Abstract
During (bacterio)chlorophyll biosynthesis of many photosynthetically active organisms, dark operative protochlorophyllide oxidoreductase (DPOR) catalyzes the two-electron reduction of ring D of protochlorophyllide to form chlorophyllide. DPOR is composed of the subunits ChlL, ChlN, and ChlB. Homodimeric ChlL2 bearing an intersubunit [4Fe-4S] cluster is an ATP-dependent reductase transferring single electrons to the heterotetrameric (ChlN/ChlB)2 complex. The latter contains two intersubunit [4Fe-4S] clusters and two protochlorophyllide binding sites, respectively. Here we present the crystal structure of the catalytic (ChlN/ChlB)2 complex of DPOR from the cyanobacterium Thermosynechococcus elongatus at a resolution of 2.4 Å. Subunits ChlN and ChlB exhibit a related architecture of three subdomains each built around a central, parallel β-sheet surrounded by α-helices. The (ChlN/ChlB)2 crystal structure reveals a [4Fe-4S] cluster coordinated by an aspartate oxygen alongside three cysteine ligands. Two equivalent substrate binding sites enriched in aromatic residues for protochlorophyllide substrate binding are located at the interface of each ChlN/ChlB half-tetramer. The complete octameric (ChlN/ChlB)2(ChlL2)2 complex of DPOR was modeled based on the crystal structure and earlier functional studies. The electron transfer pathway via the various redox centers of DPOR to the substrate is proposed.
Keywords: Crystallography, Iron-Sulfur Protein, Nitrogenase, Photosynthetic Pigments, Protein-Protein Interactions, Chlorophyll Biosynthesis, Electron Transfer, Protochlorophyllide
Introduction
Photosynthesis represents the fundamental strategy of nature to convert solar radiation into biochemically accessible energy. Chlorophylls and bacteriochlorophylls constitute the pigments employed in both harvesting and utilizing photons of visible light. Biosynthesis of these complex tetrapyrroles, of which more than 6 billion tons are produced annually, utilizes a chain of enzymatic conversions, many of which delve deep into the biochemical treasure trove of early life on earth (1).
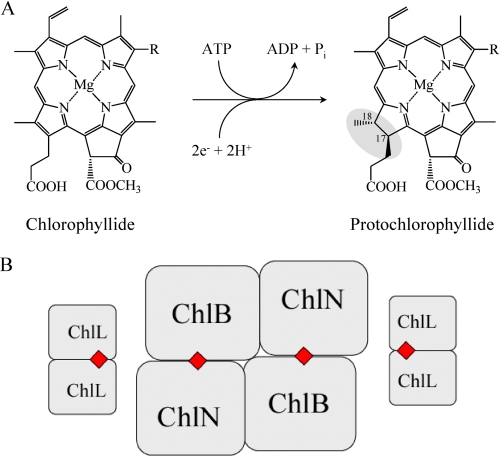
One of the more unusual steps in (bacterio)chlorophyll biosynthesis involves the chemically challenging, stereospecific reduction of the C17=C18 double bond of ring D of the porphyrin protochlorophyllide (Pchlide)2 to form the chlorin chlorophyllide (Chlide, Fig. 1A). Two unrelated pathways have evolved for this two-electron reduction (2–5). In angiosperms, a monomeric, light-dependent protochlorophyllide oxidoreductase (LPOR) (NADPH Pchlide oxidoreductase, EC 1.3.1.33) catalyzes the reaction. The bound substrate Pchlide needs to be activated by a photon to drive the NADPH-dependent reduction step (6–10).
FIGURE 1.
Catalytic reaction of the DPOR and schematic representation of the subunit architecture. A, ATP-dependent DPOR catalysis involves the two-electron reduction of the C17=C18 double bond of Pchlide to form Chlide. R is either an ethyl or a vinyl residue. B, two ChlL2 dimers interact with the heterotetrameric (ChlN/ChlB)2 complex to form hetero-octameric DPOR during catalysis. The four intersubunit [4Fe-4S] clusters are marked as red squares.
Anoxygenic, photosynthetic bacteria, by contrast, make use of an ATP-dependent process catalyzed by the dark operative protochlorophyllide oxidoreductase (DPOR). Other photosynthetic organisms such as cyanobacteria, algae, or gymnosperms encode both LPOR and DPOR (4).
DPOR consists of three subunits. In chlorophyll-synthesizing organisms, these are termed ChlN, ChlB, and ChlL (1, 11, 12); in bacteriochlorophyll synthesizers, they are BchN, BchB, and BchL (11, 13). ChlL (BchL) forms the homodimer ChlL2 (BchL2) that functions as an ATP-dependent electron shuttle carrying an intersubunit [4Fe-4S] cluster and two ATP binding sites (13–15). Recently, the crystal structure of the BchL2 complex from Rhodobacter sphaeroides was solved (16). Subunits ChlN and ChlB instead constitute a heterotetrameric complex here denoted (ChlN/ChlB)2 that bears two [4Fe-4S] clusters and two substrate binding sites (14).
Some details of the catalytic mechanism of DPOR have been established biochemically. Upon binding of two molecules of ATP, ChlL2 interacts with the catalytic, substrate binding (ChlN/ChlB)2 complex. Ferredoxin provides a single electron to ChlL2 (13), which in turn transfers an electron to (ChlN/ChlB)2. Hydrolysis of the two ATP molecules results in the dissociation of ChlL2 from reduced (ChlN/ChlB)2. Pchlide reduction is completed after two sequential catalytic redox cycles. Substrate recognition by (ChlN/ChlB)2 essentially involves all functional groups of the substrate (14). Two ChlL2 dimers simultaneously interact with the (ChlN/ChlB)2 tetramer, giving rise to a hetero-octameric holoenzyme (15) (Fig. 1B). Residues from three DPOR subunits are presumably involved in the interaction of ChlL2 with (ChlN/ChlB)2 (17).
Here, we describe the crystal structure of the heterotetrameric (ChlN/ChlB)2 complex of DPOR. The structure inter alia reveals an unusual coordination of the intersubunit [4Fe-4S] clusters by 3 cysteine residues of ChlN and a unique aspartate residue of ChlB. The active site cavity is proposed to lie at the ChlN/ChlB interface of each half-dimer. It is characterized by aromatic amino acid residues involved in substrate coordination, placing the substrate ∼14 Å from the respective [4Fe-4S] cluster. The interaction of the dynamic switch protein ChlL2 with subcomplex (ChlN/ChlB)2 was modeled, placing the [4Fe-4S] cluster of ChlL2 19 Å from the corresponding cluster of (ChlN/ChlB)2, ensuring rapid electron transfer during catalysis. Comparing (ChlN/ChlB)2 with the nitrogenase MoFe protein (NifD/NifK)2 reveals significant structural homologies as well as conservation of functional residues.
EXPERIMENTAL PROCEDURES
Heterologous Production and Purification of Thermosynechococcus elongatus DPOR
T. elongatus DPOR (ChlN/ChlB)2 subcomplexes were recombinantly produced under anaerobic conditions (oxygen partial pressure <1 ppm) in an anaerobic chamber (Coy Laboratories, Grass Lake, MI) as described previously (14, 17). Selenomethionine-labeled (ChlN/ChlB)2 was produced as described before (18). Purification of proteins was carried out as outlined earlier (14, 17). Lysis buffer contained 100 mm HEPES-NaOH, pH 7.5, 150 mm NaCl, and 10 mm MgCl2. Glutathione S-transferase tags N-terminally fused to ChlN were used for the affinity chromatographic purification of 80 mg of (ChlN/ChlB)2 complexes (370 nmol) via 10 ml of Protino® glutathione agarose (Macherey-Nagel, Düren, Germany). PreScissionTM protease (GE Healthcare, Uppsala, Sweden) treatment was employed to liberate and elute (ChlN/ChlB)2 from the matrix (14). Gel permeation chromatography using a Superdex 200 HR 26/60 column (GE Healthcare) equilibrated with lysis buffer in an anaerobic chamber (Coy laboratories) followed. The column was calibrated with protein standards (molecular weight marker kit MW-GF 1000, Sigma).
N-terminal Amino Acid Sequence Determination
Automated Edman degradation was used to confirm the identity of purified proteins and for quantification of individual protein subunits.
DPOR Enzyme Assay
To validate the quality of purified (ChlN/ChlB)2, proteins were tested for catalytic activity in the standard DPOR assay containing 13 μm Pchlide, 2 mm dithionite, 2 mm ATP, and an ATP-regenerating system. 100 pmol of T. elongatus (ChlN/ChlB)2 were supplemented with 200 pmol of purified Prochlorococcus marinus ChlL2 (in a final volume of 125 μl) and analyzed as described earlier (14, 17).
Protein Crystallization
Purified (ChlN/ChlB)2 was concentrated to 10 mg/ml (45 μm) in lysis buffer using a stirred Amicon ultrafiltration cell (Millipore) with a 50-kDa cutoff. (ChlN/ChlB)2 was crystallized by hanging drop vapor diffusion at 17 °C in an anaerobic chamber by mixing 3 μl of protein with 3 μl of reservoir solution consisting of 9.5% polyethylene glycol 6000, 85 mm HEPES-NaOH, pH 7.1, 14.3% 2-methyl pentane-2,4-diol, and 15% glycerol as cryoprotectants or 10.5% polyethylene glycol 6000, 85 mm HEPES-NaOH, pH 7.5, 14.3% 2-methyl pentane-2,4-diol, and 15% glycerol as cryoprotectants for selenomethionine-labeled protein. Crystals grew within 3–5 days and were shock-cooled in liquid nitrogen.
Data Collection, Structure Determination, and Refinement
Synchrotron diffraction data were collected to 2.4 Å resolution at beam line ID29 of the European Synchrotron Radiation Facility (ESRF) (Grenoble, France) using non-derivatized, wild-type (ChlN/ChlB)2 crystals. Anomalous data of selenomethionine-derivatized (ChlN/ChlB)2 crystals were collected to 2.8 Å resolution at beamline PROXIMA 1 of the Soleil synchrotron (Paris, France) at wavelengths 0.9790 (peak) and 0.9795 Å (inflection). For data collection statistics, see Table 1.
TABLE 1.
Data collection, phasing, and refinement
| Native | Selenium-methionine Derivative |
||
|---|---|---|---|
| Peak | Inflection | ||
| Data collection | |||
| Synchrotron source, beamline | ESRF, ID29 | Soleil, Proxima 1 | |
| Space group | P6322 | P6322 | |
| Cell parameters (Å) | a = b = 192.3, c = 132.6 | a = b = 191.7 132.3 | |
| No. of molecules per asymmetric unit | ½ (ChlNB)2 | ||
| Matthews coefficient | 3.31 | ||
| Solvent content (%) | 62 | ||
| Wavelength (Å) | 0.91 | 0.9790 | 0.9795 |
| Resolutiona (Å) | 25-2.4 (2.49-2.40) | 34.6-2.8 (2.98-2.80) | 35.1-2.8 (2.98-2.81) |
| Completenessa (%) | 99.1 (100.0) | 99.6 (98.1)b | 99.7 (98.4)b |
| R-mergea (%) | 7.8 (46.9) | 8.5 (54.5) | 8.3 (62.7) |
| Redundancya | 8.3 (8.6) | 22.8 (22.4) | 22.8 (22.5) |
| I/σIa | 20.0 (3.5) | 32.5 (6.8) | 32.5 (6.0) |
| No. of unique reflectionsa | 55 771 (5518) | 67 172 (10637)b | 67 262 (10 678)b |
| Data reduction/scaling program | HKL2000 | XDS | XDS |
| Phasing | |||
| Anomalous resolution limitc (Å) | n.a. | 3.6 | 3.8 |
| No. of selenium sites (theoretical) | 22 | ||
| No. of selenium sites found | 21 + [4Fe-4S]-cluster | ||
| Refinement | |||
| Resolution (Å)a | 103.7-2.4 (2.5-2.4) | ||
| No. of reflections | 52744 (3765) | ||
| No. of residues theoretical/observed/disordered | 968/865/103 | ||
| Rworka (%) | 20 (34) | ||
| Rfreea (%) | 26 (39) | ||
| Figure of merit | 0.80 | ||
| r.m.s.d. deviations from ideality: bonds (Å)/angles (°) | 0.2/2.0 | ||
a Values in parentheses indicate resolution shell of highest resolution.
b Anomalous data.
c ShelxC d″/σd″ ≥ 1.
Data reduction suites HKL2000 (19) and XDS (20, 21) were used for data reduction of the non-derivatized and selenomethionine-derivatized crystals, respectively (Table 1). Programs ShelxC, ShelxD, and ShelxE were used to locate anomalous scatterers and to determine initial phases (22). Phases were further improved by solvent-flattening and histogram-matching routines using DM (23). The protein model was built manually using COOT (24) and refined using REFMAC5 (25). All other manual interventions were performed using the CCP4 suite of programs (26, 27). PyMOL was used for all molecular depictions (55).
RESULTS AND DISCUSSION
Protein Crystal Analyses
Crystals of the DPOR (ChlN/ChlB)2 complex belong to the space group P6322 with one half-tetramer (ChlN/ChlB) per asymmetric unit resulting in a Matthews coefficient of 3.3 Å3/Da (solvent content of ∼62%). The crystal packing of (ChlN/ChlB)2 thus incorporates a crystallographic two-fold axis, making the tetramer perfectly symmetric.
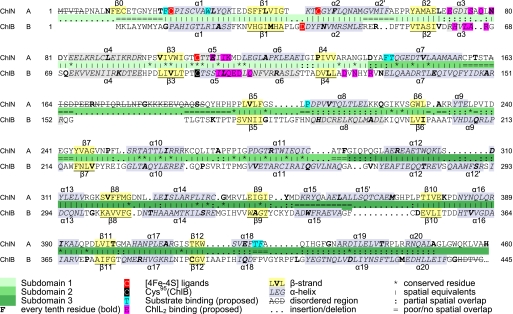
Following the application of selenium-multiwavelength anomalous dispersion phasing and solvent-flattening techniques (see “Experimental Procedures”), RESOLVE was used for automated model building (28). However, phases were clearly of insufficient quality for a complete structure. By overlaying independently phased maps, the RESOLVE model was rebuilt and completed manually in Coot (24). This resulted in a final model consisting of 865 residues in ChlN and ChlB (Fig. 2). Overall, the structure is well defined, apart from a disordered loop of ChlN (residues 163–191) as well as 5 N-terminal and 1 C-terminal residues of ChlN and 65 C-terminal residues of ChlB that are not discernable in the electron density (Fig. 3).
FIGURE 2.
Structure of the (ChlN/ChlB)2 heterotetramer of DPOR. A ribbon diagram of the (ChlN/ChlB)2 structure in two mutually perpendicular views, with subunits ChlN and ChlB, respectively, depicted in green and orange is shown. Note that the asymmetric unit contains only one ChlN/ChlB half-tetramer, the heterotetramer involving a crystallographic two-fold rotation axis indicated by appropriate spindle and dashed line. The total accessible surface area between ChlN and ChlB in the heterodimer is 2485 Å2, whereas those between ChlB/ChlB′ and ChlN/ChlB′ are 1560 and 1074 Å2. ChlN and ChlN′ do not share a common interface. The [4Fe-4S] centers are indicated by clusters of red spheres.
FIGURE 3.
Structure-based sequence alignment of ChlN/ChlB. The structure-based amino acid sequence alignment of ChlN and ChlB results in 366 aligned residues and a sequence identity of 13.7%. α-Helices and β-strands are marked as α1–α20 and β1–β12, and the three subdomains are marked by progressively darker shades of green. Ligands coordinating the [4Fe-4S] cluster (Cys22, Cys47, and Cys107 of ChlN and Asp36 of ChlB) are indicated by red shading. Cys95 of ChlB involved in stabilizing the cluster is marked in black. Residues proposed to be involved in substrate and ChlL2 binding are indicated by cyan and magenta boxes.
Structure of the DPOR (ChlN/ChlB)2 Heterotetramer
The ChlN/ChlB interface within the asymmetric unit is quite extensive at 2485 Å2. The interaction surface of ChlB with its symmetry-related counterpart covers 1560 Å2, whereas that between ChlN and the second ChlB subunit of the tetramer is 1074 Å2.
ChlN and ChlB share a related tertiary structure. The (ChlN/ChlB)2 heterotetramer would thus appear to have evolved from a symmetric tetrameric or even dimeric predecessor. Superimposing the two subunits results in a root mean square deviation of 3.3 Å for 336 aligned residues, whereas the sequence identity based on structural sequence alignments is 13.7% (24).
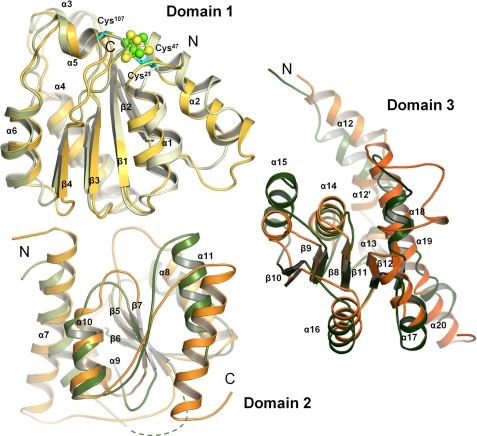
ChlN and ChlB each essentially consist of three compact subdomains each bearing a central, parallel β-sheet surrounded by α-helices (Fig. 4). The first subdomain is built around a four-stranded, parallel β-sheet covered by α-helices on either side. In ChlN, this domain (residues 21–139) provides all 3 cysteine ligands for the coordination of the [4Fe-4S] cluster. The same domain of ChlB (residues 10–127) provides the fourth aspartate ligand (Asp36, see below) to the [4Fe-4S] cluster. The strand order of the β-sheet is 2-1-3-4, reminiscent of a C-terminally deleted Rossmann fold. The similarity of these two domains from alternate chains is underscored by a root mean square deviation (r.m.s.d.) of 1.76 Å for 89 Cα atoms despite their asymmetric roles in FeS cluster coordination. The second compact domain is built around a three-stranded parallel β-sheet (strand order 2-1-3). In both ChlN (residues 140–289) and ChlB (residues 128–270), this domain appears to largely have a structural role in orienting the other two domains. It is neither involved in interdimer contacts nor in forming the tetramer. Correspondingly, some significant differences in secondary structure are noticeable. Thus both the first and the last α-helices of ChlN extend outward by an additional turn when compared with ChlB. A 40-residue insertion in ChlB following the N-terminal α-helix of this domain is disordered in the current crystal structure, and its precise role thus remains unclear. The third domain comprises residues 290–450 of ChlN and residues 271–494 of ChlB. It consists of five parallel β-strands covered by five α-helices. ChlN contains an additional sixth α-helical element (residues 361–370). Two alternative regions in this third domain appear to be involved in creating the active site channel (see below). An N-terminal stretch preceding the first subdomain of ChlN(residues 5–20) and ChlB(residues 1–10) associates most closely with the third, C-terminal domain. Although the association is not particularly pronounced in ChlB, it includes a β-strand in ChlN that extends the five-stranded β-sheet of the third domain of ChlN by a sixth parallel strand.
FIGURE 4.
Comparison of subdomains of ChlN and ChlB. ChlN (shades of green) and ChlB (yellow to orange) each consist of three similar subdomains. Each subunit bears a central, parallel β-sheet surrounded by α-helices. The first subdomain of ChlN and ChlB serves to coordinate the [4Fe-4S] and is involved in ChlL2 binding. The second subdomain appears to have a largely structural role in positioning the remaining two domains but may also be involved in substrate recognition. The third subdomain of ChlN and ChlB is involved in forming the active site channel and in substrate recognition.
Iron-Sulfur Cluster
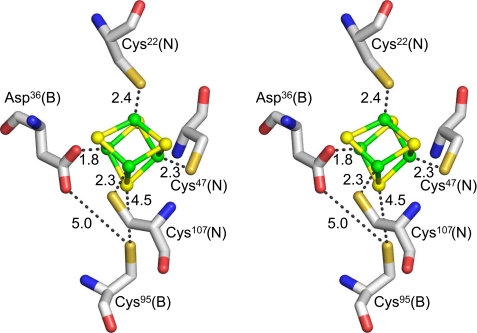
Each (ChlN/ChlB) half-tetramer binds a [4Fe-4S] cluster at the interface of ChlN/ChlB (Fig. 5). Overall, the cluster is four-fold-coordinated. Three coordinating cysteine ligands, Cys22, Cys47 and Cys107, are provided by ChlN, whereas ChlB provides a single unusual aspartate ligand, Asp36, to coordinate the fourth iron of the cluster. Coordination distances between sulfur and iron are between 2.3 and 2.5 Å, whereas that between oxygen and iron is 1.8 Å. Again this is a standard distance for oxygen-coordinated iron; however, an aspartate as a ligand for a [4Fe-4S] cluster is rather unusual.
FIGURE 5.
Coordination of the [4Fe-4S] cluster. The crystal structure reveals one intersubunit [4Fe-4S] cluster per heterodimer in the asymmetric unit or two [4Fe-4S] clusters per (ChlN/ChlB)2 complex. Residues coordinating the cluster are 3 cysteine residues (Cys22, Cys47, Cys107) of subunit ChlN and a unique aspartate residue (Asp36) of subunit ChlB. Coordinating distances are indicated in Angstroms.
In fact, [4Fe-4S] clusters have long been known to be coordinated by aspartate residues in some bacterial-type ferredoxins (29, 30), supported by NMR studies (31, 32). In addition, it could be shown that cysteine can functionally be substituted by aspartate to coordinate [4Fe-4S] clusters (33, 34). However, no structural model demonstrating the coordination of iron of a [4Fe-4S] cluster by aspartate is currently available through the Protein Data Bank. In fact, the only crystal structures showing the coordination of a [4Fe-4S] cluster by an oxygen include those of aconitase (ligand either a hydroxyl or a substrate oxygen), dihydropyrimidine dehydrogenase (glutamine ligand) (35), and the recently described radical SAM enzymes in which the amide and carboxylate group of the methionine moiety directly coordinate the fourth iron of a [4Fe-4S] cluster (36–38). Although the coordination of iron by nitrogen does not appear to destabilize [4Fe-4S] clusters significantly, as witnessed by the crystal structures of 4-hydroxybutyryl-CoA dehydratase (39), nitrate reductase A (40), and ethylbenzene dehydrogenase (41), to name but a few, coordination by oxygen appears to have a dramatically destabilizing effect, rendering these clusters highly sensitive to molecular oxygen. In fact, it has been speculated that some crystal structures bearing [3Fe-4S] clusters, all of which are exclusively coordinated through cysteines, may represent oxidized [4Fe-4S] clusters in which the fourth ligand may be oxygen (42).
The identity of the 3 cysteines of ChlN coordinating the [4Fe-4S] cluster at the ChlN/ChlB interface confirms our earlier mutational analysis study that had implicated these cysteines in cluster coordination in DPOR from Chlorobaculum tepidum (13). By contrast, we did not identify the involvement of an aspartate residue in cluster coordination through EPR spectroscopy (14). Instead, we observed the highly conserved residue Cys95 of ChlB (T. elongatus numbering) to be crucial for sustained DPOR activity and [4Fe-4S] cluster formation. In the crystal structure, Cys95 is located near the [4Fe-4S] center with its sulfur atom 4.5 Å from the closest sulfur atom of the cluster (Fig. 5). At 5.0 Å, it is also in van der Waals contact with the side-chain oxygen of the iron ligand Asp36 as well as other neighboring oxygen atoms (3.1 Å to ChlN-Thr46-Oγ1, 3.2 Å to ChlN-Thr44-Oγ1, 4.1 Å to ChlB-Thr96-Oγ1). Consequently, the observed inactivation of DPOR as a result of an exchange of Cys95 with serine or alanine might be due to destabilization of the [4Fe-4S] cluster environment (13).
Interestingly, Asp36 (ChlB) is an approximate structural counterpart of cluster ligand Cys47 (ChlN), whereas Cys95 of ChlB, involved in the stabilization of Asp36, is the structural equivalent of cluster ligand Cys107 of ChlN (Fig. 3; Table 2). This symmetry in residues involved in cluster coordination in the structurally related subunits would indicate that the observed [4Fe-4S] cluster is the remnant of a larger symmetrical cluster of an ancestral homodimeric protein complex, as has survived inter alia in the structurally related heterodimeric nitrogenase complex (see below).
TABLE 2.
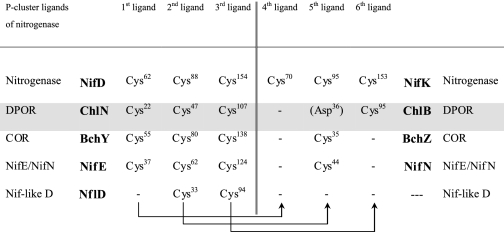
Conservation of nitrogenase MoFe-protein P-cluster ligands in nitrogenase-like enzymes
Residues are numbered according to: NifD, NifK, NifE, NifN from A. vinelandii, ChlN, ChlB from T. elongatus, BchY, BchZ from R. sphaeroides, NflD from M. jannaschii. Arrows indicate equivalent residues within the (hetero)dimers. Structural superposition confirms the equivalence for ChlN/ChlB and NifD/NifK.
Modeling of the Substrate Binding Site of (ChlN/ChlB)2
DPOR from P. marinus was recently described to bind essentially stoichiometric amounts of Pchlide (14). Co-crystallization and soaking experiments for the T. elongatus protein, although resulting in dark green DPOR crystals, did not yield (ChlN/ChlB)2 crystals with specifically bound substrate. Binding experiments with T. elongatus DPOR indicate that nonspecific binding of the natural substrate might influence this type of experiment (data not shown).
Nevertheless, the sheer physical size of the substrate Pchlide requires an appropriately sized binding cavity. Analyzing the (ChlN/ChlB)2 tetramer for voids ideally linked to the molecular surface identifies a single prominent invagination located at the interface of the two ChlN/ChlB half-tetramers (Fig. 6). Residues from one ChlN and both ChlB chains of the complex participate in creating this tunnel. In particular, mainly α-helices line the channel. Residues involved thus emanate from subdomains 1 and 3 of both ChlN and ChlB as well as from subdomain 3 of the second ChlB in the complex.
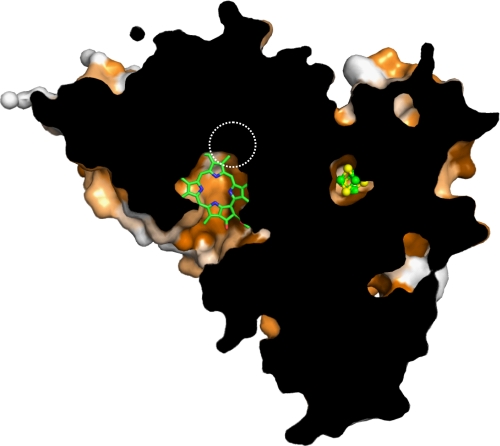
FIGURE 6.
Cut-away of the proposed binding site of DPOR. The surface representation of the (ChlN/ChlB)2 complex was cut away to reveal one of the two equivalent proposed substrate binding sites and its spatial positioning relative to the iron-sulfur cluster. A substrate molecule has been included for purely qualitative purposes; only a co-crystal structure would provide all details of interaction with a high degree of reliability. Conserved residues are indicated by shades of orange (dark orange, conserved; white, not conserved). Note that most residues in the immediate vicinity of the cluster and of the proposed binding site are highly conserved. The active site entrance is created by residues from ChlN as well as both symmetry-related ChlB subunits. The approximate position of the MoFe cofactor in the related nitrogenase structure is indicated by a dashed circle.
The tip of the proposed binding pocket is significantly enriched in aromatic and hydrophobic residues from ChlN that would appear ideal in orienting the substrate. In particular, Trp29 and Phe423 of ChlN could sandwich the substrate from below and above through π-π-stacking interactions, whereas Phe21, Phe143, and Phe421 surround it laterally. Other hydrophobic interactions would include Pro206 and Thr422 of ChlN. Manually placing the substrate into this pocket results in an edge-to-edge distance of ∼14 Å between iron-sulfur cluster and substrate, compatible with rapid electron transfer from the one to the other. The postulated substrate channel supports the results of a previous biochemical study in which 19 Pchlide analogs were tested as DPOR substrates, indicating the active site to cover large parts of the Pchlide molecule. All individual pyrrole rings A, B, C, and D were shown to be relevant for the specific substrate binding of DPOR (14). The current, theoretical model concurs with these data in the sense that the size of the binding pocket would ensure tight binding of the substrate, significantly limiting the possible modifications at any position.
Structural Similarity of DPOR and Nitrogenase
Sequence-based analyses of the DPOR subunits ChlN and ChlB have indicated a low but significant relationship to the subunits NifD and NifK of tetrameric nitrogenase MoFe protein (43), with an amino acid sequence identity between ChlN and NifD of ∼20% and between ChlB and NifK of ∼17%. Both DPOR and the nitrogenase serve to chemically reduce their specific substrates. However, the size of the substrates differs appreciably with nitrogenase reducing a small dinitrogen molecule, whereas DPOR reduces the large Pchlide molecule producing chlorophyllide.
Comparing the quaternary structure of nitrogenase (44, 45) to that of DPOR reveals that both form equivalent heterotetramers (or similar dimers of heterodimers) in which ChlN is equivalent to NifD and ChlB matches NifK. Despite a low sequence identity, a DALI search (46) clearly identifies the nearest structural neighbor of ChlB to be NifK of nitrogenase from Clostridium pasteurianum (Protein Data Bank (PDB) code 1MIO, chain D). A DALI Z-score of 34.0 and an r.m.s.d. of 2.8 Å for 408 Cα atoms confirm the structures to indeed be remarkably similar. Interestingly, an equivalent search for ChlN also identifies chain D (NifK) of 1MIO as the closest structural neighbor (a Z-score of 29.3 and an r.m.s.d. of 3.6 Å for 383 Cα atoms) instead of the structural equivalent NifD (a Z-score of 24.1 and an r.m.s.d. of 3.7 again for 383 comparable Cα atoms). Overall, these values indicate that ChlN and ChlB are both more similar to NifK than either is to NifD. This observation could imply that both DPOR and nitrogenase derive from a more highly symmetric, possibly homotetrameric progenitor.
As implied by the structural comparisons above, the overall fold and subdomain structure of ChlN and ChlB is equivalent to that of NifK and NifD. The crystal structure of DPOR nevertheless reveals distinct features not observed in nitrogenase. 1) The N-terminal extension of nitrogenase NifD extends up to residue Arg51 and is thus slightly longer than the 20 residues in ChlN. However, although it also contains a β-strand that complements the β-sheet of domain three, the arrangement is antiparallel in NifD in contrast to the parallel arrangement in ChlN. 2) In domain one of nitrogenase NifD, an insertion of ∼16 residues creates a loop that fills the active site cleft of DPOR ChlN. 3) In ChlN a short loop of only 4 amino acids connects β-strand 10 to α-helix 16. In NifD, instead, an insertion of 55 residues (His371 to Ser426) creates an extended loop with little secondary structure that wraps around domain 2.
Iron-Sulfur Cluster Coordination in DPOR and Nitrogenase
Both DPOR and nitrogenase bind iron sulfur cofactors. However, although DPOR binds two symmetrically positioned, intersubunit [4Fe-4S] clusters, nitrogenase binds two intersubunit [8Fe-7S] clusters referred to as P-clusters and two iron-molybdenum cofactors (MoFe cofactor) (47, 48). The MoFe cofactor is without equivalent in DPOR, but it partly overlaps the proposed substrate binding pocket (Fig. 6). Whereas the [4Fe-4S] cluster is coordinated by 3 cysteines (ChlN) and an aspartate (ChlB), the P-cluster of nitrogenase is symmetrically coordinated by 3 cysteine residues from NifD (Cys62, Cys88 Cys154, numbering from Azotobacter vinelandii) and three from NifK (Cys70, Cys95, Cys153) (47, 48). Of these, Cys62, Cys88 and Cys154 of NifD correspond spatially and by sequence to Cys22, Cys47, and Cys107 of ChlN from DPOR, making half the P-cluster equivalent to the [4Fe-4S] cluster in DPOR. Of the P-cluster ligands of NifK, only Cys153 has a direct equivalent in ChlB in the form of Cys95 that appears to have a role in stabilizing the [4Fe-4S] cluster rather than direct coordination (Table 1 and above). Cys95 of nitrogenase NifK to some degree matches Asp36 of DPOR ChlB, the fourth iron ligand of the [4Fe-4S] cluster. An equivalent of Cys70 of NifK is, however, not conserved within ChlB.
The asymmetry of the [4Fe-4S] cluster position in DPOR when compared with the pseudo symmetry of the dicubane [8Fe-7S] cluster of nitrogenase once more supports the idea of both enzymes deriving from a common symmetrical homotetramer (or even a homodimer) that may have borne a symmetric P-cluster-like arrangement of two [4Fe-4S] clusters sharing a central sulfur atom. The two symmetrically positioned MoFe cofactors of nitrogenase, coordinated by Cys275 and His442 of NifD, are thought to bind dinitrogen (47, 48). Although DPOR does not retain an equivalent cofactor, the MoFe binding site of NifD is structurally within 6–7 Å (center-to-center) of the proposed Pchlide binding pocket of ChlN. In fact, due to the large size of Pchlide, it laterally partly overlaps with the position of the MoFe cofactor of nitrogenase, indicating that the substrate binding site is conserved in this enzyme family.
Comparison of (ChN/ChlB)2 to Other DPOR-like Enzymes
Apart from nitrogenase, other iron-sulfur cofactor binding enzymes share significant sequence similarities with DPOR. The structure of DPOR thus allows important conclusions to be drawn for these enzymes (Fig. 6).
COR, (BchY/BchZ)2
The chlorophyllide oxidoreductase COR catalyzes the enzymatic reaction following that of DPOR in the biosynthetic pathway of bacteriochlorophyll. In analogy to DPOR, COR facilitates the two-electron reduction of the C7=C8 double bond of Chlide to yield bacteriochlorophyllide. The catalytic mechanism presumably closely resembles that of DPOR (17). The catalytic subcomplex of COR is composed of a (BchY/BchZ)2 heterotetramer homologous to the DPOR (ChlN/ChlB)2 complex and purportedly bearing two equivalent [4Fe-4S] clusters (17). The overall amino acid sequence identity of ChlN/ChlB to BchY/BchZ is in the range of 22%. The similarity is thus more pronounced than that of DPOR and nitrogenase and might reflect a more recent divergence of these two enzymes from a precursor that would potentially have catalyzed both reactions (49).
The tertiary and quaternary structure of COR may thus be expected to be very similar to that of (ChlN/ChlB)2. In particular, the structural similarity of the substrates between DPOR and COR would imply that the substrate binding pocket in subunit BchY would largely be conserved and match that of ChlN. Differences in the details of substrate recognition are to be expected to ensure discrimination between protochlorophyllide and chlorophyllide by both enzymes.
As expected, the three iron-sulfur cluster coordinating cysteine residues of ChlN are conserved within BchY. Asp36 of ChlB by contrast aligns with a conserved cysteine (Cys35 R. sphaeroides) of BchZ, indicating that the unusual ligation of a [4Fe-4S] cluster by aspartate is a unique feature of DPOR. Cys95 of ChlB is without counterpart in BchZ (Table 2), reflecting the fact that the stabilization of an unconventional iron-sulfur cluster ligand is not required in this case. Based on these observations, Cys35 of BchZ would appear to be the uncontested fourth ligand of an asymmetrical [4Fe-4S] cluster of BchY/BchZ.
NifE/NifN
Another enzyme of the nitrogenase/DPOR family is the tetrameric (NifE/NifN)2 complex that serves as scaffold during nitrogenase cofactor assembly and maturation. The sequence identity of NifE and NifN relative to ChlN and ChlB is in the order of 17% (50, 51). Again a quaternary “dimer of heterodimers” arrangement of subunits is to be expected. Subunit NifE bears 3 conserved cysteine residues (Cys37, Cys62, Cys125 A. vinelandii numbering) that align with the cluster ligands of ChlN. Similar to COR and in contrast to ChlB, NifN has a highly conserved Cys44 in place of Asp36 of ChlB, whereas Cys95 of ChlB is not conserved in NifN. These findings suggest that the [4Fe-4S] cluster of NifE/NifN is asymmetrically coordinated by 3 cysteine ligands of NifE and one cysteine ligand of NifN, analogously to COR.
NflD
Another potential member of the nitrogenase/DPOR family is NflD, an as yet largely uncharacterized enzyme proposed to be involved in cofactor F430 biosynthesis in some methanogens such as Methanococcus jannaschii (52). NflD has been proposed to form an NflD2 homodimer and to contain an intrasubunit iron-sulfur cluster. In contrast to all other enzymes of this family, NflD bears only 2 conserved cysteine residues (Cys33 and Cys94 M. jannaschii) that align with cysteine ligands Cys47 and Cys107 of ChlN (Table 2). Cys22 of ChlN is without equivalent in NflD. If NflD indeed forms a homodimer, the retention of only two cysteine ligands could suggest that it may symmetrically coordinate a [4Fe-4S] cluster at the subunit interface, deviating significantly from all DPOR-like members of the group as well as nitrogenase. The plasticity of the protein fold as witnessed by the coordination of a [4Fe-4S] cluster by DPOR but a [8Fe-7S] cluster by nitrogenase would, however, presumably also accommodate a symmetrical [4Fe-4S] cluster.
Clearly, a symmetrical NflD dimer significantly widens the debate on the evolution of the iron-sulfur cofactors of this enzyme family. Did the (presumably homodimeric) common ancestor of this family bear a symmetrically coordinated [4Fe-4S] cluster? Would the next step then have been a homodimeric complex bearing a symmetric [8Fe-7S] nitrogenase P-like cluster from which a homotetrameric or heterodimeric intermediate would have led to the extant group of heterotetrameric family of enzymes? Overall, DPOR appears to be unique within the family of nitrogenase/DPOR enzymes in using an aspartate as a fourth [4Fe-4S] cluster ligand.
Functional, Ternary Complex of DPOR
The initial steps of the ATP-dependent reduction of Pchlide as catalyzed by DPOR shows some resemblance to nitrogenase catalysis. The transient, ternary complex (ChlL2/ChlN/ChlB)2 analogous to the nitrogenase (NifH2/NifD/NifK)2 was recently trapped by replacing ATP by its non-hydrolysable analogue AMPPNP (15).
To investigate the factors affecting complex formation, heterologous DPOR complexes with subunits deriving from T. elongatus, P. marinus, and C. tepidum were tested for their ability to reduce the substrate Pchlide (17). Of the six possible combinations, five resulted in significant activity. Interestingly, combining (ChlN/ChlB)2 from a range of organisms with BchX2 (the ChlL2 analogue of COR) from C. tepidum and Roseobacter denitrificans resulted in chimeric enzymes still able to support Pchlide reduction (17). Perhaps surprisingly, protein-protein recognition has thus been preserved during the divergent evolution of the (bacterio)chlorophyll biosynthetic enzymes DPOR and COR, which raises the question as to the need of the distinct entities ChlL2 and BchX2.
Combining the available crystal structures of ternary nitrogenase (NifH2/NifD/NifK)2 complexes with sequence alignments of the DPOR subunits, we previously identified amino acid residues potentially located at the interface of ChlL2 and (ChlN/ChlB)2 (17). Substituting these residues by suitably chosen alternatives revealed Tyr127 of ChlL2, Leu70, Val107 and Lys109 of ChlN, and both Gly66 and Gln101 of ChlB (P. marinus numbering) to be crucial for the mutual recognition of the subcomplexes and for electron transfer (17).
Based on the overall structural sequence identity of 33% between ChlL and NifH (1, 43), we previously generated a structural model of the C. tepidum BchL2 dimer (13). A recent 1.6 Å crystal structure of the (R. sphaeroides BchL2 dimer clearly confirms the structural similarity to NifH2 (16). We have combined this crystal structure, sharing a 65% sequence identity with ChlL2 from T. elongatus, with the (ChlN/ChlB)2 complex presented in this study to create a qualitative model of the (ChlN/ChlB)2(ChlL2)2 ternary complex (Fig. 7). Indeed the contact surface between the subcomplexes ChlL2 and (ChlN/ChlB)2 is remarkably similar to the corresponding nitrogenase complex.
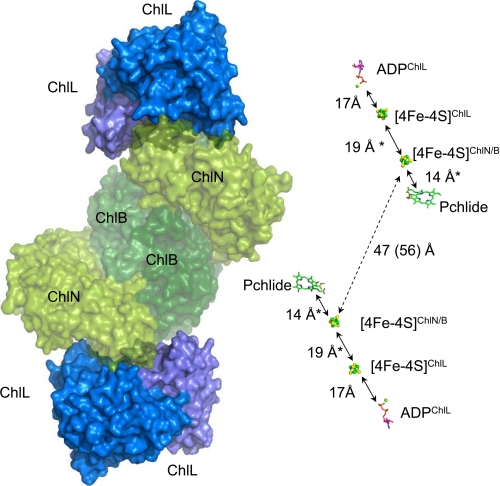
FIGURE 7.
Model of the complete hetero-octameric DPOR and distance between cofactors. Left, a qualitative, theoretical model of the complete DPOR was created by combining the heterotetrameric (ChlN/ChlB)2 core complex from T. elongatus (this study) and ChlL2 from R. sphaeroides (PDB code 3FWY) (16), based on the crystal structure of hetero-octameric nitrogenase (PDB code 1G21) (54). ChlN (ChlN′) and ChlB (ChlB′) are shown in shades of green (partly transparent), ChlL2 is shown in shades of blue. Note the good surface complementarity between the subcomplexes. Right, the cofactors and the substrate of DPOR derived from the models on the left and Fig. 5 are shown in stick or ball-and-stick representation. Edge-to-edge distances are indicated in Angstroms, and distances from theoretical models are indicated by asterisks.
This model confirms the crucial role of Tyr98 (T. elongatus numbering, Uniprot entry Q8DGH0; −11 residues relative to NP_683137.1) of ChlL2 for complex formation. Other residues in intersubunit recognition include the preceding residues Pro90, Gly92, Gly94, Cys95 as well as Asp62, Phe63, His64, all highly conserved within ChlL. In the second subunit, the interactions appear not as tight, with more poorly conserved residues being involved in binding. These include Arg168. The edge-to-edge distance between the two [4Fe-4S] clusters of ChlL2 and (ChlN/ChlB)2 derived from this model is ∼19 Å (Fig. 7). This distance is sufficiently close to allow for rapid electron transfer between the two subcomplexes.
For the two-electron reduction of the Pchlide molecule, two consecutive electron transfer steps are required. The present crystal structure reveals an edge-to-edge distance of 56 Å for the two independent [4Fe-4S] clusters of the (ChlN/ChlB)2 complex, whereas the two Pchlide molecules are located at a distance of ∼47 Å. From these data, we infer that the electron transfer processes occur independently for the two (ChlN/ChlB) half-tetramers (Fig. 7).
Interaction of ChlL2 with one ChlN/ChlB half-tetramer places the respective [4Fe-4S] clusters in a minimal distance of 19 Å. Following a nucleotide-dependent switch mechanism, ChlL2 communicates binding and hydrolysis of ATP over a distance of 17 Å to its [4Fe-4S] cluster and induces the electron transfer to the [4Fe-4S] cluster at the ChlN/ChlB interface (15). Subsequently, electrons are further transferred to the Pchlide molecule over a distance of 14 Å.
Theoretical considerations based on Markus theory would imply a microsecond timescale for an electron transfer over a distance of 19 Å (53). Taking into account the observed turnover numbers for DPOR catalysis (32–45 s−1) under in vitro assay conditions, it becomes apparent that electron transfer would not be rate-limiting. Instead, conformational changes associated with ATP hydrolysis as well as association/dissociation of the different subcomplexes could instead limit the overall rate of the system.
The two-electron transfer pathways deduced from the octameric model demonstrate that DPOR comprises two catalytic entities. However, it is not currently clear whether synchronization of electron transfer processes and dynamic protein-protein interaction of those entities is required for catalytic activity.
Supplementary Material
Acknowledgment
We gratefully acknowledge beam title at ID29, European Synchrotron Radiation Facility, Grenoble, France and Proxima 1. Soleil, St. Aubin, France.
This work was supported by grants from the Deutsche Forschungsgemeinschaft.
This article was selected as a Paper of the Week.
The atomic coordinates and structure factors (code 2XDQ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- Pchlide
- protochlorophyllide
- Chlide
- chlorophyllide
- COR
- chlorophyllide oxidoreductase
- DPOR
- dark operative protochlorophyllide oxidoreductase
- r.m.s.d.
- root mean square deviation
- AMPPNP
- 5′-adenylyl-β,γ-imidodiphosphate
- LPOR
- light-dependent protochlorophyllide oxidoreductase.
REFERENCES
- 1.Burke D. H., Hearst J. E., Sidow A. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 7134–7138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beale S. I. (1999) Photosynth. Res. 60, 43–73 [Google Scholar]
- 3.Apel K. (2001) in Regulation of Photosynthesis (Aro E. M., Anderson B. ed) Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 4.Fujita Y. (1996) Plant Cell Physiol. 37, 411–421 [DOI] [PubMed] [Google Scholar]
- 5.Schoefs B. (2001) Photosynth Res. 70, 257–271 [DOI] [PubMed] [Google Scholar]
- 6.Belyaeva O. B., Griffiths W. T., Kovalev J. V., Timofeev K. N., Litvin F. F. (2001) Biochemistry 66, 173–177 [DOI] [PubMed] [Google Scholar]
- 7.Heyes D. J., Hunter C. N., van Stokkum I. H., van Grondelle R., Groot M. L. (2003) Nat. Struct. Biol. 10, 491–492 [DOI] [PubMed] [Google Scholar]
- 8.Heyes D. J., Ruban A. V., Wilks H. M., Hunter C. N. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11145–11150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rüdiger W. (2003) in Porphyrin Handbook, Chlorophylls and Bilins: Biosynthesis, Synthesis, and Degradation (Kadish K. M., Smith K. M., Guilard R. eds) pp. 71–108, Academic Press, New York [Google Scholar]
- 10.Masuda T., Takamiya K. (2004) Photosynth. Res. 81, 1–29 [DOI] [PubMed] [Google Scholar]
- 11.Suzuki J. Y., Bollivar D. W., Bauer C. E. (1997) Annu. Rev. Genet. 31, 61–89 [DOI] [PubMed] [Google Scholar]
- 12.Bollivar D. W., Suzuki J. Y., Beatty J. T., Dobrowolski J. M., Bauer C. E. (1994) J. Mol. Biol. 237, 622–640 [DOI] [PubMed] [Google Scholar]
- 13.Bröcker M. J., Virus S., Ganskow S., Heathcote P., Heinz D. W., Schubert W. D., Jahn D., Moser J. (2008) J. Biol. Chem. 283, 10559–10567 [DOI] [PubMed] [Google Scholar]
- 14.Bröcker M. J., Wätzlich D., Uliczka F., Virus S., Saggu M., Lendzian F., Scheer H., Rüdiger W., Moser J., Jahn D. (2008) J. Biol. Chem. 283, 29873–29881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bröcker M. J., Wätzlich D., Saggu M., Lendzian F., Moser J., Jahn D. (2010) J. Biol. Chem. 285, 8268–8277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarma R., Barney B. M., Hamilton T. L., Jones A., Seefeldt L. C., Peters J. W. (2008) Biochemistry 47, 13004–13015 [DOI] [PubMed] [Google Scholar]
- 17.Wätzlich D., Bröcker M. J., Uliczka F., Ribbe M., Virus S., Jahn D., Moser J. (2009) J. Biol. Chem. 284, 15530–15540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guerrero S. A., Hecht H. J., Hofmann B., Biebl H., Singh M. (2001) Appl. Microbiol. Biotechnol. 56, 718–723 [DOI] [PubMed] [Google Scholar]
- 19.Minor W., Cymborowski M., Otwinowski Z. (2002) Acta Physica Polonica 101, 613–619 [Google Scholar]
- 20.Kabsch W. (2010) Acta Cryst. D66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kabsch W. (2010) Acta Cryst. D66, 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheldrick G. M. (2008) Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 23.Cowtan K. (1994) Joint CCP4 ESF-EACBM Newsletter 31, 34–38 [Google Scholar]
- 24.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 25.Murshudov G. N., Vagin A. A., Lebedev A., Wilson K. S., Dodson E. J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 247–255 [DOI] [PubMed] [Google Scholar]
- 26.Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 27.Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G. N., Cohen S., Perrakis A., Noble M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2288–2294 [DOI] [PubMed] [Google Scholar]
- 28.Terwilliger T. C. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moura J. J., Moura I., Kent T. A., Lipscomb J. D., Huynh B. H., LeGall J., Xavier A. V., Münck E. (1982) J. Biol. Chem. 257, 6259–6267 [PubMed] [Google Scholar]
- 30.Busch J. L., Breton J. L., Bartlett B. M., Armstrong F. A., James R., Thomson A. J. (1997) Biochem. J. 323, 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sham S., Calzolai L., Wang P. L., Bren K., Haarklau H., Brereton P. S., Adams M. W., La Mar G. N. (2002) Biochemistry 41, 12498–12508 [DOI] [PubMed] [Google Scholar]
- 32.Calzolai L., Gorst C. M., Zhao Z. H., Teng Q., Adams M. W., La Mar G. N. (1995) Biochemistry 34, 11373–11384 [DOI] [PubMed] [Google Scholar]
- 33.Zhao J., Li N., Warren P. V., Golbeck J. H., Bryant D. A. (1992) Biochemistry 31, 5093–5099 [DOI] [PubMed] [Google Scholar]
- 34.Mannan R. M., He W. Z., Metzger S. U., Whitmarsh J., Malkin R., Pakrasi H. B. (1996) EMBO J. 15, 1826–1833 [PMC free article] [PubMed] [Google Scholar]
- 35.Dobritzsch D., Schneider G., Schnackerz K. D., Lindqvist Y. (2001) EMBO J. 20, 650–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sofia H. J., Chen G., Hetzler B. G., Reyes-Spindola J. F., Miller N. E. (2001) Nucleic Acids Res. 29, 1097–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Layer G., Kervio E., Morlock G., Heinz D. W., Jahn D., Retey J., Schubert W. D. (2005) Biol. Chem. 386, 971–980 [DOI] [PubMed] [Google Scholar]
- 38.Frey P. A., Hegeman A. D., Ruzicka F. J. (2008) Crit. Rev. Biochem. Mol. Biol. 43, 63–88 [DOI] [PubMed] [Google Scholar]
- 39.Dobbek H., Svetlitchnyi V., Liss J., Meyer O. (2004) J. Am. Chem. Soc. 126, 5382–5387 [DOI] [PubMed] [Google Scholar]
- 40.Bertero M. G., Rothery R. A., Boroumand N., Palak M., Blasco F., Ginet N., Weiner J. H., Strynadka N. C. (2005) J. Biol. Chem. 280, 14836–14843 [DOI] [PubMed] [Google Scholar]
- 41.Kloer D. P., Hagel C., Heider J., Schulz G. E. (2006) Structure 14, 1377–1388 [DOI] [PubMed] [Google Scholar]
- 42.Rousset C., Fontecave M., Ollagnier de Choudens S. (2008) FEBS Lett. 582, 2937–2944 [DOI] [PubMed] [Google Scholar]
- 43.Fujita Y., Matsumoto H., Takahashi Y., Matsubara H. (1993) Plant Cell Physiol. 34, 305–314 [PubMed] [Google Scholar]
- 44.Schindelin H., Kisker C., Schlessman J. L., Howard J. B., Rees D. C. (1997) Nature 387, 370–376 [DOI] [PubMed] [Google Scholar]
- 45.Rees D. C., Akif Tezcan F., Haynes C. A., Walton M. Y., Andrade S., Einsle O., Howard J. B. (2005) Philos. Transact. A Math. Phys. Eng. Sci. 363, 971–984; discussion 1035–1040 [DOI] [PubMed] [Google Scholar]
- 46.Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Howard J. B., Rees D. C. (1994) Annu. Rev. Biochem. 63, 235–264 [DOI] [PubMed] [Google Scholar]
- 48.Igarashi R. Y., Seefeldt L. C. (2003) Crit. Rev. Biochem. Mol. Biol. 38, 351–384 [DOI] [PubMed] [Google Scholar]
- 49.Nomata J., Mizoguchi T., Tamiaki H., Fujita Y. (2006) J. Biol. Chem. 281, 15021–15028 [DOI] [PubMed] [Google Scholar]
- 50.Hu Y., Yoshizawa J. M., Fay A. W., Lee C. C., Wiig J. A., Ribbe M. W. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16962–16966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu Y., Corbett M. C., Fay A. W., Webber J. A., Hodgson K. O., Hedman B., Ribbe M. W. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17119–17124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Staples C. R., Lahiri S., Raymond J., Von Herbulis L., Mukhophadhyay B., Blankenship R. E. (2007) J. Bacteriol. 189, 7392–7398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moser C. C., Keske J. M., Warncke K., Farid R. S., Dutton P. L. (1992) Nature 355, 796–802 [DOI] [PubMed] [Google Scholar]
- 54.Chiu H., Peters J. W., Lanzilotta W. N., Ryle M. J., Seefeldt L. C., Howard J. B., Rees D. C. (2001) Biochemistry 40, 641–650 [DOI] [PubMed] [Google Scholar]
- 55.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.