV total RNA isolation system (Promega) and used as template for cDNA synthesis by SuperScript III (Invitrogen). Based on the N-terminal sequence and tandem MS experiments, degenerated primers RhaAfor 5′-AARYTITGYGARAARCCIWSIGTNACNTGG-3′ and RhaArev 5′-GGRTTYTTNGTNGGRTCRCARTCRAAGTAGCA-3′ were designed. The sequence information of the resulting PCR fragment served to design the primer RhaB for 5′-GATAAGTGTGACAAGCGGTGTATAGARTGGGAGGG-3′ used in RACE-PCR with the SMART RACE3 cDNA amplification kit (Clontech). Four distinct clones were obtained, corresponding to the plasmids pGEMT-RhaC1 to pGEMT-RhaC4 as they comprised only the C-terminal albeit largest part of the protein. To retrieve the authentic N-terminal nucleotide sequences, a 5′-RACE was performed (Smarter RACE cDNA amplification kit, Clontech). For this experiment, cDNA from flowers of 100 different ragweed plants was employed. The 5′-untranslated region of the new ragweed allergen was obtained by nested RACE-PCR with the primers CCGGAGGAGGCGGTCCATCACCTCCTTCGCCACCTGACGGAGGTGG and GGAGCAGGAGCCTTTCCCTTCGGGGCCCCTGGTGGCGGC. The 3′-untranslated region of ragweed allergen was obtained by a nested RACE PCR with the primers GCCGCCACCAGGGGCCCCGAAGGGAAAGGCTCCTGCTCC and CCACCTCCGTCAGGTGGCGAAGGAGGTGATGGACCGCCTCCTCCGG. The full sequence of ragweed homologue was then amplified with the forward primer ATTCTAATACGACTCACTATAGGGC and the reverse primer TTGAACTCATGAGGTTTGAT issued from the respective untranslated regions.
Clones were sequenced using a BIG Dye Termination Cycle sequencing kit (Applied Biosystems). Predicted amino acid sequences were sorted with the help of MultAlign (21).
Expression of Amb a 4
Using the forward primer Rha4forA (5′-AGTGCAAGGTGAAGCAGACCGATAAGTGTGACAAGCGGTG-3′) and Rha4revA (5′-GGAATTCCTCAACTTTTACCACCTCCTTCACC-3′) as reverse primer and the plasmid pGEMT-Rha4 (encoding Amb a 4 isoform 1) as a template, a fragment was amplified. Consecutively, PCR was performed with the forward primers Rha4forB (5′-TCCGTGACTTGGTCCGGCAAGTGCAAGGTGAAGCAGACC-3′), Rha4forC (5′-AAGCTGTGCGAGAAGCCGTCCGTGACTTGGTCCGGCA-3′) and Rha4forD (5′-GGGTACCAAGCTGTGCGAGAAGCCG-3′) in order to obtain a DNA encoding the complete allergen. The fragment was ligated into pET30a(+) (Novagen), and the resulting construct was transferred into Escherichia coli BL21 cells for expression. His-tagged recombinant Amb a 4 was purified with Ni-NTA agarose gel (Qiagen). Amb a 4 was further purified by reversed-phase HPLC with a C18 wide pore column (150 × 4.6 mm, 5 μm, 300-nm pore size, Phenomenex) using 0.05% TFA as starting solvent and a 5 to 35% acetonitrile gradient. Protein elution was monitored at 280 nm.
Other Analytical Techniques
Amino acids were determined using either o-phthaldehyde and 2-mercaptoethanol for primary amino acids or o-phthaldehyde, 3-mercaptopropionic acid and 9-fluorenylmethyl-chloroformate for secondary amino acids as described (16). To obtain N-terminal sequence information purified natural Amb a 4 was separated by SDS-PAGE and electroblotted onto polyvinyl difluoride membranes (Millipore, Bedford, MA). Bands were excised and sequenced on a Procise 491 sequencer (Applied Biosystems, Carlsbad, CA), using the pulsed-liquid program. Internal peptide sequences were obtained from proteinase K peptides. The peptides were analyzed by reversed-phase HPLC coupled to an ESI source and a Q-TOF hybrid mass spectrometer operating in the MS/MS mode as described recently (12). C-terminal sequencing was attempted with carboxypeptidase Y (Sigma-Aldrich) in pyridine acetate buffer (22).
The molecular mass of native allergens was determined by MALDI-TOF-MS using a sinapinic acid matrix. Recombinant Amb a 4 was subjected to LC-ESI-MS via a Discovery® BIO Wide Pore C5 reversed-phase column (50 × 0.32 mm, 3 μm, Supelco) at a flow rate of 5 μl/min using 0.1% formic acid in water and a 10 to 50% acetonitrile gradient. The spray voltage was set to 3000 V, the RF lens value was set to 60, and the MS profile was set to 450/750/950. Deconvolution of spectra was performed with the MaxEnt1 procedure of MassLynx.
Patient Sera
Patients allergic to ragweed (and mugwort) were selected according to typical case history, positive skin prick test, and radioallergosorbent test classes >3.5. Eight sera were obtained from Canada (Dr. Alain Didierlaurent, Research Laboratory, Stallergenes S.A., Antony, France), 81 from Italy (Dr. Riccardo Asero, Ambulatorio di Allergologia, Clinica San Carlo, Paderno Dugnano, Italy), and 74 from Austria (Dr. Christof Ebner, Allergieambulatorium Reumannplatz, Vienna, Austria) and stored at −20 °C. Informed written consent was obtained from all subjects included in the study.
Immunoblot Inhibition Experiments
For immunoblot analysis, crude ragweed pollen extract was electroblotted onto nitrocellulose membrane (Protran, Schleicher & Schuell, Dassel, Germany) and incubated with patient IgE. Serum (0.6 ml) from an Austrian mugwort allergic patient was preincubated overnight with 0.00 μg, 0.01 μg, 0.10 μg, 1.00 μg, and 10.00 μg of natural Art v 1 and 10 μg recombinant Bet v 1 as a control. After an overnight incubation, the serum was added to the blotted membrane. Bound IgE antibodies were detected using 125I-labeled rabbit anti-human IgE (MALT Allergy system, IBL GmbH, Hamburg, Germany) on a phosphorimaging screen (Fujifilm BAS-1800 II, Tokyo, Japan).
ELISA and Cross-inhibition Experiment
To detect whether mugwort or ragweed allergic sera reacted with Amb a 4, the IgE reactivity to the recombinant and natural allergen was measured. Maxisorp ELISA plates (NUNC, Roskilde, Denmark) were coated with purified allergens (4 μg/ml) overnight at 4 °C. Plates were washed three times with Tris-buffered saline (TBS), pH 7.4, 0.05% Tween (TT buffer) and remaining binding sites were saturated by incubation with TT buffer plus 1% bovine serum albumin for 2 h at room temperature. Coated plates were incubated with patient sera overnight at 4 °C. All sera were diluted 1:3 in TT buffer plus 0.5% bovine serum albumin. After 5 TT buffer washes, plates were incubated (2 h at room temperature) with alkaline phosphatase conjugated monoclonal anti-human IgE antibodies (BD Biosciences) and developed with the substrate 4-nitrophenyl phosphate (Sigma-Aldrich). The optical density was measured at 405 nm, reference wavelength 492 nm, using a microplate reader (SLT Spectra 1.1, TECAN, Salzburg, Austria). Adequate ELISA controls, including omission of primary and/or secondary antibodies, were performed. The negative cut-off values were 0.2. For cross-inhibition experiments, recombinant Amb a 4 was coated to ELISA plates for inhibition experiments. In each case, six sera were used (four Austrian and two Canadian), diluted 1:5 and preincubated with increasing amounts of inhibitory proteins. Inhibition was performed at protein concentrations of 0.000, 0.002, 0.020, 0.200, and 2.000 μg/ml in 50 μl using natural and recombinant Art v 1. Proteins were incubated with patients IgE at 4 °C overnight. Values at protein concentration 0.000 μg/ml were used for data normalization.
RESULTS
Detection of a New Allergen in Ragweed Pollen Extracts
Separation of mugwort and ragweed pollen extracts by SDS-PAGE demonstrated a strong protein band of about the size of Art v 1 in ragweed pollen (Fig. 1A). Cross-reactivity between these bands of similar size was demonstrated using a patient reactive with mugwort pollen Art v 1 and its presumed ragweed homologue Amb a 4.
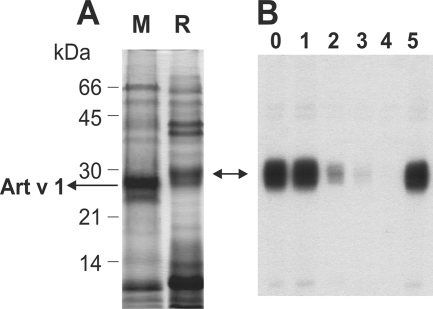
FIGURE 1.
Pollen extracts and IgE inhibition. A, crude extracts of mugwort (M) and ragweed (R) pollen were separated by SDS-PAGE followed by Coomassie Blue staining. The bands of Art v 1 in lane M are identified. B, ragweed pollen extract was separated by SDS-PAGE, transferred to nitrocellulose, and developed with serum IgE from a mugwort pollen-allergic patient. The patient serum was further preincubated with increasing concentrations of purified natural Art v 1 (lanes 0–4 indicate none, 0.01, 0.1, 1.0, and 10.0 μg, respectively) and with 10 μg of recombinant Bet v 1 as a control (lane 5).
Natural Art v 1 was able to inhibit binding of IgE to Amb a 4 in ragweed (Fig. 1B). Complete inhibition of IgE binding to Amb a 4 in ragweed pollen extract occurred only at higher Art v 1 concentrations (0.2 mg/ml), but partial inhibition was already observed at low protein concentrations. No inhibition was observed upon preincubation with recombinant Bet v 1 the major birch pollen allergen or with serum alone (Fig. 1B).
Natural Amb a 4 was purified by cation-exchange and size-exclusion chromatography, similar to Art v 1. Initially the purification was monitored using sera of allergic patients. Further on, SDS-PAGE and LC-ESI-MS were used to detect the fractions containing the 30-kDa protein with the relevant N-terminal tryptic peptide. Purified natural Amb a 4 migrated on SDS-PAGE as one very diffuse band at ∼30 kDa but did not show the double band typical for natural Art v 1 (Fig. 2). In line with this observation, MALDI-TOF MS of Amb a 4 gave only one broad peak (Fig. 2).
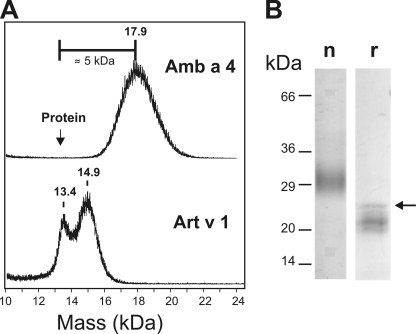
FIGURE 2.
Analysis of natural Amb a 4. A, MALDI-TOF MS analysis of natural ragweed Amb a 4 in comparison with mugwort Art v 1. The arrow indicates the calculated mass of the (large) isoform 1. The mass difference resulting from glycosylation of Amb a 4 is indicated. Depending on the prevailing isoforms this difference may even be larger. B, reducing SDS-PAGE of natural (n) and recombinant (r) Amb a 4. The arrow points at the polypeptide representing the complete amino acid sequence of Amb a 4 (isoform 1 with N-terminal His-tag), whereas the bands below are C-terminally truncated versions.
Amino acid analysis of Amb a 4 revealed an unusually high amount of glycine and proline residues, of which 78% were oxidized to Hyp. A parallel analysis of Art v 1 from mugwort revealed an even higher amount of 93% oxidized proline.
Edman sequencing of the Art v 1 cross-reactive protein revealed the sequence of its first nineteen N-terminal residues as KLCEKPSVTWSGKCKVKQT. Sequence comparison revealed high homology to the defensin-like mugwort allergen Art v 1 and a protein from Helianthus annuus (Fig. 3). The predominating role of Art v 1 in mugwort allergy prompted us to investigate the molecular and immunological properties of its homologue in ragweed.
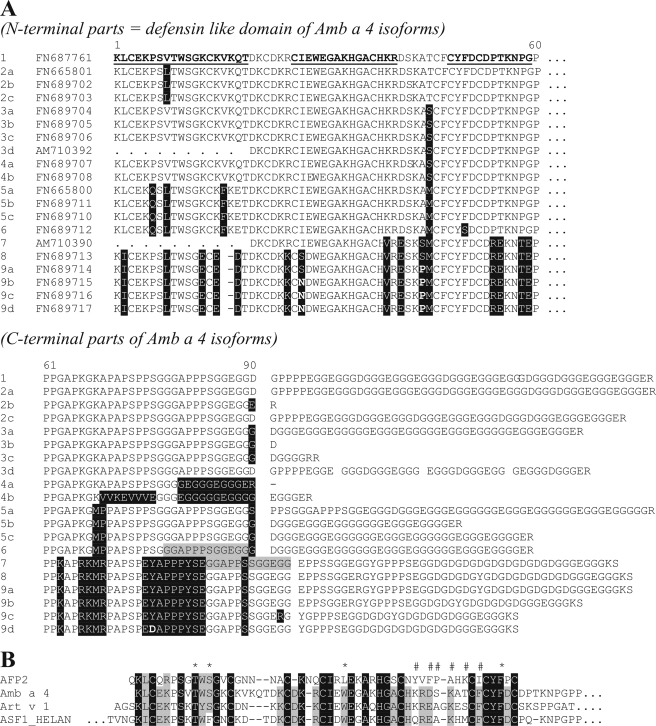
FIGURE 3.
Amb a 4 sequence and sequence alignments. A, deduced protein sequences encoded by the various cDNAs obtained from the gene rha1 (ragweed homolog of Art v 1). Isoform 1 was expressed recombinantly and used for IgE binding studies. The parts of its sequence, which were found on the protein level, are emphasized by bold and underlined font. Isoforms were grouped on the basis of amino acid differences in the defensin like domain (roughly the first 60 amino acids in analogy to Art v 1) and subgrouped according to their C-terminal parts. The gray background highlights a homologous but slightly displaced peptide stretch from isoforms 7 onwards. B, amino acid sequence comparison of a radish defensin, Amb a 4, Art v 1 and the sunflower protein ASF1_HELAN. Fully and partially conserved residues are emphasized by dark or light gray background, respectively. # and *, residues found to be involved in the defensin role of the Raphanus protein (30). Apparently, only the second group (second site) identified by an asterisk is conserved to some degree.
Determination of Internal Amino Acid Sequence and Cloning of Amb a 4
Several attempts to obtain internal sequence information by mass spectrometric analysis of peptides digested with either trypsin, chymotrypsin, pepsin, or Pronase failed as well as RT-PCR amplification of Amb a 4 based on the N-terminal sequence. Finally, a proteinase K digest revealed the internal sequence CYFDCDPTKNPG. Its partial homology to Art v 1 and the sunflower protein P22357 indicated that this peptide originated from the C-terminal end of the allergens defensin domain (Fig. 3A).
Next, degenerate primers were designed and, with the help of the already available 5′-primer a 49-bp nucleotide sequence could be cloned (Fig. 3B). Based on this sequence the cloning of the complete cDNA of Amb a 4 was accomplished with the exception of the 5′-end encoding the first 19 N-terminal amino acids as degenerate primers were used for this part. Four cDNA clones encoding three different protein sequences were obtained for the C-terminal part.
Retrieval of the entire sequence of the allergen was hampered severely by the occurrence of large amounts of cDNA with some homology to our allergen but lacking the C-terminal Pro- and Gly-rich extension (data not shown). Such putative defensins are known from other plants, e.g. Arabidopsis. Finally, nested 5′-RACE based on the polyproline domain led to the amplification of the entire coding sequence of the allergen. 20 different cDNA clones were obtained from ∼50 colonies exemplifying a considerable genetic variability of this allergen.
Knowing the possible protein sequences of Amb a 4 allowed us to re-evaluate the ESI-MS results. This retrieved the tryptic peptide CIEWEGAKHGACHKR, which, together with the already known two peptides, indicated isoforms 1, 3, and/or 4 to be a major variant on the protein level (Fig. 3C). A smaller signal for the N terminus of isoform 2 was found, whereas no indication for the presence of isoforms 5 to 9 in pollens was obtained. (Note that the cDNA was isolated from whole flowers.)
Unfortunately, information on the C terminus of the protein could not be obtained, as no amino acids were released from Amb a 4 by carboxypeptidase Y even at a very high enzyme to substrate ratio, which may indicate an unidentified C-terminal modification.
Also, trials to chemically deglycosylate Amb a 4 as done with Art v 1 inevitably led to degradation of the protein backbone and thus the actual size of the C-terminal moiety must remain elusive. Similarly, application of α-arabinofuranosidase as for Art v 1 (11) led to degradation, which could not be prevented by a protease inhibitor (Complete Mini inhibitor mixture, Roche Applied Science; supplemented with EDTA).
Carbohydrate Moieties of Amb a 4
Amb a 4 contained galactose and arabinose in a ratio of 1 to 24. A comparison of the results of amino acid and of monosaccharide analysis indicated a galactose content of 2.6 mol per mol protein (±20%). This translates into a carbohydrate mass of ∼6 kDa, which explains the large discrepancy between the theoretical mass of the polypeptide chains (12.6 to 13.5 kDa depending on isoform) and the results obtained by MALDI-TOF MS (∼18 ± 1 kDa) (Fig. 2).
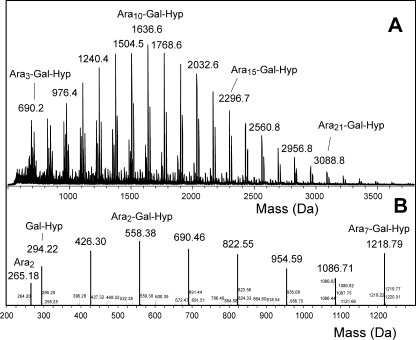
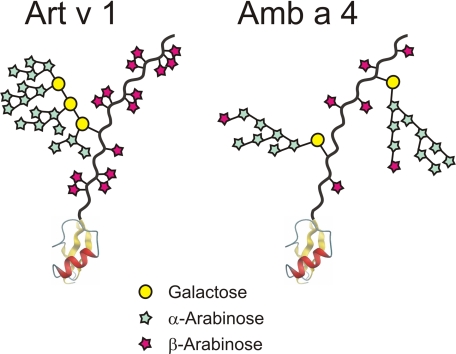
To detect whether Amb a 4 contained an arabinogalactan polysaccharide, the polymer fraction obtained upon alkaline digestion of Amb a 4 was analyzed by MALDI-TOF MS. This identified a Hyp-linked polysaccharide containing 5 to more than 20 arabinose but only 1 galactose residue (Fig. 4A). ESI-MS/MS of this sample revealed the galactose to be bound to hydroxyproline (Fig. 4B). The C-terminal part of Art v 1 contained a number of adjacent Pro residues, of which a large part was oxidized to Hyp (10). Although such contiguous Hyp residues are considered usually as attachment sites for arabinan oligosaccharides (23, 24), Art v 1 only contained single β-linked arabinose residues (see Fig. 6) (11).
FIGURE 4.
Mass spectra of the Hyp-linked arabinogalactan of Amb a 4. A, MALDI spectrum with mainly [M+Na]+ ions. [M+K]+ and [M+H]+ ions are not annotated. (B) Fragment spectrum obtained by ESI-MSMS of the [M+H]+ ion of the Ara7-Gal-Hyp glycopeptide.
FIGURE 6.
Scheme of the ragweed allergen Amb a 4 in comparison with the major mugwort allergen Art v 1. The globular defensin-like domain is shown at the bottom with the hydroxyproline-rich region extending. Yellow circles represent galactose, turquoise and red pentagons represent α- and β-arabinosyl residues, respectively.
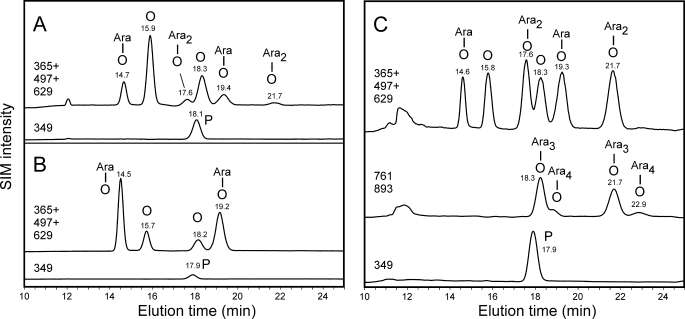
The Ara-Hyp glycopeptides obtained upon alkaline hydrolysis from Art v 1 and Amb a 4 were analyzed as dansyl derivatives by LC-ESI-MS with a graphitic carbon column. Amb a 4 also contained single Hyp-linked Ara residues (Fig. 5A). In addition, some Ara-Ara-Hyp was found in Amb a 4. Longer arabinan chains would have been detected by the chosen methodology if present, as a control experiment with gum arabic revealed a distribution of di- to tetra-arabinans similar to the literature (25) (Fig. 5B). A rough estimation of all Pro-derivatives found by LC-ESI-MS (Fig. 5) yields 4.2 mol Hyp-linked Ara per mol protein in Amb a 4 versus 14.8 mol in Art v 1.
FIGURE 5.
LC-ESI-MS analysis of dansylated glycopeptides. The results for Amb a 4 (A), Art v 1 (B), and gum arabic (C) are shown. The chromatograms of each sample are depicted as two or three SIM traces from the same run to allow presentation of overlapping peaks. SIM traces truly reflect the relative signal intensities. Hyp (shown by the letter O) and its glycopeptides give two peaks according to its trans and allo-d form (31).
Glycosylation Analysis by NMR
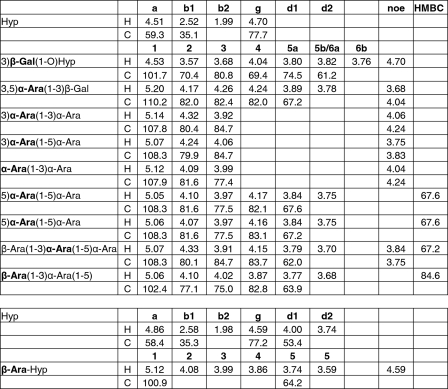
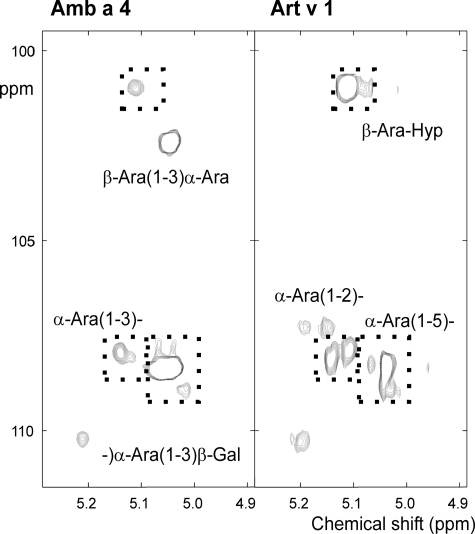
The assignment of the glyco part of the Amb a 4 glycoprotein was done by homo- and heteronuclear experiments similar to our recent work on Art v 1 (11). At first glance, the glycosylation of Amb a 4 appeared similar to that of Art v 1 consisting of β-galactose, α-arabinose, and β-arabinose residues. However, unlike Art v 1, only one type of β-galactose was found in Amb a 4 (Table 1 and Fig. 6). Further notable differences were observed in the anomeric parts of the arabinose residues of Amb a 4 and Art v 1 when compared in an HSQC spectrum (Fig. 7). The Amb a 4 signals exhibited more -5)-α-Ara-(1–5)- stretches, less -3)-α-Ara(1- branching and no -2)-α-Ara(1- residues.
TABLE 1.
NMR analysis of the Hyp-polysaccharide from Amb a 4
FIGURE 7.
NMR analysis of arabinose linkages in Amb a 4 and Art v 1. The HSQC spectra of the anomeric part of the arabinose signals show differences for the α-linked arabinoses (signals in the lower part of the plot) and the β-linked arabinoses (upper part). Note the presence of β-Ara to α-Ara linkages, the absence of two-linked arabinoses in Amb a 4 and also the quantitative difference for the Hyp-linked β-arabinoses.
As in Art v 1, a single β-arabinose linked to hydroxyprolin was found in Amb a 4 (5.12 ppm for 1H and 100.9 ppm for 13C), but in a smaller amount (Fig. 7). Amb a 4 on the other hand contained terminal β-arabinose linked to an α-arabinose residue (5.05 ppm for 1H and 102.4 ppm for 13C), which are not found in Art v 1 (Fig. 7).
Amb a 4 Carbohydrate Epitopes
For a previous study on the major mugwort allergen rabbit antibody were raised against natural Art v 1. IgG binding to Art v 1 was absorbed with the recombinant nonglycosylated protein. The remaining serum was found to bind to Art v 1 containing β-Ara residues (11). Interestingly this serum, did not bind to natural Amb a 4, demonstrating the difference in the carbohydrate moiety of the two homologous proteins (data not shown).
Expression of Recombinant Amb a 4
The cDNA-encoding isoform 1 of Amb a 4, which contains all the peptide sequences found on the protein level (Fig. 3), was expressed in E. coli with an N-terminal His-tag. Metal-affinity purification gave a strong signal in Western blot experiments using anti-His antibody. Further purification by reversed-phase HPLC resulted in essentially one peak, which again migrated as a set of bands of apparently 21–26 kDa (Fig. 2B). Analysis by ESI-MS revealed the presence of the expected translation product of 17,733 Da but also of peaks indicative of premature termination of translation (data not shown). The truncation sites hinted at depletion of tRNAs, mainly that for Gly, despite expression in an E. coli Rosetta strain. Regardless, the apparent mass of recombinant Amb a 4 was much higher than the theoretical value of 13.8 kDa. This is reminiscent of Art v 1, where the rigid, rod-like conformation of the Pro/Hyp-rich C-terminal moiety likewise results in an aberrant mass estimation by SDS-PAGE (11, 26).
IgE Binding by Natural and Recombinant Amb a 4
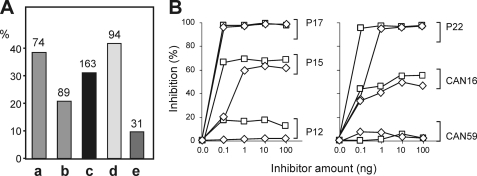
ELISA experiments with sera from three geographic regions were performed to evaluate the IgE-binding activity of purified natural and recombinant Amb a 4. Italian and Canadian patients have been confronted mainly with ragweed pollen, whereas Austrians mostly encounter mugwort pollen and an increasing concentration of ragweed pollen. Hence, Austrian patients are mainly mugwort-sensitized and showed strong cross-reactivity with ragweed pollen, whereas Italian and Canadian patients mostly have been ragweed-sensitized. Remarkably, many more Austrian than Italian/Canadian sera reacted with Amb a 4 (Fig. 8A). The natural and recombinant protein demonstrated comparable IgE binding reactivity. Only minor differences were observed. 38/48 of the tested patients showed the same IgE reactivity, five patients showed stronger reactivity to natural Amb a 4, and five patients with recombinant Amb a 4. In total, in this preliminary survey on the role of Amb a 4 as an allergen, we could demonstrate that ∼31% of the mugwort- and ragweed-sensitized patients displayed IgE antibodies to Amb a 4. Art v 1 and Amb a 4 exhibit considerable cross-reactivity because ∼42% of the Art v 1 sensitized patients also reacted with Amb a 4 (Fig. 8A). In the Art v 1 non-binding group, three patients (∼10% of the 31 sera-tested) reacted with Amb a 4 but not with Art v 1 from mugwort.
FIGURE 8.
IgE binding and inhibition studies. Sera (n = 163) from patients allergic to ragweed (and mugwort) were used to test the IgE-reactivity of purified recombinant Amb a 4 in ELISA. A, the percentage of sera reacting with recombinant Amb a 4 of the Austrian (a), Canadian plus Italian (b) and of the total study population (c) are shown. d gives the frequency of Amb a 4 recognition by patients sera reacting with Art v 1, whereas e demonstrates the percentage of Amb a 4 reactive sera in the Art v 1-negative group (not all of these 163–94 = 69 sera could be tested). Numbers above bars indicate the number of sera in each group. B, native Amb a 4 was coated on ELISA plates, and cross-inhibition was performed with sera from four Austrian (P12, P15, P17, and P22) and two Canadian (CAN16 and CAN59) weed allergic patients using increasing concentrations (0.0 to 100 ng in 50 μl) of either natural Art v 1 (□) or recombinant Art v 1 (◇).
Quantitative evaluation of IgE cross-reactivity was performed with dose-related inhibition experiments using six patients (four Austrian, two Canadian), previously shown to react with Art v 1 and Amb a 4 in ELISA studies. Cross-inhibition experiments using recombinant Amb a 4 as the coat antigen demonstrated that natural and recombinant Art v 1 exerted a rather similar inhibitory effect on IgE binding (Fig. 8B). The degree of inhibition varied considerably between patients. Complete IgE-inhibition was observed using two Austrian patients; only partial inhibition was achieved with two more patients and hardly any interference by Art v 1 was found for another two sera (Fig. 8B).
DISCUSSION
The presence of a protein in ragweed pollen cross-reactive and similar in size to the major mugwort pollen allergen Art v 1 could be assumed from immunoblots of ragweed pollen extracts using mugwort allergic patients sera (32). In this work, we show that this ragweed pollen protein, now designated Amb a 4, indeed contains a defensin-like domain with a high homology to Art v 1, the sunflower protein P22357 and the feverfew allergen Par h 1 (27). Many smaller, homologous proteins with a proven or assumed function in pathogen defense are found throughout the plant kingdom, but the four proteins listed above are the only known examples, where the defensin domain is found in combination with a proline-rich and glycosylated part (for the sunflower homologue, the latter is only assumed). Smaller defensins are also expressed in ragweed flowers as their mRNAs/cDNAs severely complicated attempts at obtaining full length clones of Amb a 4 by 5′-RACE.
The proline-rich region of Amb a 4 differs from that of Art v 1 in four ways. First, it is C-terminally elongated by a low complexity sequence rich in glycine and acidic amino acids. Second, the arabinogalactan polysaccharide consists of only one galactose residue and, almost as a consequence, of less arabinoses. These are attached to the C3 of the galactose as in Art v 1 (Fig. 7). Thus, the Amb a 4 polysaccharide can be regarded as the second known member of the type III arabinogalactans (11). Third, some arabinan side chains bear terminal β-arabinosyl residues. The fourth and maybe most important difference concerns the Hyp-linked Ara residues, of which Amb a 4 contains 3.5-times less than Art v 1 as revealed by the analysis of dansylated glycopeptides. This translates into a ratio of Ara to (Hyp and Pro) of 1 to 4.3 in Amb a 4 versus 1 to 1.4 in Art v 1. Irrespective of the uncertainties arising from measurement errors and isoform distribution, these figures point at a significant difference in the occurrence of patches of adjacent Ara residues (Fig. 7). Adjacent Ara-Hyp dimers or trimers constitute IgE-binding determinants in Art v 1 (11). Therefore, this structural difference explains the finding that a fractionated rabbit serum against the glycoprotein part of Art v 1 did not react with Amb a 4.
It remains to be demonstrated whether the different degree of arabinosylation resides in the glycosylation capacity of these two related weeds or in the glycosylation motifs present in Art v 1 and Amb a 4. Notably, both allergens contain several spots with contiguous Hyp (or at least Pro) residues, which according to Shpak et al. (24) promote the attachment of arabinan oligosaccharides.
In a recent study by Asero and colleagues (28), some ragweed allergic patients did not show any IgE reactivity with the available tested recombinant ragweed allergens, which indicated that an important ragweed allergen was still unknown. Probably, the missing ragweed allergen was the Art v 1 homologue Amb a 4, which is recognized by ∼30% of ragweed sensitized patients. Thus, Amb a 4 can be considered to be a major allergen in ragweed pollen. Remarkably, despite the high similarity and cross-reactivity between Art v 1 and Amb a 4, their relative role in sensitization to, respectively, mugwort and ragweed appears to differ. While Art v 1 is recognized by >90% of mugwort allergic patients (26), Amb a 4 is bound by IgE from 39% Austrian and 20% of Italian/Canadian patients only. Given the differing load of mugwort and ragweed pollens in these areas, one could speculate that IgE production against Amb a 4 often follows or accompanies sensitization against mugwort pollen, whereas isolated ragweed sensitization involves other allergens. This possibly region-specific IgE binding pattern reminds of the Rosaceae allergy, where birch pollen confrontation in middle and northern Europe provokes allergies based on Bet v 1 homologues, whereas in Spain or Italy lipid-transfer proteins are the relevant components (29). Although certainly more work is needed to clarify this point, the inhibition characteristics of different patients and the occurrence of patients reacting only with Amb a 4 indicate that in some cases Amb a 4 binding is just cross-reactivity to Art v 1, whereas in others, it is the consequence of true sensitization or co-sensitization to ragweed pollen. Inhibition studies, however, cannot be included in routine diagnosis and thus, Amb a 4 joins the list of ragweed allergens with homology and cross-reactivity to mugwort pollen proteins (3, 4, 6, 7). The presence of β-Ara patches in Art v 1 but not in Amb a 4 indicates the occurrence of a possibly unique epitope in mugwort pollen that would allow to differentiate sensitization to these pollens, at least for some patients.
The newly identified Amb a 4 will round off the panel of ragweed allergens used for component-resolved diagnosis of weed pollen allergy. The results thus obtained will help to decide whether recombinant Amb a 4 should be considered in the development of allergy vaccines for therapy of patients suffering from weed pollen allergy.
Acknowledgments
We thank Thomas Dalik for amino acid analyses. NMR spectra were obtained at the 800 MHz Bruker Avance spectrometer of the Danish Instrument Center for NMR Spectroscopy of Biological Macromolecules. We especially thank Andreas Thader for help with isoform sequencing.
This work was supported in part by the Austrian Science Fund (S8802 and P19172).
- RACE
- rapid amplification of cDNA ends
- ESI
- electrospray ionization
- Q-TOF
- quadrupole time of flight.
REFERENCES
- 1.Oberhuber C., Ma Y., Wopfner N., Gadermaier G., Dedic A., Niggemann B., Maderegger B., Gruber P., Ferreira F., Scheiner O., Hoffmann-Sommergruber K. (2008) Int. Arch. Allergy Immunol. 145, 94–101 [DOI] [PubMed] [Google Scholar]
- 2.Hirschwehr R., Heppner C., Spitzauer S., Sperr W. R., Valent P., Berger U., Horak F., Jäger S., Kraft D., Valenta R. (1998) J. Allergy Clin. Immunol. 101, 196–206 [DOI] [PubMed] [Google Scholar]
- 3.Gadermaier G., Wopfner N., Wallner M., Egger M., Didierlaurent A., Regl G., Aberger F., Lang R., Ferreira F., Hawranek T. (2008) Allergy 63, 1543–1549 [DOI] [PubMed] [Google Scholar]
- 4.Gadermaier G., Dedic A., Obermeyer G., Frank S., Himly M., Ferreira F. (2004) Curr. Allergy Asthma Rep. 4, 391–400 [DOI] [PubMed] [Google Scholar]
- 5.Subiza J., Subiza J. L., Hinojosa M., Garcia R., Jerez M., Valdivieso R., Subiza E. (1989) J. Allergy Clin. Immunol. 84, 353–358 [DOI] [PubMed] [Google Scholar]
- 6.Wopfner N., Gadermaier G., Egger M., Asero R., Ebner C., Jahn-Schmid B., Ferreira F. (2005) Int. Arch. Allergy Immunol. 138, 337–346 [DOI] [PubMed] [Google Scholar]
- 7.Wopfner N., Gruber P., Wallner M., Briza P., Ebner C., Mari A., Richter K., Vogel L., Ferreira F. (2008) Allergy 63, 872–881 [DOI] [PubMed] [Google Scholar]
- 8.Radauer C., Breiteneder H. (2007) J. Allergy Clin. Immunol. 120, 518–525 [DOI] [PubMed] [Google Scholar]
- 9.Roebber M., Marsch D. G. (1991) J. Allergy Clin. Immunol. 87, 324–324 [Google Scholar]
- 10.Himly M., Jahn-Schmid B., Pittertschatscher K., Bohle B., Grubmayr K., Ferreira F., Ebner H., Ebner C. (2003) J. Allergy Clin. Immunol. 111, 882–888 [DOI] [PubMed] [Google Scholar]
- 11.Leonard R., Petersen B. O., Himly M., Kaar W., Wopfner N., Kolarich D., van Ree R., Ebner C., Duus J. Ø., Ferreira F., Altmann F. (2005) J. Biol. Chem. 280, 7932–7940 [DOI] [PubMed] [Google Scholar]
- 12.Dragosits M., Stadlmann J., Albiol J., Baumann K., Maurer M., Gasser B., Sauer M., Altmann F., Ferrer P., Mattanovich D. (2009) J. Proteome Res. 8, 1380–1392 [DOI] [PubMed] [Google Scholar]
- 13.Altmann F., Lomonossoff G. P. (2000) J. Gen Virol. 81, 1111–1114 [DOI] [PubMed] [Google Scholar]
- 14.Fu D., O'Neill R. A. (1995) Anal. Biochem. 227, 377–384 [DOI] [PubMed] [Google Scholar]
- 15.Anumula K. R. (1994) Anal. Biochem. 220, 275–283 [DOI] [PubMed] [Google Scholar]
- 16.Altmann F. (1992) Anal. Biochem. 204, 215–219 [DOI] [PubMed] [Google Scholar]
- 17.Lamport D. T. (1969) Biochemistry 8, 1155–1163 [DOI] [PubMed] [Google Scholar]
- 18.Tapuhi Y., Schmidt D. E., Lindner W., Karger B. L. (1981) Anal. Biochem. 115, 123–129 [DOI] [PubMed] [Google Scholar]
- 19.Duus J., Gotfredsen C. H., Bock K. (2000) Chem. Rev. 100, 4589–4614 [DOI] [PubMed] [Google Scholar]
- 20.Kjaer M., Andersen K. V., Poulsen F. M. (1994) Methods Enzymol. 239, 288–307 [DOI] [PubMed] [Google Scholar]
- 21.Corpet F. (1988) Nucleic Acids Res. 16, 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winder J. S., Walker J. M. (1993) Methods Mol. Biol. 16, 313–318 [DOI] [PubMed] [Google Scholar]
- 23.Kieliszewski M. J. (2001) Phytochemistry 57, 319–323 [DOI] [PubMed] [Google Scholar]
- 24.Shpak E., Barbar E., Leykam J. F., Kieliszewski M. J. (2001) J. Biol. Chem. 276, 11272–11278 [DOI] [PubMed] [Google Scholar]
- 25.Goodrum L. J., Patel A., Leykam J. F., Kieliszewski M. J. (2000) Phytochemistry 54, 99–106 [DOI] [PubMed] [Google Scholar]
- 26.Himly M., Jahn-Schmid B., Dedic A., Kelemen P., Wopfner N., Altmann F., van Ree R., Briza P., Richter K., Ebner C., Ferreira F. (2003) Faseb. J. 17, 106–108 [DOI] [PubMed] [Google Scholar]
- 27.Gupta N., Martin B. M., Metcalfe D. D., Rao P. V. (1996) J. Allergy Clin. Immunol. 98, 903–912 [DOI] [PubMed] [Google Scholar]
- 28.Asero R., Wopfner N., Gruber P., Gadermaier G., Ferreira F. (2006) Clin. Exp. Allergy 36, 658–665 [DOI] [PubMed] [Google Scholar]
- 29.Fernández-Rivas M., Bolhaar S., González-Mancebo E., Asero R., van Leeuwen A., Bohle B., Ma Y., Ebner C., Rigby N., Sancho A. I., Miles S., Zuidmeer L., Knulst A., Breiteneder H., Mills C., Hoffmann-Sommergruber K., van Ree R. (2006) J. Allergy Clin. Immunol. 118, 481–488 [DOI] [PubMed] [Google Scholar]
- 30.De Samblanx G. W., Goderis I. J., Thevissen K., Raemaekers R., Fant F., Borremans F., Acland D. P., Osborn R. W., Patel S., Broekaert W. F. (1997) J. Biol. Chem. 272, 1171–1179 [DOI] [PubMed] [Google Scholar]
- 31.Bollig K., Lamshöft M., Schweimer K., Marner F. J., Budzikiewicz H., Waffenschmidt S. (2007) Carbohydr. Res. 342, 2557–2566 [DOI] [PubMed] [Google Scholar]
- 32.Mutschlechner S., Mari A., Van Ree R., Didier-Laurent A., Ebner C., Ferreira F., Wopfner N. (2003) Patterns of Art v 1 recognition in mugwort and ragweed-sensitized patients abstract nr. 384 at the XXII Congress of the European Academy of Allergy and Clinical Immunology, Paris, June 7–11, 2003, 2003:384 [Google Scholar]