Abstract
Cellular supply of dNTPs is essential in the DNA replication and repair processes. Here we investigated the regulation of thymidine kinase 1 (TK1) in response to DNA damage and found that genotoxic insults in tumor cells cause up-regulation and nuclear localization of TK1. During recovery from DNA damage, TK1 accumulates in p53-null cells due to a lack of mitotic proteolysis as these cells are arrested in the G2 phase by checkpoint activation. We show that in p53-proficient cells, p21 expression in response to DNA damage prohibits G1/S progression, resulting in a smaller G2 fraction and less TK1 accumulation. Thus, the p53 status of tumor cells affects the level of TK1 after DNA damage through differential cell cycle control. Furthermore, it was shown that in HCT-116 p53−/− cells, TK1 is dispensable for cell proliferation but crucial for dTTP supply during recovery from DNA damage, leading to better survival. Depletion of TK1 decreases the efficiency of DNA repair during recovery from DNA damage and generates more cell death. Altogether, our data suggest that more dTTP synthesis via TK1 take place after genotoxic insults in tumor cells, improving DNA repair during G2 arrest.
Keywords: Cell Cycle, Checkpoint Control, DNA Damage, DNA Repair, Nucleoside Nucleotide Metabolism, p53, Thymidine Kinase 1
Introduction
The synthesis of dTTP is highly regulated and is important for DNA replication and repair in all living cells (1, 2). There are two pathways for dTTP synthesis in cells. In the de novo pathway, ribonucleotide reductase, which is composed of two pairs of R1 and R2 subunits, converts CDP and UDP to dCDP and dUDP, respectively. Both dCDP and dUDP can be metabolically converted to dUMP, and thymidylate synthase (TS)3 catalyzes the reaction of dTMP formation from dUMP. In the salvage pathway, thymidine kinases (TKs), TK1 in cytosol and TK2 in mitochondria (3, 4), are responsible for dTMP production from thymidine (5). Phosphorylation of dTMP from either the salvage or de novo pathway to dTDP is catalyzed by thymidylate kinase, and nucleoside diphosphate kinase then converts dTDP to dTTP (6). Unlike TK2, the expression of TK1, TS, and thymidylate kinase is cell cycle-dependent (7–13).
In response to DNA damage, cells trigger multifaceted responses such as cell cycle arrest, DNA repair, or apoptosis (14, 15). In Saccharomyces cerevisiae, it has been reported that DNA damage by γ-irradiation, UV, or methyl methane sulfonate leads to increases in the levels of the four dNTPs through ribonucleotide reductase-mediated de novo synthesis, indicating a close relationship between the regulation of dNTP synthesis and DNA damage response (16, 17). In mammalian cells, expression of p53-inducible R2 (p53R2), a homolog of the R2 subunit, is increased due to p53-dependent transcriptional activation upon DNA damage, suggesting a master role of p53 in integrating regulation of dNTP pools through the de novo pathway (18–21). However, it is known that more than 50% human cancer cells harbor mutated or deleted p53, and these tumors are more resistant to chemotherapy due to the loss of p53-dependent apoptosis (22, 23). This evoked the question as how p53-deficient cancer cells carry out the synthesis of dNTPs for the repair process after DNA damage and survive after genotoxic insults by chemotherapy. In this regard, one report showed that the level of R2 subunit increased in colon cancer HCT-116 p53−/− cells after cisplatin treatment. It was suggested that up-regulation of R2 might be sufficient to replace the role of p53R2 and contribute to the increases in dNTPs needed for efficient DNA repair (24, 25). Because the R2 subunit of ribonucleotide reductase and TK1 share similar regulations in terms of transcriptional activation and proteolytic control in the cell cycle (10, 26–32), we were interested in addressing the questions of whether TK1 is regulated in response to DNA damage and whether p53 participates in this process.
In this study we show that DNA damage causes up-regulation of TK1 in different tumor cell types. Our investigation into the molecular mechanism of TK1 induction indicates that the involvement of checkpoint activation in G2 phase prevents TK1 mitotic proteolysis in p53-deficient cells. In the case of p53-proficient tumor cells, p21-mediated G1 arrest in response to DNA damage causes less induction of TK1. A number of studies have shown that the decrease in the dTTP level due to TK deficiency is correlated with the increase in DNA damage-induced cell death and mutagenesis (33–38). It was previously shown that reducing the cellular level of dTTP by silencing thymidylate kinase expression significantly sensitizes tumor cells to a chemotherapeutic agent, demonstrating the importance of dTTP supply in tumor cell survival after genotoxic insults (39). Importantly, blocking TS does not necessarily lead to chemosensitization unless thymidine is deprived from the culture medium (39). These findings prompt us to further investigate the functional contribution of TK1-mediated dTTP synthesis in DNA damage response. The results show that TK1 up-regulation endows p53-deficient tumor cells the expansion of the dTTP pool during the recovery from DNA damage in the G2 phase for improved repair and survival.
EXPERIMENTAL PROCEDURES
Materials and Antibodies
Anti-human TK1 polyclonal antibody (13, 40) and the anti-human TK1 monoclonal antibody (XPA 210) (41) were described previously. Anti-human TS antibody (clone 4H4B1) was obtained from Zymed Laboratories Inc., anti-R2 (N18), anti-cyclin B1 (GNS1), anti-chk1 (G-4), anti-p21(187), and anti-p53R2(N16) antibodies were from Santa Cruz, anti-phospho-chk1 (Ser-345) antibody was from Cell Signaling, anti-phospho-histone 3 (H3; Ser-10) and anti-phospho-H2A.X (Ser-139) antibodies were from Upstate, anti-p53 (Ab-6) antibody was from Calbiochem. Anti-β-tubulin, anti-β-actin, anti-rabbit IgG-TRITC, anti-mouse IgG-FITC antibodies, doxorubicin, etoposide, aphidicolin, hydroxyurea, actinomycin D, cycloheximide, and caffeine were from Sigma. Human TK1 small interfering RNA (siRNA) was purchased from Dharmacon siGenome SMART pools, and Lipofectamine 2000 was from Invitrogen.
Cell Culture
Colon carcinoma cell lines HCT-116 p53+/+, p53−/−, p21+/+, and p21−/− were kindly provided by Bert Vogelstein (John Hopkins University Medical Institutions, Baltimore, MD). HCT-116 and osteosarcoma U2OS cells were maintained in McCoy's 5A medium (Invitrogen) supplemented with 10% fetal bovine serum plus 100 μg/ml streptomycin and 100 units/ml penicillin (Invitrogen) at 37 °C under 5% CO2. Colon adenocarcinoma SW480 cells were maintained in Dulbecco's modified Eagle's medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum plus 100 μg/ml streptomycin and 100 units/ml penicillin (Invitrogen) at 37 °C under 5% CO2. Lung carcinoma H1299 cells were maintained in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum plus 100 μg/ml streptomycin and 100 units/ml penicillin.
Cell Cytotoxicity Assay
Cells were seeded onto 96-well-plates at 2.5 × 103 cells per well and followed by the indicated treatment. Cell viability was then measured by an MTS assay (Promega).
Whole-cell dNTP Pools Extraction and Pool Size Determination
Cells (1 × 106) were washed twice with 10 ml of cold PBS and extracted with 1 ml of ice-cold 60% methanol at −20 °C for overnight followed by centrifugation for 30 min at 16,000 × g. The supernatant was transferred to a fresh tube and dried under vacuum. The residue was dissolved in sterile water and store at −20 °C for later analysis. Determination of the dNTP pool sizes in each extract was based on DNA polymerase-catalyzed incorporation of radioactive dATP or dTTP into the synthetic oligonucleotide template method described by Sherman and Fyfe (42).
Immunofluorescence Staining
Cells on the coverslip were washed twice with PBS and fixed with 3% paraformaldehyde, PBS for 30 min. After fixation, cells were permeabilized with 0.3% Triton X-100, TBST (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.1% Triton X-100) for 5 min and blocked with 5.5% normal goat serum in TBST for 1 h at room temperature followed by staining with anti-γH2AX antibody at a 1:500 dilution, anti-human TK1 antibody at a 1:100 dilution, or anti-phospho-H3 (Ser-10) antibody at 1:500 dilution in TBST containing 3% BSA for 2 h at room temperature. After TBST washing, cells were stained with FITC-conjugated goat anti-mouse IgG antibody at a 1:100 dilution or TRITC-conjugated goat anti-rabbit IgG antibody at a 1:200 dilution in TBST containing 3% BSA and Hoechst 33342 for 1 h at room temperature. After mounting overnight, endogenous TK1 or phospho-H3 (Ser-10) was observed with Leica TCS SP2 confocal spectral microscope by using a 63× oil immersion objective, and γH2AX foci were observed with Olympus BX51 microscope by using a 100× oil immersion objective.
Immunoblotting
Fifteen micrograms of cell lysate proteins were resolved on SDS-PAGE (11% (w/v) gel) followed by electrophoretic transfer to polyvinylidene fluoride membranes (Millipore). After blocking with 5% (w/v) powdered nonfat milk, the membrane was incubated with different antibodies for 16 h and treated for 1 h with horseradish peroxidase-conjugated goat anti-rabbit IgG, goat anti-mouse, and donkey anti-goat antibodies (Santa Cruz). ECL detection for the horseradish peroxidase reaction was performed according to the manufacturer's instructions (PerkinElmer Life Sciences).
Cell Synchronization
For synchronization, cells were treated twice with 1 mm thymidine (Sigma) for 15 h intermitted by 9 h of release. Nocodazole (Sigma) was added to the medium at 200 ng/ml after cell release from the double thymidine block for mitotic arrest.
RESULTS
Up-regulation of TK1 Expression in Response to Genotoxic Insults
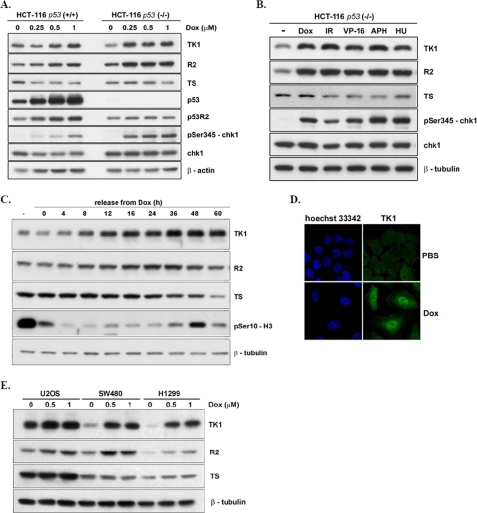
Initially, HCT-116 p53-proficient and -deficient cells were treated with increasing concentrations of doxorubicin (Dox), a topoisomerase II inhibitor, to induce DNA double-strand breaks. In HCT-116 p53+/+ cells, the levels of TK1 and R2 were increased by a Dox dosage of 0.5 μm or higher (Fig. 1A). There were clear increases in p53R2, p53, and chk1 phosphorylation at Ser-345 at 0.25 μm Dox treatment. In p53−/− cells, the levels of TK1 and R2 were simultaneously increased after treatment with 0.25 μm Dox, and the levels were higher than those in p53+/+ cells. We further treated HCT-116 p53-deficient cells with various DNA damage agents including etoposide (VP-16), γ-irradiation, aphidicolin, a DNA polymerase-α inhibitor, and hydroxyurea, a ribonucleotide reductase inhibitor. After treatment for 24 h, the steady-state levels of TK1 and R2 were increased in response to these different treatments (Fig. 1B), indicating that up-regulation of TK1 is a general DNA damage-associated event. In contrast, TS expression in all case was not increased by DNA damage.
FIGURE 1.
Up-regulation of TK1 in response to DNA damage. A, HCT-116 p53+/+ and p53−/− cells were treated with increasing concentrations of Dox for 24 h. Total lysates containing 15 μg of proteins were separated by SDS-PAGE and analyzed by Western blotting. B, HCT-116 p53−/− cells were treated with the indicated DNA damage agents, 0.5 μm Dox, 5 gray of γ-irradiation, 10 μm etoposide (VP-16), 2 μg/ml aphidicolin (APH), and 1 mm hydroxyurea (HU), for 24 h and harvested for Western blot analysis. C, HCT-116 p53−/− cells were treated with 1 μm Dox for 4 h, after which Dox was washed out, and fresh medium was added to the cultures. Cells were harvested at the indicated times after release from Dox for Western blot analysis. D, shown is immunofluorescence staining of TK1 in HCT-116 p53−/− cells that were released from Dox for 15 h as described above. Cells were fixed and stained with anti-human TK1 (XPA210)/FITC antibody and Hoechst 33342 for confocal microscopy observation. E, U2OS, SW480, and H1299 cells were treated with various dosages of Dox for 4 h followed by release from Dox-induced DNA damage for 24 h for Western blot analysis.
We further looked into whether up-regulation of TK1 can be observed in cells recovering from DNA damage. HCT-116 p53−/− cells were given 1 μm Dox treatment for 4 h followed by extensive washout of Dox by incubation in fresh media. After release from Dox, the TK1 levels increased from 8 to 36 h and stayed high for another 24 h (Fig. 1C). The expression of R2 showed a similar trend but a lesser degree of increment. The level of TS expression was relatively constant but dropped 60 h after release from Dox treatment. Ser-10 phosphorylation of H3 was also examined (Fig. 1C), a marker of mitotic chromatin condensation, and a rapid reduction of H3Ser-10 phosphorylation after release from Dox was found due to a block in mitotic entry after DNA damage. However, after release form Dox for 48 h, H3Ser-10 phosphorylation was restored, indicating mitotic re-entry after spontaneous recovery from DNA damage. Immunofluorescence staining of cells with the MPM2 antibody, which recognizes a phosphorylated epitope (S/T)P of phosphoproteins at the onset of mitosis, confirmed that cells indeed re-entered the mitotic phase around 42–48 h after recovery from Dox-induced DNA damage (supplemental Fig. S1). We also found that endogenous TK1 accumulated in the nucleus 15 h after release (Fig. 1D). Thus, DNA damage causes an increase in TK1 expression with a nuclear localization.
The expression of TK1 in other cell lines was also determined, including colon cancer SW480, lung cancer H1299, and osteoblastoma U2OS cells, which were released from Dox treatment. Despite the differences in the basal level of TK1 protein in each cell line, up-regulation of TK1 was observed 24 h after release from Dox treatment in the different cell lines (Fig. 1E), indicating that TK1 induction during recovery from DNA damage appears to be a general event. In addition, the induction of TK1 was higher in p53-defective SW480 or H1299 cells than that in U2OS cells, which have a functional p53. Altogether, TK1 expression is induced by DNA damage in different tumor cells, and the extent of TK1 induction is related to the cellular p53 context.
The Association of TK1 Induction with DNA Damage-induced G2 Phase Arrest
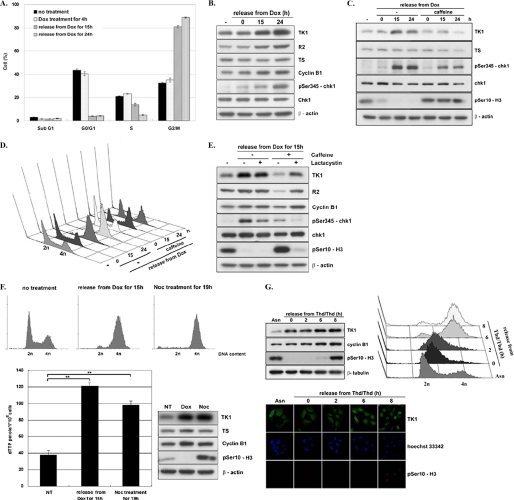
It is known that expression of TK1 is regulated in a cell cycle-dependent manner, reaching maximum during G2/M transition due to transcriptional activation. Furthermore, TK1 is degraded by APC/C targeted ubiquitination during mitotic exit (8, 10, 31). To assess whether TK1 up-regulation by DNA damage is related to the cell cycle control of TK1, the cell cycle distribution during release from Dox treatment was determined. Results from the flow cytometric analysis showed that after release from Dox treatment for 15–24 h, up to 90% HCT-116 p53−/− cells were arrested in the G2/M phase with increased levels of TK1, R2, cyclin B1, and chk1 S345 phosphorylation (Fig. 2, A and B). Previous studies have already shown that the addition of caffeine, which inhibits ATM and ATR kinases, to the culture medium can suppress checkpoint activation, preventing cells from checkpoint-induced G2 arrest and allowing mitotic entry (43–45). We then examined the effect of caffeine addition on TK1 expression during release from Dox treatment. It turned out that TK1 induction was abolished by caffeine treatment, coinciding with reduced levels of chk1 Ser-345 phosphorylation and retention of H3Ser-10 phosphorylation because of checkpoint inactivation (Fig. 2C). Flow cytometric analysis confirmed that caffeine treatment prevented G2 accumulation (Fig. 2D). These results suggest that DNA damage-induced TK1 induction is dependent on checkpoint activation and G2 arrest.
FIGURE 2.
TK1 induction as a result of DNA damage-induced G2 arrest. HCT-116 p53−/− cells were released from Dox treatment as described in the legend to Fig. 1C and harvested at the indicated times for flow cytometric analysis (A) and Western blot analysis (B). Data are the mean ± S.D. from three independent experiments. C, HCT-116 p53−/− cells were treated with 1 μm Dox for 4 h in the presence or absence of 1 mm caffeine. After washing out Dox, cells were incubated in fresh medium with or without caffeine. Cells were harvested at the indicated times for Western blot analysis. D, parallel sets of cells were subjected to flow cytometric analysis. E, HCT-116 p53−/− cells treated as described above were co-treated with or without 10 μm lactacystin. After release for 15 h, total lysates containing 15 μg of proteins were analyzed by Western blotting. F, HCT-116 p53−/− cells were either treated with 200 ng/ml nocodazole (Noc) for 19 h or with 1 μm Dox for 4 h and released for 15 h as described above. NT, no treatment. Cells were harvested for flow cytometric and Western blot analysis. Data are the mean ± S.D. from three independent experiments. **, p < 0.01 relative to samples without treatment based on a Student's t test. G, HCT-116 p53−/− cells were synchronized by a double thymidine (Thd) block and released by replacement with fresh medium containing 200 ng/ml nocodazole, allowing S/G2 progression. Cells at the indicated time point were fixed and stained with the anti-human TK1 (XPA210)/FITC antibody, anti-human phospho-H3 (Ser-10)/TRITC, and Hoechst 33342 for confocal microscopy observation. The remaining cells were harvested for Western blot and flow cytometric analysis.
Because TK1 is degraded via APC/C-mediated proteolysis during mitotic exit (10), we then tested whether TK1 induction inhibited by caffeine treatment during DNA damage recovery involves protein destabilization. To this end, we treated cells with lactacystin, a specific proteasome inhibitor, during recovery from DNA damage. Without caffeine co-treatment, the addition of lactacystin did not affect the level of either TK1 induction or H3Ser-10 phosphorylation during release from Dox treatment (Fig. 2E). In contrast, the addition of lactacystin in cells co-treated with caffeine restored TK1 and R2 expression, whereas H3Ser-10 phosphorylation was reduced. These results suggest that lactacystin treatment prevents checkpoint inactivation-induced mitotic progression, thereby resulting in TK1 accumulation. Moreover, the level of TK1 expression or the dTTP pool in cells arrested in prometaphase by nocodazole treatment, a microtubule-depolymerizing drug, was similar to what was observed in cells arrested in G2 by releasing from DNA damage for 15 h (Fig. 2F), demonstrating that the lack of proteolysis during mitotic exit contributes to the increased level of TK1 during recovery from DNA damage. We also examined the nuclear localization of TK1 during G2 progression by releasing cells from a double thymidine block. Mitotic rounding and chromatin condensation in cells were observed at 8 h after releasing from the double thymidine block. At 6 h, cells showed clear nuclear accumulation of TK1 when reaching the G2/M transition before mitotic rounding (Fig. 2G). In conclusion, there is an expansion of dTTP associated with up-regulation of TK1 after DNA damage as a result of G2 arrest by checkpoint activation in HCT-116 p53−/− cells.
p53 Affects the Magnitude of DNA Damage-induced TK1 Expression through p21-mediated Cell Cycle Control
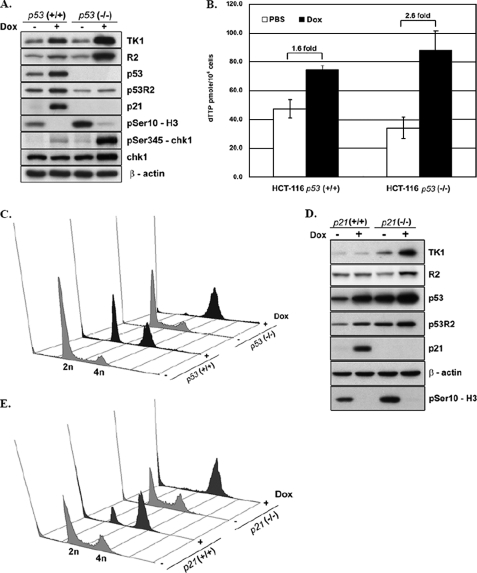
Given that less amount of TK1 was induced in HCT-116 p53+/+ isogenic line, we then investigated the functional involvement of p53 in regulating the level of TK1 in response to DNA damage. The effects of Dox treatment on protein expression and cell cycle profiles were compared in p53−/− cells and p53+/+ cells after release from Dox for 24 h. As expected, the levels of p53R2 and p21, a universal cyclin-dependent kinase inhibitor (46, 47), was induced in p53+/+ but not p53−/− cells. The induced levels of TK1 and R2 and the extent of chk1 activation were consistently higher in p53−/− than that in p53+/+ cells after release for 24 h. H3Ser-10 phosphorylation was undetectable in both cell lines after DNA damage, indicating a lack of mitotic progression during recovery (Fig. 3A). The cellular level of dTTP increased 2.6- and 1.6-fold in p53−/− and p53+/+ cells, respectively, indicating a correlation of R2 and TK1 induction with dTTP synthesis in DNA-damaged cells (Fig. 3B). The flow cytometric analysis showed that unlike p53−/− cells, which were only enriched in G2 phase, p53+/+ cells were distributed in both G1 and G2 phases after DNA damage (Fig. 3C). Because p53-mediated elevation of p21 due to DNA damage could prevent p53+/+ cells from G1/S progression, we speculated that lower extent of TK1 induction in p53+/+ cells might be due to the p21-mediated G1 checkpoint, resulting in less G2 accumulation. To test this hypothesis, we then compared the expression level of TK1 and cell cycle distribution after DNA damage in p21+/+ and its isogenic p21−/− HCT-116 line. The results showed a 4.7-fold induction of TK1 in p21−/− cells in contrast to a minor change in TK1 levels in p21+/+ cells (Fig. 3D). Results from the flow cytometric analysis showed that p21+/+ cells exhibited a G1 and G2 phase distribution at 24 h after release from DNA damage, whereas p21−/− cells accumulated in G2 phase (Fig. 3E). These results supported the hypothesis that p21-mediated cell cycle inhibition affects the level of TK1 in response to DNA damage in the presence of a functional p53 protein. Conversely, in the absence of p53, cells are blocked in the G2 phase, allowing more TK1 accumulation.
FIGURE 3.
p53-induced p21 affects the cell cycle progression and the extent of TK1 induction in response to DNA damage. HCT-116 p53+/+ and p53−/− cells were released from Dox treatment as described in the legend to Fig. 1C. After release from Dox for 24 h, cells were harvested for Western blotting (A), dTTP pool determination (B), and flow cytometric analysis (C). Data are the mean ± S.D. from three independent experiments. D, HCT-116 p21−/− or p21+/+ cells were treated as above. After recovery from DNA damage for 24 h, cells were harvested for Western blot and flow cytometric analysis (E).
TK1 Contributes to the Increase in dTTP Pool during Recovery from DNA Damage
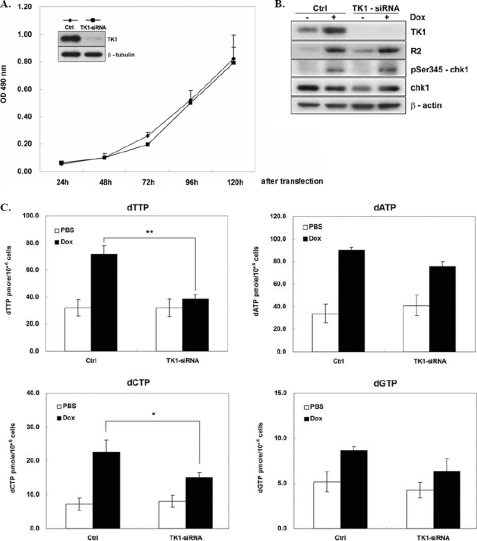
To find the relationship between the cellular levels of dTTP and TK1 in response to DNA damage, we determined the effect of silencing TK1 expression on the cellular dTTP pool by TK1-siRNA transfection of HCT-116 p53−/− cells. Results from MTS assays showed that TK1 knockdown did not affect the growth rate of transfected cells over a 4-day period (Fig. 4A). Apparently, the expression of TK1 is dispensable for proliferation of HCT-116 p53−/− cells. In cells treated with Dox, TK1 knockdown did not have an effect on the induction of R2 or chk1 phosphorylation during recovery from Dox-induced DNA damage (Fig. 4B). Meanwhile, the cellular level of dTTP increased 2.25-fold, and TK1 depletion greatly diminished (Fig. 4C). The levels of dATP, dCTP, and dGTP were also increased during recovery from Dox-induced DNA damage. TK1 knockdown did not significantly affect the levels of dATP and dGTP, whereas the induction of the dCTP pool was reduced by TK1 depletion. Because dTTP allosterically inhibits reduction of CDP by ribonucleotide reductase, it is unlikely that less dCDP production in TK1-depleted cells occurred via ribonucleotide reductase. Probably, other dCTP regulators in DNA damage response are sensitive to the TK1 knockdown. It should be noted that TK1 depletion did not affect the level of dTTP in the cells without Dox treatment, which imply that in growing cells the de novo pathway is sufficient for dTTP synthesis. However, in DNA damage-stressed cells, the TK1-mediated pathway may become essential in generating sufficient dTTP during the recovery from DNA damage.
FIGURE 4.
Depletion of TK1 suppressed the cellular level of dTTP during recovery from DNA damage. HCT-116 p53−/− cells transfected with 100 pmol of TK1-siRNA were trypsinized and re-plated into a 96-well plate at 1 × 103 cells per well. The growth rate of cells with TK1 knockdown was measured by the MTS assay over a 4-day period. Data are the mean ± S.D. from three independent experiments (A). The inset shows the Western blot of cell lysates harvested on 2 days. After transfection with TK1 and control (Ctrl) siRNA for 48 h, cells were treated with Dox for 4 h and released as described in the legend to Fig. 1C. After 24 h, cells were harvested for Western blot analysis (B) and dNTP pool determinations (C). Data are the mean ± S.D. from three independent experiments. p < 0.05 (*) and p < 0.01 (**) relative to control samples with Dox treatment based on Student's t test.
TK1 Affects the Efficiency of DNA Repair during Recovery for G2/M Progression
Because the dNTP supply is essential for the repair process of damaged DNA, we asked whether TK1 expression is important in the DNA repair during recovery from DNA damage and for cell cycle progression. To address this question, γ-H2AX foci staining was used to examine the extent of DNA double-strand breaks induced by Dox treatment. According to immunofluorescence quantification (Fig. 5A), pulse-exposure of cells to Dox caused widespread γ-H2AX foci formation, in marked contrast to minimal staining in cells without Dox treatment. After release for 24–42 h, the intensity of γ-H2AX foci staining decreased, but it did not vanish in the control cells. Depletion of TK1 by siRNA transfection did not enhance the intensity of γ-H2AX foci after Dox treatment, but the cells had persistent γ-H2AX focal staining during recovery from DNA damage for 24–42 h. These results demonstrate the importance of TK1-mediated dTTP synthesis for DNA repair. As mentioned earlier, mitotic entry was detected in cells after recovery from DNA damage for 48 h, as indicated by a marked increase in H3Ser-10 phosphorylation and MPM2 immunofluorescence staining. Although DNA repair was retarded by TK1 depletion, we found that the timing of mitotic entry was unaffected (Fig. 5B), indicating that TK1 depletion did not disturb checkpoint recovery for the M phase entry. Notably, DNA lesions were still present in most of the cells during mitotic entry, an indication of cell adaptation to DNA damage (supplemental Fig. S2). The MTS assay was performed in cells treated for 4 h with Dox, and the results showed that TK1 depletion significantly decreased cell survival (Fig. 5C). Probably, TK1 depletion retards the repair of Dox-induced DNA lesions, thus promoting cell death. Altogether, our results suggest that TK1-mediated dTTP synthesis in p53-deficient tumor cells provides better DNA repair efficiency during recovery from genome lesions.
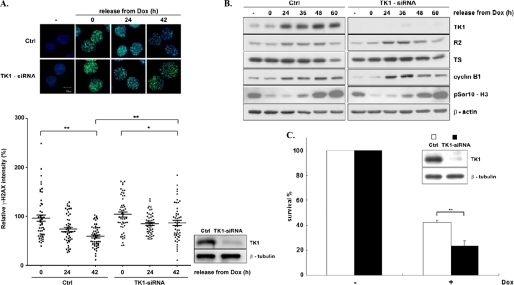
FIGURE 5.
The contribution of TK1 to the DNA repair efficiency during recovery from DNA damage. A, HCT-116 p53−/− cells plated on the coverslip were transfected with 100 pmol of control and TK1-siRNA. After 48 h cells were treated with Dox and released as described previously. At the indicated times cells were stained with anti-γ-H2AX/FITC and Hoechst 33342 for immunofluorescence microscope observations. The immunofluorescence intensity of γ-H2AX staining was quantified by the ImageJ software (n = 50). The mean value of γ-H2AX immunofluorescence staining in control cells with Dox pulse-treatment for 4 h was set to 100%. The graph shown in the right panel represents relative intensity of γ-H2AX staining in nuclei. p < 0.05 (*) and p < 0.01 5 (**) is based on Student's t test. Ctrl, control. B, total proteins were harvested at the indicated times during recovery from DNA damage and analyzed by Western blotting. C, cells after siRNA transfection for 6 h were trypsinized and re-plated onto a 96-well-plate at 2.5 × 103 cells per well. These cells were allowed to grow additionally for 48 h before 5 μm Dox treatment and released as described previously. After 48 h, cell survival was measured by MTS assay; data are the mean ± S.D. from three independent experiments. **, p < 0.01 relative to control samples based on a Student's t test.
DISCUSSION
In response to DNA damage, coordination of dNTP synthesis is required for the DNA repair process. In light of the importance of up-regulation of key proteins, p53R2 and R2, in the de novo pathway of dNTP synthesis in cells after DNA damage, this study evaluated the contribution of salvage synthesis of dTTP in tumor cells during recovery from genotoxic insults. Several novel findings are presented here. DNA damage in p53-deficient tumor cells increases the expression of TK1 as a result of G2 arrest due to checkpoint activation. Second, less TK1 is induced by DNA damage in tumor cell proficient in functional p53 because of p21-mediated G1 arrest. Third, TK1 expression in p53-deficient HCT-116 cells is dispensable for growth but important for providing sufficient dTTP pools for DNA repair and cell survival.
Sartorelli and co-workers (24) have previously shown that treatment of HCT-116 p53−/− cells with cisplatin caused an increase in dNTP levels. Even though R2 knockdown was able to reduce the amounts of dNTPs in these cells, the level of dTTP remained elevated after DNA damage in R2-depleted cells, indicating an alternative pathway for dTTP supply after DNA damage. The findings in this report show that genotoxic insults increase TK1 expression, which contributes to the expansion of the dTTP pool. Notably, HCT-116 p53−/− cells arrest in the G2 phase during recovery from DNA damage, and DNA lesions diminish gradually before mitotic re-entry. Significantly, depletion of TK1 increases the DNA lesion intensity, as revealed by sustained γ-H2AX foci staining, without affecting the timing of mitotic re-entry. Thus, checkpoint inactivation takes place in TK1-depleted cells, so that these tumor cells apparently adapt to the remaining DNA lesions, enhancing Dox-induced cell death.
Very interestingly, neither the steady-state level of dTTP nor cell proliferation under normal growth condition was affected by TK1 depletion. Because high levels of TK1 are often associated with many tumors, these results evoke a reinterpretation of the main role for TK1 in DNA repair in tumor cells rather than in providing dTTP for replication and growth. In agreement with our findings, a number of studies have observed that decreased dTTP levels due to TK deficiency correlate with an increase in DNA damage-induced cell death (33–36, 38).
In this study we also investigated the molecular mechanism responsible for TK1 induction in response to DNA damage. Our data indicate that TK1 up-regulation by DNA damage appears to be a consequence of its cell cycle control. In HCT-116 p53+/+ cells, DNA damage results in a sub-G1 population, a block in the G1/S transition, and a G2 cell accumulation. The magnitude of TK1 induction was not as pronounced as that observed in p53-deficient cells. Our results suggest that p53-dependent p21 expression causes G1 arrest, thus reducing G2 cell accumulation. As a result, TK1 levels were lower in p53-proficient cells. Consequently, more TK1 induction together with a significant increase in G2 accumulation was observed in DNA-damaged HCT-116 p21−/− as compared with their parental HCT-116 p21+/+ cells. Checkpoint inactivation by caffeine treatment allowed the DNA-damaged cells to undergo mitotic progression, thus diminishing TK1 expression. Consistent with TK1 degradation via APC/C-mediated ubiquitination during mitotic exit, we found that inhibition of the proteasome by lactacystin treatment during checkpoint inactivation by caffeine restored high TK1 expression. Thus, DNA damage-induced checkpoint activation in G2 phase controls the TK1 level by blocking the mitotic entry and subsequent TK1 proteolysis. Moreover, the level of TK1 expression, the dTTP pool, or nuclear localization in cells arrested in prometaphase by nocodazole was all quite similar to what was observed in Dox-treated cells. All together, our results suggest that DNA damage-induced up-regulation of TK1 is a consequence of DNA damage-induced G2 accumulation.
Recently, one report has demonstrated that chk1 activation by Top1-mediated DNA damage increases R2 mRNA and protein levels via E2F1-mediated transcriptional activation (48). E2F1 is a known transcription activator of R2 and required for S phase progression. In our study we also found an increase in the TK1 mRNA level in response to DNA damage (data not shown). Therefore, it is possible that transcriptional activation and protein stabilization during G2 arrest act together to increase the TK1 level.
Here, we observed a similar expression pattern of R2 and TK1 in response to DNA damage, probably because these genes share common control mechanisms for cell cycle-regulated transcription, translation, and proteolysis. In subcellular localization experiments, we found a nuclear translocation of TK1 in response to DNA damage, similar to what has been reported for p53R2 or R2 proteins (19, 48), although the nuclear localization of the ribonucleotide reductase subunits has been debated (49). A nuclear form of TK1 was recently described in non-small lung cancer cells (50), but the role and mechanism of the translocation process remain to be determined. Relevantly, it has been shown that UV damage is able to induce nuclear translocation of TS via SUMOylation (51, 52). In sum, we conclude that checkpoint activation after DNA damage allows tumor cells to integrate de novo synthesis of dNTP via ribonucleotide reductase and salvage synthesis of dTTP via TK1 to achieve an appropriate level of nucleotides for efficient DNA repair and survival. Nuclear localization of TK1 and TS might further provide an efficient means to support dTTP supply for DNA repair. Because conventional chemotherapeutic agents kill tumor cells through DNA double-stand breaks, the results presented here imply that blocking dTTP synthesis would enhance the genotoxic insult-induced lethality in p53-defective tumor cells by preventing efficient repair of DNA lesions during G2/M progression. Although inhibition of TS by fluorouracil has long been used for chemotherapy (53), tumors devoid of p53 function often develop drug resistance (22, 54), and the anti-TS agents by itself cause general cytotoxicity due to misincorporation of dUTP and 5′-fluoro-2′-deoxyuridine triphosphate into DNA (55, 56). Therefore, inhibition of alternative targets such as TK1 or thymidylate kinase to reduce dTTP levels would be non-toxic to normal cycling cells but lead to chemosensitization and elimination of tumor cells.
Supplementary Material
This study was supported by Grants NHRI-EX99-9701B and NSC99-3112-B-010-021 from National Health Research Institute and National Science Council, Taiwan, respectively, and a grant from Aim for the Top University plan in National Yang-Ming University supported by the Ministry of Education, Taiwan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- TS
- thymidylate synthase
- TK1
- thymidine kinase 1
- TRITC
- tetramethylrhodamine isothiocyanate
- Dox
- doxorubicin
- H3
- histone 3
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
- γH2AX
- Ser-139 phosphorylated H2AX.
REFERENCES
- 1.Reichard P. (1988) Annu. Rev. Biochem. 57, 349–374 [DOI] [PubMed] [Google Scholar]
- 2.Mathews C. K. (2006) FASEB J. 20, 1300–1314 [DOI] [PubMed] [Google Scholar]
- 3.Johansson M., Karlsson A. (1997) J. Biol. Chem. 272, 8454–8458 [DOI] [PubMed] [Google Scholar]
- 4.Wang L., Eriksson S. (2000) Biochem. J. 351, 469–476 [PMC free article] [PubMed] [Google Scholar]
- 5.Pontarin G., Gallinaro L., Ferraro P., Reichard P., Bianchi V. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12159–12164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnér E. S., Eriksson S. (1995) Pharmacol. Ther. 67, 155–186 [DOI] [PubMed] [Google Scholar]
- 7.Bello L. J. (1974) Exp. Cell Res. 89, 263–274 [DOI] [PubMed] [Google Scholar]
- 8.Coppock D. L., Pardee A. B. (1987) Mol. Cell. Biol. 7, 2925–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sherley J. L., Kelly T. J. (1988) J. Biol. Chem. 263, 8350–8358 [PubMed] [Google Scholar]
- 10.Ke P. Y., Chang Z. F. (2004) Mol. Cell. Biol. 24, 514–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Navalgund L. G., Rossana C., Muench A. J., Johnson L. F. (1980) J. Biol. Chem. 255, 7386–7390 [PubMed] [Google Scholar]
- 12.Huang S. H., Tang A., Drisco B., Zhang S. Q., Seeger R., Li C., Jong A. (1994) DNA Cell Biol. 13, 461–471 [DOI] [PubMed] [Google Scholar]
- 13.Ke P. Y., Kuo Y. Y., Hu C. M., Chang Z. F. (2005) Genes Dev. 19, 1920–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harper J. W., Elledge S. J. (2007) Mol. Cell 28, 739–745 [DOI] [PubMed] [Google Scholar]
- 15.Jackson S. P., Bartek J. (2009) Nature 461, 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eckstein H., Ahnefeld S., Albietz-Loges K. (1974) Z. Naturforsch C 29, 272–282 [DOI] [PubMed] [Google Scholar]
- 17.Chabes A., Georgieva B., Domkin V., Zhao X., Rothstein R., Thelander L. (2003) Cell 112, 391–401 [DOI] [PubMed] [Google Scholar]
- 18.Nakano K., Bálint E., Ashcroft M., Vousden K. H. (2000) Oncogene 19, 4283–4289 [DOI] [PubMed] [Google Scholar]
- 19.Tanaka H., Arakawa H., Yamaguchi T., Shiraishi K., Fukuda S., Matsui K., Takei Y., Nakamura Y. (2000) Nature 404, 42–49 [DOI] [PubMed] [Google Scholar]
- 20.Yamaguchi T., Matsuda K., Sagiya Y., Iwadate M., Fujino M. A., Nakamura Y., Arakawa H. (2001) Cancer Res. 61, 8256–8262 [PubMed] [Google Scholar]
- 21.Guittet O., Håkansson P., Voevodskaya N., Fridd S., Gräslund A., Arakawa H., Nakamura Y., Thelander L. (2001) J. Biol. Chem. 276, 40647–40651 [DOI] [PubMed] [Google Scholar]
- 22.Lowe S. W., Ruley H. E., Jacks T., Housman D. E. (1993) Cell 74, 957–967 [DOI] [PubMed] [Google Scholar]
- 23.Lowe S. W., Bodis S., McClatchey A., Remington L., Ruley H. E., Fisher D. E., Housman D. E., Jacks T. (1994) Science 266, 807–810 [DOI] [PubMed] [Google Scholar]
- 24.Lin Z. P., Belcourt M. F., Cory J. G., Sartorelli A. C. (2004) J. Biol. Chem. 279, 27030–27038 [DOI] [PubMed] [Google Scholar]
- 25.Lin Z. P., Belcourt M. F., Carbone R., Eaton J. S., Penketh P. G., Shadel G. S., Cory J. G., Sartorelli A. C. (2007) Biochem. Pharmacol. 73, 760–772 [DOI] [PubMed] [Google Scholar]
- 26.Nordlund P., Reichard P. (2006) Annu. Rev. Biochem. 75, 681–706 [DOI] [PubMed] [Google Scholar]
- 27.Björklund S., Skog S., Tribukait B., Thelander L. (1990) Biochemistry 29, 5452–5458 [DOI] [PubMed] [Google Scholar]
- 28.Chabes A., Thelander L. (2000) J. Biol. Chem. 275, 17747–17753 [DOI] [PubMed] [Google Scholar]
- 29.Chabes A. L., Pfleger C. M., Kirschner M. W., Thelander L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3925–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeGregori J., Kowalik T., Nevins J. R. (1995) Mol. Cell. Biol. 15, 4215–4224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roehl H. H., Conrad S. E. (1990) Mol. Cell. Biol. 10, 3834–3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang Z. F., Huang D. Y., Hu S. F. (1999) J. Cell. Biochem. 75, 300–309 [PubMed] [Google Scholar]
- 33.McKenna P. G., Hickey I. (1981) Cell Biol. Int. Rep. 5, 555–561 [DOI] [PubMed] [Google Scholar]
- 34.McKenna P. G., Yasseen A. A., McKelvey V. J. (1985) Somat. Cell Mol. Genet. 11, 239–246 [DOI] [PubMed] [Google Scholar]
- 35.McKenna P. G., McKelvey V. J., Frew T. L. (1988) Mutat. Res. 200, 231–242 [DOI] [PubMed] [Google Scholar]
- 36.al-Nabulsi I., Takamiya Y., Voloshin Y., Dritschilo A., Martuza R. L., Jorgensen T. J. (1994) Cancer Res. 54, 5614–5617 [PubMed] [Google Scholar]
- 37.Boothman D. A., Davis T. W., Sahijdak W. M. (1994) Int. J. Radiat. Oncol. Biol. Phys. 30, 391–398 [DOI] [PubMed] [Google Scholar]
- 38.Wakazono Y., Kubota M., Furusho K., Liu L., Gerson S. L. (1996) Mutat. Res. 362, 119–125 [DOI] [PubMed] [Google Scholar]
- 39.Hu C. M., Chang Z. F. (2008) Cancer Res. 68, 2831–2840 [DOI] [PubMed] [Google Scholar]
- 40.Chang Z. F., Huang D. Y., Hsue N. C. (1994) J. Biol. Chem. 269, 21249–21254 [PubMed] [Google Scholar]
- 41.Gasparri F., Wang N., Skog S., Galvani A., Eriksson S. (2009) Eur. J. Cell Biol. 88, 779–785 [DOI] [PubMed] [Google Scholar]
- 42.Sherman P. A., Fyfe J. A. (1989) Anal. Biochem. 180, 222–226 [DOI] [PubMed] [Google Scholar]
- 43.Sarkaria J. N., Busby E. C., Tibbetts R. S., Roos P., Taya Y., Karnitz L. M., Abraham R. T. (1999) Cancer Res. 59, 4375–4382 [PubMed] [Google Scholar]
- 44.van Vugt M. A., Brás A., Medema R. H. (2004) Mol. Cell 15, 799–811 [DOI] [PubMed] [Google Scholar]
- 45.Mamely I., van Vugt M. A., Smits V. A., Semple J. I., Lemmens B., Perrakis A., Medema R. H., Freire R. (2006) Curr. Biol. 16, 1950–1955 [DOI] [PubMed] [Google Scholar]
- 46.Xiong Y., Hannon G. J., Zhang H., Casso D., Kobayashi R., Beach D. (1993) Nature 366, 701–704 [DOI] [PubMed] [Google Scholar]
- 47.Espinosa J. M., Emerson B. M. (2001) Mol. Cell 8, 57–69 [DOI] [PubMed] [Google Scholar]
- 48.Zhang Y. W., Jones T. L., Martin S. E., Caplen N. J., Pommier Y. (2009) J. Biol. Chem. 284, 18085–18095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pontarin G., Fijolek A., Pizzo P., Ferraro P., Rampazzo C., Pozzan T., Thelander L., Reichard P. A., Bianchi V. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17801–17806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brockenbrough J. S., Morihara J. K., Hawes S. E., Stern J. E., Rasey J. S., Wiens L. W., Feng Q., Vesselle H. (2009) J. Histochem. Cytochem. 57, 1087–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woeller C. F., Anderson D. D., Szebenyi D. M., Stover P. J. (2007) J. Biol. Chem. 282, 17623–17631 [DOI] [PubMed] [Google Scholar]
- 52.Fox J. T., Shin W. K., Caudill M. A., Stover P. J. (2009) J. Biol. Chem. 284, 31097–31108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Longley D. B., Harkin D. P., Johnston P. G. (2003) Nat. Rev. Cancer 3, 330–338 [DOI] [PubMed] [Google Scholar]
- 54.Bunz F., Hwang P. M., Torrance C., Waldman T., Zhang Y., Dillehay L., Williams J., Lengauer C., Kinzler K. W., Vogelstein B. (1999) J. Clin. Investig. 104, 263–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Longley D. B., Boyer J., Allen W. L., Latif T., Ferguson P. R., Maxwell P. J., McDermott U., Lynch M., Harkin D. P., Johnston P. G. (2002) Cancer Res. 62, 2644–2649 [PubMed] [Google Scholar]
- 56.Goulian M., Bleile B., Tseng B. Y. (1980) J. Biol. Chem. 255, 10630–10637 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.