Abstract
The addition of α2,8-polysialic acid to the N-glycans of the neural cell adhesion molecule, NCAM, is critical for brain development and plays roles in synaptic plasticity, learning and memory, neuronal regeneration, and the growth and invasiveness of cancer cells. Our previous work indicates that the polysialylation of two N-glycans located on the fifth immunoglobulin domain (Ig5) of NCAM requires the presence of specific sequences in the adjacent fibronectin type III repeat (FN1). To understand the relationship of these two domains, we have solved the crystal structure of the NCAM Ig5-FN1 tandem. Unexpectedly, the structure reveals that the sites of Ig5 polysialylation are on the opposite face from the FN1 residues previously found to be critical for N-glycan polysialylation, suggesting that the Ig5-FN1 domain relationship may be flexible and/or that there is flexibility in the placement of Ig5 glycosylation sites for polysialylation. To test the latter possibility, new Ig5 glycosylation sites were engineered and their polysialylation tested. We observed some flexibility in glycosylation site location for polysialylation and demonstrate that the lack of polysialylation of a glycan attached to Asn-423 may be in part related to a lack of terminal processing. The data also suggest that, although the polysialyltransferases do not require the Ig5 domain for NCAM recognition, their ability to engage with this domain is necessary for polysialylation to occur on Ig5 N-glycans.
Keywords: Adhesion, Glycoprotein Biosynthesis, Glycoprotein Structure, Glycosylation, Protein Structure, Fibronectin Repeat, Immunoglobulin Domain, Polysialic Acid
Introduction
In mammalian cells, polysialylation is a protein-specific glycosylation event involving the addition of multiple α2,8-linked sialic acid residues to N- or O-linked glycans. Two enzymes, ST8Sia II and ST8Sia IV (PST),2 catalyze the polysialylation reaction, which takes place in the Golgi apparatus as proteins progress through the secretory pathway (1–4). The polysialyltransferases (polySTs) have a very limited number of substrates, including the neural cell adhesion molecule (NCAM) (5, 6), neuropilin-2 (7), the CD36 scavenger receptor in milk (8), the α subunit of the voltage dependent sodium channel (9), and the enzymes themselves (autopolysialylation) (10, 11). NCAM is by far the most abundant polysialylated protein in mammalian cells. Addition of large, negatively charged, highly hydrated polysialic acid chains generally results in a switch in the properties of NCAM, from being an adhesive protein involved in cell-cell interactions to having a global anti-adhesive function (12–14).
NCAM is heavily polysialylated during embryogenesis and early postnatal development, due to abundant polyST expression (15, 16). Mice completely devoid of all NCAM isoforms have specific defects, including a smaller olfactory bulb, decreased mossy fiber fasciculation, and deficits in spatial learning (17, 18). In contrast to the relatively mild phenotype of NCAM knock-out mice, mice that lack both polySTs and are therefore completely devoid of polysialic acid, demonstrate postnatal growth retardation and severe neurological defects, and usually die within 4 weeks of birth (19). Surprisingly, simultaneous deletion of NCAM and the polySTs results in a similar mild phenotype as the NCAM single knock-out mice. This suggests that the effects observed in polysialic acid-null mice are caused by uncontrolled NCAM adhesion during development (19). In most locations in the adult brain NCAM is not polysialylated (5, 6). However, polysialylated NCAM is detected in specific regions that require continuous cell migration and display synaptic plasticity, such as the olfactory bulb and the hippocampus (20–23). Moreover, polysialylated NCAM is re-expressed in certain cancers, where it promotes tumor cell growth and invasiveness (24–31). In neuroblastoma cells, it has been demonstrated that the presence of polysialic acid negatively regulates heterophilic NCAM signaling events, such as those that involve mitogen-activated protein kinase activation, thereby promoting tumor progression (32).
There are three mammalian NCAM isoforms. NCAM-140 and NCAM-180 are transmembrane proteins that differ in the length of their cytosolic tails, whereas NCAM-120 does not traverse the membrane but is attached to the lipid bilayer via a glycosyl phosphatidylinositol anchor (33). The three isoforms share a common extracellular domain structure, with five immunoglobulin (Ig) domains and two fibronectin type III repeats. NCAM molecules on the cell surface are postulated to engage in cis homophilic as well as trans homophilic interactions, and the cis interactions are predicted to stabilize the trans interactions and therefore cell adhesion (34).
There are six consensus N-linked glycosylation sites within the extracellular domain of NCAM. Of these, polysialic acid is added to the fifth and sixth N-glycans located on Ig5 (35). It has been demonstrated that the polySTs polysialylate N-glycans attached to NCAM far more efficiently than free N-glycans enzymatically released from NCAM, providing evidence of a protein-specific recognition event between NCAM and the polySTs (36, 37). Previous work from our laboratory and others using truncated NCAM proteins demonstrated that the first fibronectin type III repeat (FN1), adjacent to Ig5, is required for polysialylation of N-linked glycans on Ig5 (35, 38). Nelson et al. (35) determined that the minimal structural unit that can be polysialylated was a membrane-bound Ig4-Ig5-FN1 tandem. We subsequently demonstrated that an even more truncated NCAM protein with an extracellular region composed of only the Ig5 and FN1 domains was efficiently polysialylated, and that membrane attachment was not a requirement. Moreover, we found that a protein containing just the Ig5 domain was not polysialylated (38). These results highlighted the importance of the FN1 domain and suggested a model in which the polySTs recognize and bind to FN1 to position themselves for polysialylating the N-glycans on the adjacent Ig5 domain.
To identify specific regions within NCAM FN1 that may be required for enzyme recognition, we modeled and subsequently solved the structure of the FN1 domain and identified a unique surface acidic patch and α helix (39, 40). Mutation of the core residues of the acidic patch, Asp-520, Glu-521, and Glu-523 (numbering includes the signal sequence and no VASE exon), to alanine residues reduced NCAM polysialylation, whereas replacement with arginine residues eliminated polysialylation (39). Replacement of the α helix linking strands five and six of the FN1 β sandwich with two threonine or alanine residues resulted in a shift in the location of polysialic acid from N-glycans on the Ig5 domain to O-glycans on the FN1 domain (40). These and other results suggest that the acidic patch is likely part of a larger recognition region required for initial interaction with the polySTs (39), whereas the α helix may be required to correctly position the N-glycans on Ig5, or the polyST itself, for polysialylation to occur (40).
One possibility is that the α helix mediates an interaction between the Ig5 and FN1 domains and this allows polyST access to Ig5 N-glycans (40). In support of this hypothesis, early rotary shadowing electron microscopy studies demonstrated the presence of a flexible hinge in the extracellular region of NCAM, in the vicinity the polysialic acid attachment site (41, 42). The presence of a flexible hinge was later supported by Leckband and colleagues using direct force measurements (43). These authors suggested that their data were most consistent with the flexible hinge being found between the Ig5 and FN1 domains (43), as originally predicted by Becker et al. (42), whereas others suggested that the flexible hinge was more likely to be between Ig4 and Ig5 (34, 44). However, a recently solved crystal structure of the NCAM FN1–FN2 tandem indicated a large degree of flexibility between these domains, suggesting that the flexible linker demonstrated by the electron microscopy studies might actually be positioned between FN1 and FN2 (45).
In this report, we have evaluated the relationship of the Ig5 and FN1 domains by solving the crystal structure of the bacterially expressed, unglycosylated Ig5-FN1 tandem. The Ig5-FN1 tandem crystallized as a dimer. However, our data suggest that this dimer is not formed in vivo and is prevented by the presence of an N-linked glycan on Asn-423 (ASN4). In the Ig5-FN1 monomer structure we observed that the attachment sites for the two polysialylated Ig5 N-glycans (ASN5 and ASN6) are on the face opposite the FN1 acidic patch and α helix sequences implicated in polyST recognition and positioning. To test the possibility that the Ig5 domain may be able to bend and twist relative to the FN1 domain, we engineered N-glycans at different positions on the surface of the Ig5 domain and tested their ability to be polysialylated by the PST enzyme. Our results indicate some flexibility in the placement of N-glycans for polysialylation, and that the glycan on ASN4 may not be efficiently polysialylated because it is not completely processed to a sialylated, complex form.
EXPERIMENTAL PROCEDURES
Tissue culture reagents, oligonucleotides, restriction enzymes, PCR supermix, and anti-V5 epitope tag antibody were purchased from Invitrogen. The cDNA for human NCAM-140 was a gift from Dr. Nancy Kedersha (Brigham and Women's Hospital, Boston, MA). The cDNA for human ST8Sia IV/PST was obtained from Dr. Minoru Fukuda (Burnham Institute, La Jolla, CA). The QuikChangeTM site-directed mutagenesis kit and Pfu DNA polymerase were purchased from Stratagene. DNA purification kits were purchased from Qiagen. Protein A-Sepharose was purchased from Amersham Biosciences. The cDNA for yeast SUMO and SUMO protease proteins, cloned into pET14b bacterial expression vectors (Novagen) were kind gifts from Dr. Manfred Konrad (Max Planck Institute for Biophysical Chemistry, Göttingen, Germany). Peptide N-glycosidase F (PNGaseF) and T4 DNA ligase were obtained from New England Biolabs. Streptomyces plicatus endoglycosidase H (Endo H) purified from recombinant Escherichia coli, neuraminidase purified from Clostridium perfringens, and neuraminidase purified from Vibrio cholerae were purchased from Roche Applied Science. Phage PK1E endo-N-acylneuraminidase (Endo N) cloned in the pEndo-N expression plasmid was a kind gift from Dr. Eric Vimr (University of Illinois at Champagne-Urbana). Precision Plus ProteinTM Standard was purchased from Bio-Rad. Nitrocellulose membranes were purchased from Schleicher and Schuell. Horseradish peroxidase (HRP)-conjugated secondary antibodies were obtained from Jackson ImmunoResearch. SuperSignal West Pico chemiluminescence reagent was obtained from Pierce. Other chemicals and reagents were purchased from Sigma and Fisher Scientific.
Generation, Purification, and Crystallization of Ig5-FN1 for Structural Studies
The Ig5-FN1 fragment of NCAM-140 was PCR-amplified using the primers 5′-CATATGTATGCCCCAAAGCTACAGGGCCC-3′ and 5′-GGATCCCTATTCCCCTTGGACTGGCTGCGTC-3′. These primers specifically introduced NdeI and BamHI restriction sites at the 5′- and 3′-ends of the cDNA, respectively. The Ig5-FN1 cDNA was cloned into the bacterial expression vector pET14b (Novagen), downstream of the His6 tag, an inserted SUMO sequence, and the thrombin cleavage site. This vector was transformed into E. coli BL21 CodonPlus cells (Stratagene). Cells were grown to an optical density of 0.6–0.9, induced with 1 mm isopropyl-1-thio-β-d-galactopyranoside for 4 h at 37 °C, collected by centrifugation, and lysed by sonication in lysis buffer (50 mm Tris, pH 8.0, 200 mm NaCl, 1 mm phenylmethylsulfonyl fluoride (PMSF), 1% Triton X-100, 20 mm β-mercaptoethanol). The His6-SUMO-Ig5-FN1 protein was incorporated into inclusion bodies, which were solubilized with 6 m guanidine hydrochloride, containing 20 mm β-mercaptoethanol. After centrifugation, the soluble extract was loaded onto a 5-ml HiTrapTM Chelating HP column charged with nickel ions (Amersham Biosciences). After eluting with 500 mm imidazole, the protein was dialyzed in dialysis buffer (50 mm Tris-HCl, pH 8.0, 500 mm NaCl) for 3 days, changing the buffer occasionally. This allowed disulfide bond formation between Cys-426 and Cys-479 within the Ig5 domain. After dialysis, His6-SUMO was cleaved using 1 μg of SUMO protease per 200 μg of protein. The protein was reloaded onto the nickel column, and the flow-through containing Ig5-FN1 protein was collected and concentrated to 4 mg/ml. Ig5-FN1 was further purified on an S-75 gel filtration column (Amersham Biosciences), equilibrated with dialysis buffer. Single, small crystals (10–30 μm by 100–250 μm) were obtained using the hanging drop, vapor-diffusion method, with purified Ig5-FN1 (1 mg/ml) mixed with an equal volume of the reservoir solution (0.2 m ammonium sulfate, 0.1 m Bis-Tris, pH 5.5, 25% polyethylene glycol 3350) and incubated at room temperature.
X-ray Data Collection and Structure Determination and Refinement
X-ray diffraction data to 1.8-Å resolution was collected using the Southeast Regional Collaborative Access Team beamline BM-22 at the Advanced Photon Source, Argonne National Laboratories, and data were processed with XDS (46). Molecular replacement was performed using the programs MOLREP (47) and Phaser (48) and the structure of the NCAM FN1 domain (40) (PDB ID 2HAZ). However, attempts to locate the Ig5 domain using several Ig domain models were not successful. To generate a model of the Ig5 domain, we used the automatic model building program ARP (49), inputting the FN1 domain data, the Ig5-FN1 sequence, and our high resolution diffraction data. This was very successful, yielding a total of 343 of 396 amino acids. Several rebuilding and refinement cycles were subsequently performed using O (50) and REFMAC (51). Structure figures were made using the program PyMOL (52). The root mean square deviation value of the FN1 domain structure in isolation (40) and in the Ig5-FN1 tandem was determined using the program LSQMAN (53).
Construction of Soluble and Membrane-bound Ig5-FN1 for Expression in Mammalian Cells
The Ig5-FN1 fragment of NCAM-140 was amplified using the PCR primers 5′-GATATCTATGCCCCAAAGCTACAGGGC-3′ and 5′-TCTAGAGCTTCCCCTTGGACTGGCTGCGTC-3′. This resulted in EcoRV and XbaI restriction site insertions at the 5′- and 3′-ends of the Ig5-FN1 fragment, respectively. The Ig5-FN1 fragment was ligated in-frame with the pancreatic prepro-insulin signal peptide and transmembrane sequence from a previously generated construct in the pcDNA3.1 V5/HisB expression vector (38). To generate soluble Ig5-FN1 this membrane-bound form was digested with HindIII (upstream of signal peptide sequence) and XbaI (upstream of transmembrane sequence) and cloned into previously digested empty pCDNA3.1 V5/HisB, or pcDNA3.1 Myc/HisB vectors. A stop codon was inserted after the Myc epitope sequence to prevent expression of the His6 tag in the pcDNA3.1 Myc/HisB vector. The EcoRV restriction site was subsequently removed from both soluble and membrane-bound Ig5-FN1, and the XbaI site was removed from the membrane-bound form by mutagenesis PCR.
Mutagenesis of NCAM and Ig5-FN1
NCAM and Ig5-FN1 mutants were constructed using the Stratagene QuikChangeTM site-directed mutagenesis kit. DNA sequencing performed by the DNA Sequencing Facility of the Research Resources Center at the University of Illinois at Chicago confirmed the mutations. Primers used are listed in supplemental Table 1. For the glycosylation site analysis, an NCAM construct with the ASN5 and ASN6 glycosylation sites mutated from N449YS to NYA and N478CT to NCA, respectively, was generated (mut[5.6]NXA). Seven glycosylation sites were individually engineered into the Ig5 domain of mut[5.6]NXA. In three cases, an Asn residue was inserted to create an Asn-X-Ser/Thr consensus sequence (T435N, Q487N, and E465N), in one case the third position was corrected (N452IK to NIS), and in three cases both an Asn and either a Ser or Thr were simultaneously inserted (F439RD to NRS, E491FI to NFT, and Q443LL to NLS). Under “Results and Discussion” all mutants are indicated by the amino acid number of their new or existing Asn residue.
Transfection of COS-1 Cells with NCAM and PST cDNAs
COS-1 cells maintained in Dulbecco's modified Eagle's medium (DMEM), 10% fetal bovine serum (FBS), were plated on 100-mm tissue culture plates and grown at 37 °C, 5% CO2, until 50–70% confluent. Cells were transfected using 30 μl of Lipofectin in 3 ml of Opti-MEM 1, with 10 μg each of V5-tagged NCAM and PST-Myc cDNA, according to the manufacturer's protocol. The NCAM cDNAs were cloned into the pcDNA3.1 V5/HisB vector. PST cDNA was cloned upstream of the Myc tag in the pcDNA3.1 Myc/HisB vector. A stop codon was inserted before the His6 coding sequence in the pcDNA3.1 Myc/HisB vector. Cells were incubated with transfection mixture for 6 h, and then 7 ml of DMEM, 10% FBS, was added to bring the mixture to a final volume of 10 ml.
Immunoprecipitation of NCAM Proteins
Sixteen hours post-transfection, cells were washed with 10 ml of phosphate-buffered saline (PBS) and lysed in 1 ml of immunoprecipitation buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm EDTA, 0.5% Nonidet P-40, 0.1% SDS). Lysates were pre-cleared with 50 μl of protein A-Sepharose beads (50% suspension in PBS) for 1 h at 4 °C. NCAM proteins were immunoprecipitated with 3 μl of anti-V5 epitope tag antibody for 2 h at 4 °C, followed by incubation for 1 h with 50 μl of protein-A-Sepharose beads. Beads were washed four times with immunoprecipitation buffer and once with immunoprecipitation buffer containing 1% SDS. Samples were then resuspended in 50 μl of Laemmli sample buffer containing 5% β-mercaptoethanol, heated at 65 °C for 10 min, and separated on a 3% stacking/5% resolving SDS-polyacrylamide gel. To evaluate relative NCAM protein expression levels, an aliquot of cell lysate was removed prior to immunoprecipitation, and an equal volume of Laemmli sample buffer, 5% β-mercaptoethanol, was added. Samples were boiled at 110 °C for 10 min and separated on a 5% stacking/7.5% resolving SDS-polyacrylamide gel.
Glycosidase Treatment of NCAM and Ig5-FN1 Proteins
To evaluate N-linked polysialylation of NCAM proteins, transfections were performed in duplicate, and, after immunoprecipitation and washing, identical samples were resuspended in 77 μl of dH2O, 10 μl of Nonidet P-40, 10 μl of G7 buffer (0.5 m sodium phosphate, pH 7.5), with or without 3 μl of PNGaseF, and incubated with shaking at 37 °C overnight. 50 μl of Laemmli sample buffer containing 5% β-mercaptoethanol was then added to the samples, which were heated at 65 °C for 10 min, and separated on a 3% stacking/5% resolving SDS-polyacrylamide gel. To evaluate glycosylation of Ig5-FN1, COS-1 cells were transfected with 10 μg of Ig5-FN1-TM-tail or various mutants. Cell lysates were collected and subject to enzyme digestion. To cleave N-linked glycans, 70 μl of lysate was treated with 10 μl of Nonidet P-40, 10 μl of G7 buffer, 3 μl of PNGaseF, and 7 μl of dH2O. To digest α2,3-linked, α2,6-linked, and α2,8-linked sialic acid, 70 μl of lysate was treated with 10 μl of Nonidet P-40, 10 μl of 0.5 m sodium phosphate, pH 7.5, 5 μl of Vibrio cholerae neuraminidase, and 5 μl of Clostridium perfringens neuraminidase. To cleave high mannose oligosaccharides, 70 μl of lysate was treated with 100 milliunits of Endo H in a 100-μl total reaction volume of 50 mm sodium citrate, 0.1 m β-mercaptoethanol, and 0.5 mm PMSF. For untreated samples, 70 μl of lysate was mixed with 30 μl of dH2O. All samples were incubated overnight at 37 °C, with rotation, and then 100 μl of Laemmli sample buffer containing 5% β-mercaptoethanol was added. Samples were heated at 65 °C for 10 min, and separated on a 5% stacking/10% resolving SDS-polyacrylamide gel.
Co-immunoprecipitation Analysis of Soluble NCAM and Soluble Ig5-FN1
COS-1 cells maintained in advanced DMEM, 1% FBS, were transfected with 5 μg of V5-tagged soluble NCAM or soluble Ig5-FN1 and 5 μg of empty pCDNA3.1 Myc/HisB vector, or with 5 μg of both V5- and Myc-tagged soluble NCAM or soluble Ig5-FN1. Following a 6-h incubation, the transfection mixture was removed, and 4 ml of advanced DMEM, 1% FBS, was added to each plate. The next day, cell media was collected, and debris was removed by centrifugation. Myc-tagged constructs were immunoprecipitated from the cell media using 5 μl of polyclonal anti-Myc epitope tag antibody (Abcam Inc.), for 3 h at 4 °C, followed by a 2-h incubation with 75 μl of protein-A-Sepharose beads. Beads were washed four times with co-immunoprecipitation buffer (50 mm Hepes, pH 7.2, 100 mm NaCl, 1% Triton X-100), resuspended in 50 μl of Laemmli sample buffer containing 5% β-mercaptoethanol, and boiled at 110 °C. Soluble NCAM or soluble Ig5-FN1 samples were separated on 5% stacking/10% resolving, or 5% stacking/12.5% resolving SDS-polyacrylamide gels, respectively. To evaluate relative NCAM or Ig5-FN1 protein expression levels, an aliquot of cell media was removed prior to immunoprecipitation, and an equal volume of Laemmli sample buffer, 5% β-mercaptoethanol, was added. Samples were boiled at 110 °C for 10 min, and resolved by SDS-polyacrylamide electrophoresis.
Immunoblot Analysis of NCAM and Ig5-FN1 Proteins and Mutants
Following gel electrophoresis, proteins were transferred to a nitrocellulose membrane at 500 mA overnight. Membranes were blocked for 1 h at room temperature in blocking buffer (5% nonfat dry milk in Tris-buffered saline, pH 8.0, 0.1% Tween 20). To detect polysialic acid, membranes were incubated overnight with a 1:100 dilution of anti-polysialic acid antibody OL.28 in 2% nonfat dry milk in Tris-buffered saline, pH 8.0, and for 1 h with HRP-conjugated goat anti-mouse IgM, diluted 1:4000 in blocking buffer. To test self-association or relative NCAM or Ig5-FN1 expression levels, membranes were incubated for 2 h or overnight with a 1:5000 dilution of anti-V5 or anti-Myc epitope tag antibody (Cell Signaling Technologies) diluted in blocking buffer, and for 1 h with HRP-conjugated goat anti-mouse IgG, diluted 1:4000 in blocking buffer. Membranes were washed with Tris-buffered saline, pH 8.0, 0.1% Tween 20, for 15 min two times, or four times, before and after secondary antibody incubation, respectively. Immunoblots were developed using the SuperSignal West Pico chemiluminescence kit and BioExpress Blue Ultra Autorad film.
Pulse-chase Analysis of NCAM Polysialylation
COS-1 cells were transfected with V5-tagged NCAM or mutants along with PST-Myc as described above. Eighteen hours post-transfection, cell media was removed and 5 ml of Met/Cys-free DMEM added to the plates for 1 h. Cells were then labeled with 100 μCi/ml 35S-Express protein labeling mix (PerkinElmer Life Sciences) diluted in 4 ml of fresh Met/Cys-free DMEM. The labeling mixture was removed after a 1-h incubation, and 4 ml of DMEM, 10% FBS, was added to each plate. Following a 3-h chase, media was removed and cell lysates were collected. NCAM proteins were immunoprecipitated with anti-V5 epitope tag antibody as described above. Following immunoprecipitation, NCAM proteins bound to protein-A Sepharose beads were incubated on ice or subject to Endo N digestion. Endo N cleaves α2,8-linked sialic acid chains of 8 units or higher (54). Beads were incubated for 2 h at 37 °C with a 1:10 dilution of Endo N (30 μg/ml) in 20 mm Tris-HCl, pH 7.4. After enzyme digestion, 50 μl of Laemmli sample buffer containing 5% β-mercaptoethanol was added to the samples, which were then heated at 65 °C for 10 min, and resolved on a 3% stacking/5% separating SDS-PAGE gel. Gels were treated with 10% 2,5-diphenyloxazole in DMSO to visualize radiolabeled proteins by fluorography and exposure to BioExpress Blue Ultra Autorad film. Relative NCAM protein expression levels were evaluated by immunoblotting of aliquots of cell lysate collected prior to immunoprecipitation, as described above.
RESULTS AND DISCUSSION
Previous work from our laboratory has demonstrated the importance of the NCAM FN1 domain in the polysialylation of N-glycans on the adjacent Ig5 domain (38–40). Structural studies led to the identification of an acidic surface patch and α helix that appear to be involved in polyST recognition and positioning, respectively (39, 40). We observed that replacing the α helix switches polysialic acid addition from N-linked glycans on Ig5 to O-linked glycans on FN1, suggesting that the helix may be involved in an Ig5-FN1 interaction that positions the N-glycans for polysialylation (40). To evaluate the relationship between the Ig5 and FN1 domains we solved the crystal structure of the NCAM Ig5-FN1 tandem to 1.8-Å resolution.
The Crystal Structure of the NCAM Ig5-FN1 Tandem
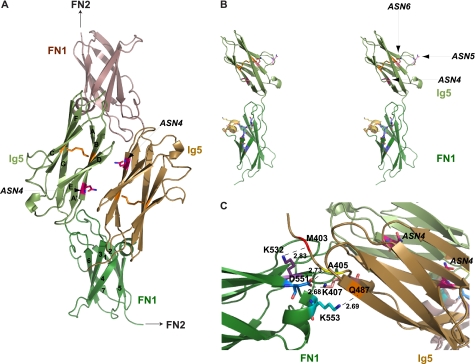
The Ig5-FN1 tandem of NCAM was expressed in E. coli and purified as described under “Experimental Procedures.” For data collection and refinement statistics see Table 1. The crystal structure of Ig5-FN1 is a dimer, with dimerization mediated by interactions between the Ig5 domain of one molecule and the FN1 domain of the other (Fig. 1A). We previously solved the structure of the NCAM FN1 domain (40). However, to our knowledge, this is the first structure of the NCAM Ig5 domain reported. A common feature of the structures of NCAM Ig domains solved to date (34, 55–57) is that they are members of the intermediate set of Ig domains, with characteristics of both constant and variable regions (58). The structure of the Ig5 domain solved in this study suggests it is also of the intermediate type, with strands ABDE and A′CFG forming the first and second sheets, respectively, of the β sandwich (Fig. 1A). The two β sheets are connected by a disulfide bond between Cys-426 on strand B and Cys-479 on strand F. A short helical turn links strands E and F. This domain has three consensus Asn glycosylation sites. Asn-423 (ASN4) is on strand B on one sheet, Asn-449 (ASN5) is situated in a loop between strands C and D, and Asn-478 (ASN6) is on strand F on the second sheet (Fig. 1B). ASN5 and ASN6 would carry the glycans that become polysialylated. The FN1 domain has the unique α helix linking strands 4 and 5 that we reported previously (40). The root mean square deviation of isolated FN1 (40) and the FN1 domain in our Ig5-FN1 tandem is 0.59 Å over 101 common Cα atoms.
TABLE 1.
Data collection and refinement statistics for the Ig5-FN1 domains of NCAM
| Data collection | |
| X-ray source | SERCAT BM-22 |
| Wavelength (Å) | 1.0 |
| Unit cell (Å) | a = 77.09 |
| b = 155.84 | |
| c = 71.75 | |
| Space group | C2221 |
| No. molecules/a.u. | 2 |
| Resolution (Å) | 30-1.79 |
| Measured reflections | 190,962 |
| Unique reflections | 40,353 |
| Completeness (%)a | 98.7 (96.3) |
| I/σIa | 14.9 (6.1) |
| Rsyma (%) | 9.5 (58.5) |
| Refinement | |
| Resolution (Å) | 30-1.80 |
| No. reflections (working/free) | 35,851/4,060 |
| Rcryst (%) | 18.3 |
| Rfree (%) | 23.0 |
| No. protein atoms/sulfates/waters | 3,018/1/340 |
| Root mean square deviation from ideal geometry | |
| Bond length (Å) | 0.014 |
| Angle distances (°) | 1.567 |
| Estimated coordinated error (Å) | 0.132 |
| Ramachandran plot statistics | |
| Residues in most favored regions | 91.4% |
| Residues in allowed regions | 8.3% |
| Residues in generously allowed regions | 0.3% |
| Residues in disallowed regions | 0.0% |
a Last shell (1.90-1.79 Å) is in parentheses.
FIGURE 1.
Bacterially expressed unglycosylated Ig5-FN1 forms a dimer, mediated by interactions between the Ig5 and FN1 domains. A, a schematic diagram of the Ig5-FN1 dimer, with one monomer colored green and the second monomer brown. The disulfide bond linking strands B and F of the Ig5 domains is shown in orange. B, stereo view of an Ig5-FN1 monomer is shown, with the three asparagines in the Ig5 domain that are part of consensus N-linked glycosylation sites displayed. Amino acids that form the α helix and surface acidic patch within the FN1 domain are colored yellow and blue, respectively. C, a close-up view of the Ig5-FN1 dimer interface showing the bond distances between specific residues in the Ig5 and FN1 domains. The ASN4 glycosylation site in the Ig5 domains is highlighted in pink.
The formation of a dimer of the Ig5-FN1 tandem was unexpected. However, analysis of purified unglycosylated Ig5-FN1 by analytical ultracentrifugation demonstrated that it forms concentration-dependent dimers (supplemental Fig. 1). Inspection of the dimer interface reveals several interactions between Ig5 and FN1 residues (Fig. 1C). The side chain of Asp-551, flanking the FN1 α helix, makes a main-chain hydrogen bond with Ala-405 in Ig5, as well as a side-chain electrostatic interaction with Lys-407 in Ig5. The side chain of Lys-553, within the FN1 α helix, makes a main-chain hydrogen bond with Gln-487 of Ig5, whereas the side chain of Lys-532 in FN1 makes a main-chain hydrogen bond with Met-403 at the N terminus of the Ig5 domain (Fig. 1C). However, we noted that the ASN4 glycosylation sites in the two Ig5 domains are positioned between these domains in the dimer in such a way that, if they are glycosylated, this could prevent dimer formation (Fig. 1, A and C). To investigate whether a similar dimer might exist in vivo, we analyzed the use of the ASN4 glycosylation site and the self-association of glycosylated Ig5-FN1 expressed in mammalian cells.
NCAM Ig5-FN1 Is Glycosylated on ASN4 When Expressed in Mammalian Cells
Various analyses of NCAM glycosylation site usage have yielded disparate results (59, 60). However, a recent mass spectrometry analysis of the glycosylation pattern of NCAM isolated from adult mouse brain demonstrated that all of the six consensus N-linked glycosylation sites are modified, including ASN4 (61). To confirm that Ig5-FN1 is glycosylated on ASN4, we expressed a truncated NCAM protein consisting of Ig5, FN1, the transmembrane region, and cytoplasmic tail (Ig5-FN1) in COS-1 cells and performed PNGaseF analysis of the unmodified protein, and mutants with either ASN5 and ASN6 mutated to Gln (Ig5-FN1 Asn[5.6]Gln) or all three glycosylation site Asn residues mutated to Gln (Ig5-FN1 Asn[4.5.6]Gln). These Asn to Gln mutations disrupted the consensus Asn-X-Ser/Thr N-linked glycosylation sites, preventing glycan addition. Immunofluorescence microscopy demonstrated that all the mutant proteins trafficked to the cell surface, like wild-type Ig5-FN1 and NCAM, indicating that the loss of glycosylation sites did not severely affect protein folding (data not shown).
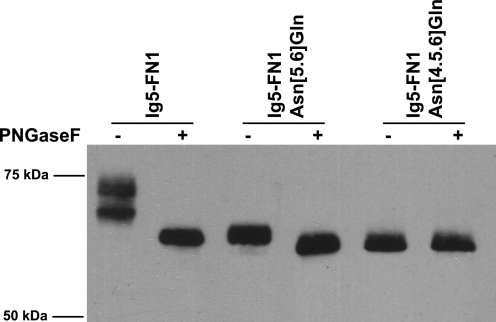
PNGaseF, a glycosidase that cleaves all types of N-linked glycans, was used to demonstrate the presence or absence of N-glycans on these proteins. As shown in Fig. 2, treatment of Ig5-FN1 with PNGaseF results in a decrease in molecular mass on SDS-PAGE from 74 to 64 kDa, thereby confirming this protein is glycosylated on N-linked glycans. Data presented below in Fig. 8 suggest that the two bands seen in the untreated Ig5-FN1 lane represent molecules with sialylated, complex type N-glycans (upper band) and those possessing high mannose type N-glycans (lower band), both of which will be removed by PNGaseF. The Ig5-FN1 Asn[5.6]Gln mutant also migrated at a lower molecular mass upon PNGaseF treatment, suggesting the presence of an additional N-linked glycan. When ASN4 was additionally replaced in the Ig5-FN1 Asn[4.5.6]Gln mutant, PNGaseF had no further effect on molecular mass, indicating no additional N-glycan attachment sites remained, and demonstrating that all three N-glycosylation sites were used. These results confirmed that ASN4 is glycosylated on membrane-associated Ig5-FN1 expressed in mammalian cells.
FIGURE 2.
The ASN4 glycosylation site is utilized in mammalian cells. Analysis of N-linked glycosylation of membrane-bound Ig5-FN1 expressed in mammalian cells. COS-1 cells were transfected with V5-tagged Ig5-FN1-TM-tail (Ig5-FN1) or mutant Ig5-FN1 with ASN5 and ASN6 or ASN4, ASN5, and ASN6 mutated to Gln (Ig5-FN1 Asn[5.6]Gln and Ig5-FN1 Asn[4.5.6]Gln, respectively). Cell lysates were subjected to PNGaseF digestion and analyzed by immunoblotting with anti-V5 epitope tag antibody.
FIGURE 8.
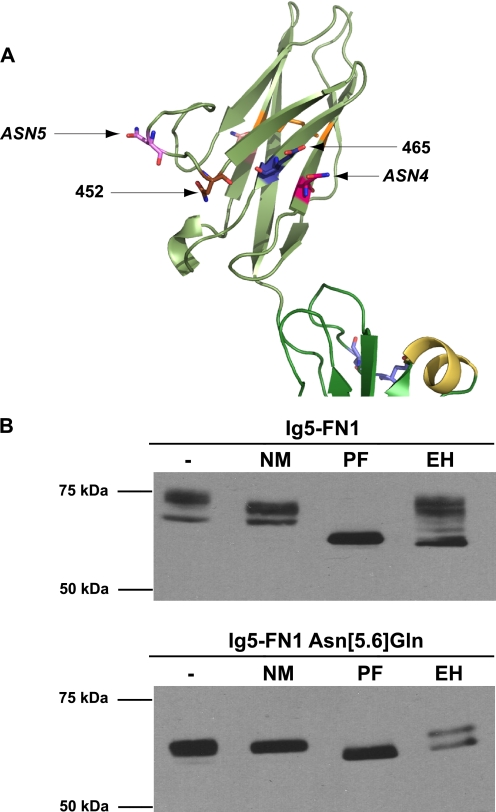
Glycosidase analysis of ASN4 glycosylation reveals less glycan processing at this site. A, a schematic diagram of the Ig5 domain of NCAM showing the relative positions of ASN4, ASN5, and the engineered glycosylation sites, 452 and 465. B, COS-1 cells were transfected with wild-type Ig5-FN1-TM-tail (Ig5-FN1, top panel) or with Ig5-FN1 carrying Asn to Gln mutations at both ASN5 and ASN6 (Ig5-FN1 Asn[5.6]Gln, bottom panel). Cell lysates were collected and either left untreated or treated with neuraminidase (NM), PNGaseF (PF), or Endo H (EH). Proteins were resolved by SDS-PAGE and analyzed by immunoblotting with anti-V5 epitope tag antibody.
Elimination of the ASN4 Glycosylation Site Allows Self-association of Soluble Ig5-FN1 and Enhanced Self-association of Soluble NCAM
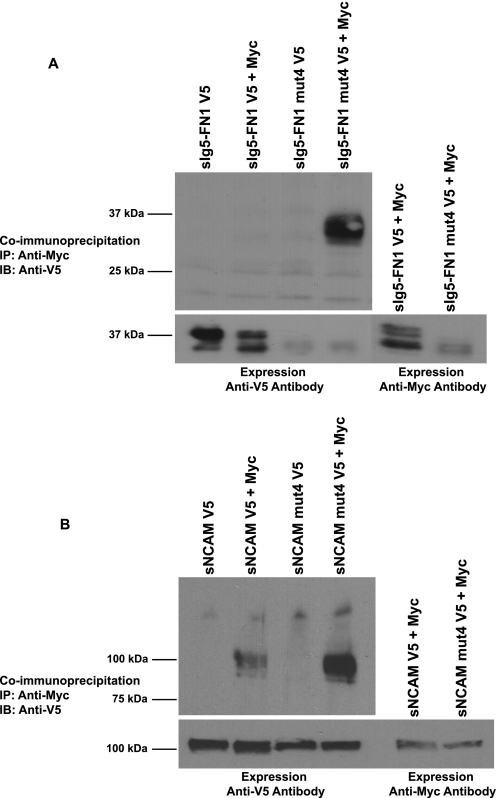
A co-immunoprecipitation approach was taken to evaluate whether the absence of N-glycans on Ig5-FN1 may have allowed the self-association of the bacterially expressed Ig5-FN1 tandem. Myc- and V5-tagged versions of soluble Ig5-FN1 (sIg5-FN1) were co-expressed in COS-1 cells and tested for interaction by co-immunoprecipitation from cell media. No self-association of glycosylated sIg5-FN1 was detected in this analysis (Fig. 3A). However, the same protein lacking the ASN4 glycosylation site (sIg5-FN1 mut4, third position of consensus sequence altered NIT to NIA) did exhibit self-association, demonstrating that the presence of a glycan on ASN4 was sufficient to prevent self-association (Fig. 3A).
FIGURE 3.
Elimination of glycosylation at ASN4 allows self-association of soluble Ig5-FN1, and enhances self-association of soluble NCAM. Differentially tagged soluble Ig5-FN1 (sIg5-FN1) or soluble NCAM (sNCAM) proteins were tested for self-association by co-immunoprecipitation analysis. To eliminate glycosylation at ASN4, the consensus glycosylation site was mutated from Asn423-Ile-Thr to Asn423-Ile-Ala (mut4). A, top panel, Myc-tagged sIg5-FN1 or sIg5-FN1 mut4 was immunoprecipitated from cell media with polyclonal anti-Myc epitope tag antibody and association with V5-tagged sIg5-FN1 or sIg5-FN1 mut4 analyzed by immunoblotting with monoclonal anti-V5 epitope tag antibody. Bottom panel, to measure relative protein expression levels, an aliquot of cell media was removed prior to immunoprecipitation and immunoblotted with anti-V5 or anti-Myc epitope tag antibodies. B, top panel, Myc-tagged sNCAM or sNCAM mut4 was immunoprecipitated from cell media and association with V5-tagged sNCAM or sNCAM mut4 measured by immunoblotting. Bottom panel, relative V5-tagged and Myc-tagged sNCAM expression levels were analyzed by immunoblotting of an aliquot of cell media.
Analysis of sIg5-FN1 and sIg5-FN1 mut4 secreted into the cell medium demonstrated that the glycosylation mutant was either secreted at very low levels or was very unstable, despite the relatively robust co-immunoprecipitation result (Fig. 3A, Expression). Notably, even secreted sIg5-FN1 exhibited incomplete glycosylation, as seen for membrane-associated Ig5-FN1 in Fig. 2, and/or instability resulting in the presence of multiple bands on the immunoblot (Fig. 3A, Expression). We reasoned that additional NCAM domains might enhance stability and glycosylation of the soluble, secreted protein. With this in mind, we tested the impact of removing the ASN4 glycosylation site on the self-association of soluble NCAM (sNCAM).
NCAM has been demonstrated to exhibit homophilic trans interactions, and these are predicted to occur via anti-parallel interactions of all its Ig domains (43, 44), Ig1-Ig2 (56, 62), and/or a variety of anti-parallel interactions involving Ig1, Ig2, and Ig3 (34). Polysialylation of the Ig5 domain disrupts these interactions (12–14). Structural analysis of the first three Ig domains suggests that a cross-like dimer of Ig1 and Ig2 may mediate homophilic cis interactions between two NCAM molecules (34). To our knowledge there is no evidence for an Ig5-Ig5 interaction in either NCAM trans or cis dimers. For this reason, we wondered whether the elimination of the ASN4 glycosylation site in the Ig5 domain of a soluble NCAM protein would impact its self-association. We observed an interaction of differentially tagged sNCAM proteins in the COS-1 cell medium, and this interaction was noticeably increased when the ASN4 glycosylation site on Ig5 was mutated (Fig. 3B). These results suggest that, whether NCAM is forming cis or trans dimers in solution, the glycosylation status of the Ig5 domain can impact this association.
Sites of Ig5 N-Glycan Polysialylation Do Not Align with FN1 Sequences Shown to Impact NCAM Polysialylation
In our working model, a polyST recognizes and binds to sequences in FN1 in such a way so that it can reach the N-linked glycans to be polysialylated on the adjacent Ig5 domain. In this scenario, one might expect that the sequences recognized by the polySTs would be on the same side of the Ig5-FN1 unit as the glycans on ASN5 and ASN6 that are polysialylated, and that the reason that the glycan on ASN4 is not polysialylated is because it does not align with the polyST recognition sequences. Surprisingly, in the Ig5-FN1 crystal structure the ASN5 and ASN6 glycosylation sites are on the opposite face from the FN1 acidic patch and α helix, whereas ASN4 comes closer to aligning with these sequences (Fig. 1B). How can this be explained? One possibility is that there may be more than one polyST recognition site. Another possibility is that the Ig5-FN1 dimer formed in the crystal may have constrained the Ig5-FN1 domain orientation, whereas under physiological conditions the inter-domain relationship is flexible allowing the ASN5 and ASN6 glycosylation sites to align with the FN1 α helix and acidic patch. Although the N-glycans on Ig5 ASN5 and ASN6 are clearly the optimal sites for polysialylation, a flexibility in the polysialylation process has been suggested by our finding that replacing the FN1 α helix sequences shifts polysialic acid addition to O-glycans on the FN1 domain (40). This observation could reflect either the change in binding and positioning of the polyST in a relatively rigid Ig5-FN1 structure or a change in the relationship between the Ig5 and FN1 domains, or both. To evaluate the importance of Ig5 N-glycan placement for polysialylation, and to obtain some understanding of why the glycan on ASN4 possesses little to no polysialic acid despite being glycosylated, we have individually introduced seven new glycosylation sites in various locations on the surface of Ig5 and analyzed their polysialylation.
The Polysialylation of Novel Glycans Engineered in Different Locations on the Surface of Ig5 Suggests Some Flexibility in the Polysialylation Process and the Importance of the Ig5 Domain in polyST Engagement
We used the structure of the Ig5 domain to guide us in the construction of consensus N-linked glycosylation sites to understand what restrictions are placed on the location of polysialylated N-glycans. First, we created an NCAM mutant that would be largely devoid of polysialic acid by mutating the consensus N-linked glycosylation sequence of ASN5 and ASN6 from Asn-X-Ser/Thr to Asn-X-Ala (mut[5.6]NXA). The mut[5.6]NXA protein localized to the Golgi and cell surface indicating the absence of glycosylation at these sites did not severely affect protein folding and trafficking (supplemental Fig. 2).
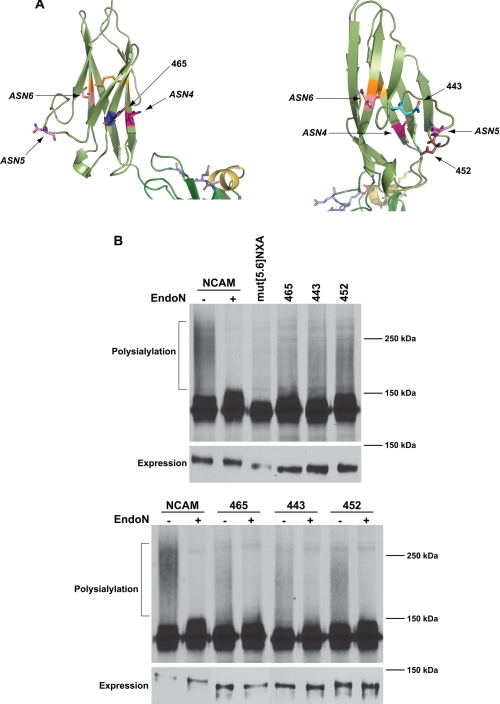
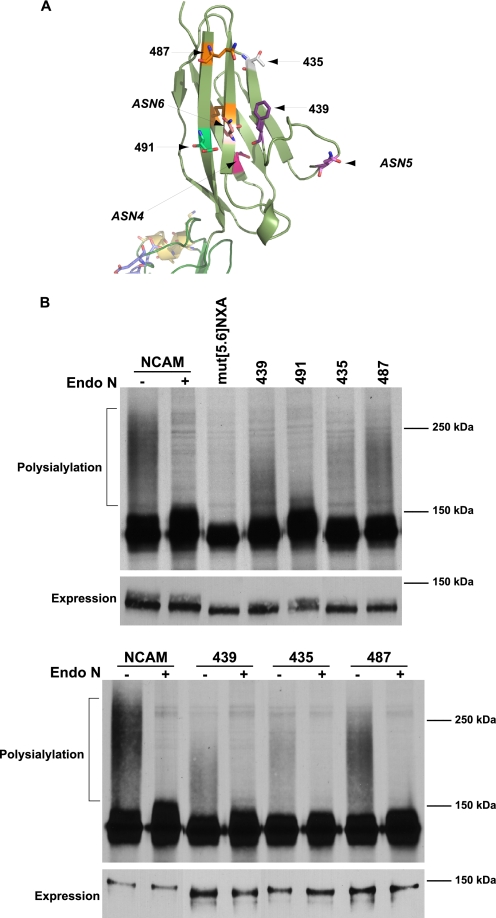
The three N-glycans on the Ig5 domain are in the mid section of the Ig5 β sandwich, with ASN4 being somewhat closer to the FN1 domain (Fig. 1B). The glycans that are the primary sites of polysialylation, found on ASN5 and ASN6, are closer together on one side of the domain, whereas the glycan that is very weakly polysialylated found on ASN4 is on the opposite side of the domain (Fig. 1B). From this observation, we predicted that new glycans placed near to ASN4 would not be polysialylated, whereas those placed closer to ASN5 and ASN6 would be polysialylated. To test this prediction, we created a series of glycosylation sites that circled the Ig5 domain (Figs. 4A and 5A). A site at position 465 was placed on strand E right next to the ASN4 site on strand B (Fig. 4A, left). N-Glycosylation sites at positions 443 and 452 flanked ASN5 (position 449) in the loop connecting the two sheets, with 443 being between ASN5 and ASN6 and 452 being between ASN4 and ASN5 (Fig. 4A, right). Finally, glycosylation sites at positions 439 and 491 on strands C and G were positioned next to ASN6, found at position 478 on strand F of the second sheet of the β sandwich (Fig. 5A). We also rationalized that glycosylation sites placed on the same face as ASN6 but closer to the Ig4 domain may not be polysialylated if Ig5 and FN1 are rigidly positioned. To test this hypothesis, we placed glycosylation sites at positions 435 and 487, located at the “top” of the Ig5 domain closer to the adjacent Ig4 domain (Fig. 5A). Each of these mutants trafficked to the cell surface following expression in COS-1 cells, similar to wild-type NCAM and mut[5.6]NXA, indicating that the changes made did not severely impact protein folding and trafficking (supplemental Fig. 2).
FIGURE 4.
Pulse-chase analysis reveals differential polysialylation of engineered N-glycans flanking ASN4 and ASN5. A, schematic diagrams of the Ig5 domain showing ASN4, ASN5, ASN6, and the positions of inserted glycosylation sites. B, top panel, COS-1 cells were co-transfected with NCAM, mut[5.6]NXA, or mut[5.6]NXA with individual engineered N-linked glycosylation sites and PST-Myc. Cells were labeled with [35S]Met/Cys for 1 h followed by a 3-h chase period in unlabeled media. NCAM proteins were immunoprecipitated from cell lysates with anti-V5 epitope tag antibody, resolved by SDS-PAGE, and visualized by fluorography. The high molecular weight broad band (NCAM, −) that collapses upon Endo N treatment (NCAM, +) indicates the presence of α2,8-linked polysialic acid on NCAM. Relative NCAM protein expression levels revealed by immunoblotting are shown below (Expression). B, bottom panel, COS-1 cells expressing NCAM or mutant proteins and PST-Myc were metabolically labeled. NCAM proteins were immunoprecipitated and either left untreated (−) or treated with Endo N (+). Immunoblot analysis of relative NCAM protein expression levels is shown in the lower panel (Expression).
FIGURE 5.
Pulse-chase analysis reveals differential polysialylation of engineered N-glycans flanking ASN6. A, schematic diagram of the Ig5 domain demonstrating the sites of inserted glycosylation sites near ASN6. B, top panel, COS-1 cells were co-transfected with NCAM, mut[5.6]NXA, or mut[5.6]NXA with individual engineered glycans, and PST-Myc. Cells were metabolically labeled with [35S]Met/Cys for 1 h and chased in unlabeled media for 3 h. Cell lysates were collected, and NCAM proteins were immunoprecipitated and visualized by SDS-PAGE and fluorography. Digestion of NCAM with Endo N results in the disappearance of the high molecular weight broad band that indicates the presence of α2,8-linked polysialic acid on NCAM. B, bottom panel, COS-1 cells expressing NCAM or mutant proteins and PST-Myc were metabolically labeled. NCAM proteins were immunoprecipitated and either left untreated (−) or subject to Endo N digestion (+). In both experiments, relative NCAM protein expression levels were analyzed by immunoblotting with anti-V5 epitope tag antibody (Expression).
To evaluate glycosylation site usage as well as polysialylation, we chose to subject these mutants to pulse-chase analysis and Endo N digestion (Figs. 4 and 5). Endo N cleaves α2,8-linked polysialic acid chains of eight residues or more, leaving a short four-unit α2,8-oligosialic acid chain at the terminus of the cleaved glycan (54). We then confirmed polysialylation by immunoblotting with the OL.28 anti-polysialic acid antibody (Fig. 6 and data not shown). In the pulse-chase analysis, polysialylated wild-type NCAM migrated as a broad, high molecular mass band of 150 kDa to over 250 kDa that collapses upon Endo N treatment to 126–150 kDa (Figs. 4B and 5B, NCAM ± Endo N). It is likely that the NCAM molecules migrating between 150 and ∼180 kDa are primarily modified by longer oligosialic acid chains, because these species are not recognized by the OL.28 antibody (compare with Fig. 6). The lower molecular mass 126–147 kDa band of unpolysialylated wild-type NCAM represents both those NCAM proteins that are expressed in cells that are not expressing PST, as well as a fraction of the protein that carries Endo H-sensitive high-mannose N-glycans, rather than complex N-glycans (data not shown). The presence of NCAM proteins with incompletely processed N-glycans likely reflects the inability of the cellular glycosylation machinery to fully modify all of the highly expressed NCAM protein. We have observed a similar situation with overexpressed polySTs (11). Notably, the unpolysialylated forms of each NCAM mutant with an additional N-glycosylation site were somewhat larger than the mut[5.6]NXA mutant suggesting that these engineered sites were indeed glycosylated and further modified (Figs. 4B and 5B, upper panels).
FIGURE 6.
Immunoblot analysis reveals different patterns of polysialylation of NCAM mutants with engineered glycosylation sites. A, top panel, COS-1 cells were transiently co-transfected with wild-type or mutated NCAM and PST-Myc. NCAM proteins were immunoprecipitated from cell lysates with anti-V5 epitope tag antibody and incubated at 37 °C overnight either with (+) or without (−) PNGaseF enzyme. Polysialylation levels were determined by immunoblotting with the anti-polysialic acid antibody, OL.28. Bottom panel, immunoblot analysis of relative NCAM protein expression levels. B, top panel, COS-1 cells transiently expressing NCAM proteins and PST-Myc were collected. NCAM proteins were immunoprecipitated and either untreated or subject to PNGaseF digestion. Bottom panel, relative NCAM protein expression levels were determined by immunoblotting.
Surprisingly, and contrary to our predictions, with the exception of the 491 mutant, all of the other mutants were polysialylated to a greater extent than mut[5.6]NXA (Figs. 4B and 5B). Immunoblotting with the OL.28 anti-polysialic acid antibody further demonstrated a low level of polysialic acid addition to mut[5.6]NXA and confirmed the presence of polysialic acid on all of the mutant NCAM proteins except 491 (Fig. 6 and data not shown). Comparison of the relative levels of polysialylation of each new glycan demonstrated that the extent of mutant protein polysialylation did not strictly mirror the proximity of the new sites to ASN5 and ASN6. For example, the glycan 491 was not polysialylated, whereas the glycan at 439 was weakly polysialylated, despite the fact that both glycosylation sites are adjacent to ASN6 (Fig. 5B). However, we did note more efficient polysialylation of sites where the structure predicts the glycan attachment site and hence the glycan would project out from the domain in the same way as either ASN5 or ASN6. For example, the glycosylation site at 487 is robustly polysialylated, whereas that found at position 435 is less polysialylated (Fig. 5B). Although both sites are found at some distance from ASN6 and closer to the Ig4 domain, only the side chain of the asparagine residue at position 487 is predicted to extend from the domain in the same direction as does that of ASN6 (Fig. 5A). Likewise, even though the asparagine residue at position 452 is closer to ASN4, its side chain is predicted to extend away from the loop structure in the same direction as the glycan on ASN5 (Fig. 4A). While it is difficult to precisely predict how an attached glycan may extend from these sites, it seems likely that the flexibility of the glycan itself could contribute significantly to polyST access.
The Polysialylation of New Glycosylation Sites at Positions 452 and 487 Suggest Flexibility in the Polysialylation Process
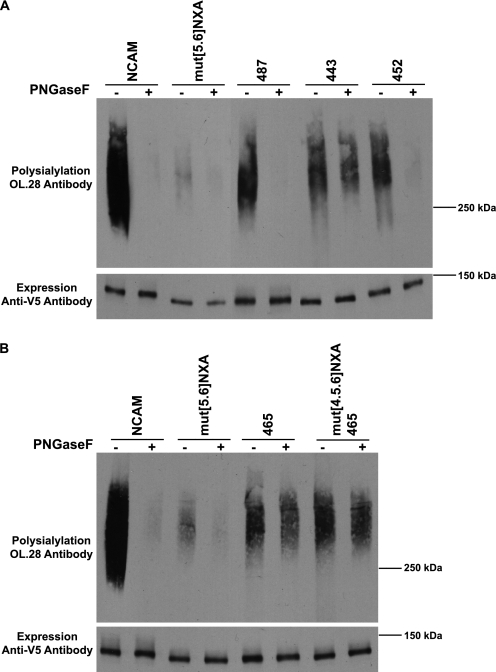
The first inclination is to presume that any polysialylation observed in the mutant proteins is occurring on the new glycan. However, we could not discount the possibility that other glycans, such as O-glycans on the FN1 domain, which are polysialylated in both a FN1 Δhelix mutant and a truncated NCAM protein lacking all Ig domains (40, 38), might be polysialylated in these mutants. To determine whether N-linked or O-linked glycans were polysialylated in the NCAM glycosylation mutants, we used PNGaseF analysis. We focused on 465, 443, 452, and 487, because these mutants were found to be the most heavily polysialylated by immunoblot analysis. We found that the 452 mutant with a new glycosylation site on the ASN5 loop and the 487 mutant placed on a strand adjacent to that carrying ASN6, but closer to the Ig4 domain, were polysialylated on N-linked glycans (Fig. 6A). The polysialylation of both these mutants in mut[5.6]NXA with glycosylation sites placed at a greater distance from the FN1 domain (487) or closer to the ASN4 face of the FN1 domain (452) suggest that there is some flexibility in the placement of the N-glycans for polysialylation.
Addition of New Glycosylation Sites at Positions 443 and 465 Leads to O-Glycan Polysialylation
In contrast to the 452 and 487 mutants, analysis of the 443 and 465 mutants demonstrated that polysialic acid was modifying predominantly PNGaseF-insensitive O-glycans in both proteins (Fig. 6, A and B). We envisioned three possible reasons for the redirection of the polyST to O-glycans, possibly on the FN1 domain. First, the addition of a new glycan may have changed the surface structure of the Ig5 domain making it difficult for the polyST to engage. This could be the case in either the 443 or 465 mutant. Interestingly, removal of the adjacent ASN4 glycosylation site in the 465 mutant continues to lead to O-glycan polysialylation, arguing that it is not the presence of these two glycans in close proximity that is leading to changes that block polyST access (Fig. 6B). Second, the presence of a new glycan may physically block the polyST from engaging with the domain. This could be the case in the 443 mutant in which a new N-glycan is placed between ASN5 and ASN6. Third, the lack of an appropriately placed Ig5 N-glycan may allow the polyST to more efficiently polysialylate O-glycans that are weaker acceptors. We favor the first two possibilities and believe that the third possibility is unlikely, because we observed that, in the absence of ASN5 and ASN6 in the mut[5.6]NXA mutant, the polyST is not redirected to O-glycans. Upon PNGaseF digestion, we observed only a low level O-glycan polysialylation in this mutant that is also observed in the wild-type NCAM protein (Fig. 6).
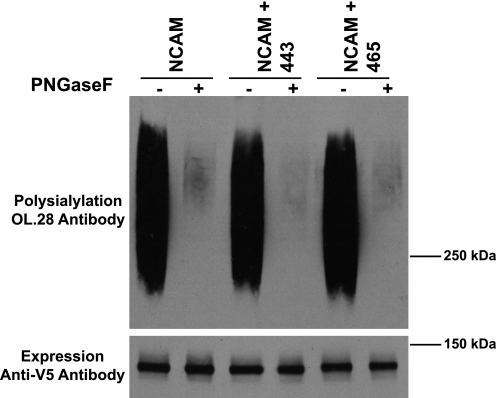
If the presence of the new glycans altered the surface structure or blocked polyST access to the Ig5 domain, then they might function in the same way if inserted into the wild-type NCAM protein possessing ASN4, ASN5, and ASN6 glycosylation sites. However, after inserting these new glycans into the fully glycosylated NCAM protein, we found that they had no effect on the N-linked polysialylation of wild-type NCAM (Fig. 7). These results suggest it is unlikely that the new glycan at position 443 is physically blocking access of the polyST to the Ig5 domain. Instead, the new glycans may be causing changes in the surface structure that interfere with the ability of the polyST to engage the Ig5 domain. However, in the presence of the naturally occurring N-glycans on ASN5 and ASN6, the effects of any alterations are negated.
FIGURE 7.
Polysialylation of fully glycosylated NCAM is unaffected by the insertion of engineered glycans. Top panel, wild-type NCAM or NCAM with either the 443 or 465 glycosylation site inserted was immunoprecipitated from COS-1 cell lysates and incubated at 37 °C overnight either with (+) or without (−) PNGaseF enzyme. Proteins were resolved by SDS-PAGE, and polysialylation was measured using the anti-polysialic acid antibody, OL.28. Bottom panel, immunoblot analysis of relative NCAM protein expression levels.
The Glycan on ASN4 Is Less Processed Than Those on ASN5 and ASN6 and This May Explain Its Low Level of Polysialylation
The weak polysialylation of the glycan on ASN4 in the mut[5.6]NXA mutant and the more robust polysialylation of the 452 mutant (Fig. 6A), could reflect the positioning of the 452 N-glycan closer to the polyST engagement site or differences in the underlying glycan structures at these sites. One possibility is that the ASN4 glycan might be more highly polysialylated if it carried a more fully processed glycan. PolySTs require a terminal α2,3- or α2,6-linked sialic acid residue terminating a complex type N-glycan to initiate synthesis of the α2,8-linked polysialic acid chain (63, 64). To determine whether the glycan on ASN4 was properly processed for polysialylation, we used an Ig5-FN1 protein lacking the ASN5 and ASN6 glycosylation sites (Ig5-FN1 Asn[5.6]Gln) and compared its glycosylation to Ig5-FN1 containing all three Ig5 N-glycosylation sites using neuraminidases to remove sialic acid, PNGaseF to remove N-glycans, and Endo H to remove high mannose N-glycans (Fig. 8).
Endo H digestion demonstrated that ∼50% of the oligosaccharides on ASN4 in the Ig5-FN1 Asn[5.6]Gln mutant are of the high mannose type, whereas a greater proportion of the glycans on Ig5-FN1 are processed to Endo H-resistant, complex forms (Fig. 8, EH). In addition, digestion with a combination of neuraminidases demonstrated that Ig5-FN1 N-glycans carry larger quantities of terminal sialic acid than does the ASN4 glycan on the Ig5-FN1 Asn[5.6]Gln protein (Fig. 8, NM). These results, combined with the polysialylation of the 452 mutant, suggest that the low level polysialylation of the ASN4 glycan may be due in part to its lack of terminal processing rather than strictly due to its location on the Ig5 domain.
In sum, we have presented the first structure of the NCAM Ig5-FN1 tandem and the Ig5 domain. Our results show that the two NCAM Ig5 glycosylation sites that are modified with polysialic acid (ASN5 and ASN6) are found in close proximity on the same side of the Ig5 domain, with the ASN4 glycosylation site that is weakly polysialylated found on the opposite side of the domain. Although the crystal structure indicates that the unglycosylated Ig5-FN1 tandem can form a dimer, our results demonstrate that the fully glycosylated Ig5-FN1 unit does not self-associate until the ASN4 glycosylation site is removed. Interestingly, a soluble form of NCAM does self-associate, and this self-association is augmented in the absence of the Ig5 ASN4 glycosylation site. We also observed that the glycans on the ASN4 site are not efficiently processed to sialylated complex forms and that they are only polysialylated to very low levels if at all. These results make it tempting to consider the possibility that NCAM forms dimers in the secretory pathway and that, whereas the Ig5-FN1 unit is not directly involved in dimer contacts, the dimerization of the whole protein still partially obscures the processing and polysialylation of the ASN4 glycan.
In the Ig5-FN1 crystal structure the ASN5 and ASN6 glycosylation sites are on the opposite face to FN1 residues previously shown to be involved in NCAM polysialylation. This observation led us to question whether the relative relationship of the Ig5 and FN1 domains observed in the crystal structure is fixed, and whether there is flexibility in the Ig5-FN1 interdomain relationship and the polysialylation process in general. Although we cannot be certain that the Ig5 and FN1 domains can twist relative to one another, we have observed some flexibility in the polysialylation process. Insertion of new glycosylation sites in the Ig5 domain demonstrated that there is some flexibility in the location of sites that can be polysialylated, in that glycans placed on the Ig5 domain further away from the FN1 domain and closer to the Ig4 domain, or those placed closer to the weakly polysialylated ASN4 glycosylation site, are polysialylated. This implied polyST flexibility might not be so surprising considering that they sequentially add sialic acid residues to a growing polysialic acid chain. This analysis also suggested the importance of polyST engagement with the Ig5 domain for the polysialylation of Ig5 N-glycans. We found that disrupting Ig5 domain surface structure by the addition of new glycans shifted polysialylation to O-glycans that are likely to be located on the FN1 domain. So, while the Ig5 domain is dispensable for polyST recognition per se, we predict that an interaction of the polyST with Ig5 glycans and/or sequences ensures Ig5 N-glycan polysialylation and prevents the redirection of the enzyme to other sites.
Supplementary Material
Acknowledgments
We thank Dr. Christine Schar for performing the analytical ultracentrifugation analysis of purified Ig5-FN1. We also gratefully acknowledge Saugata Hazra, Ming-Fong Lye, and Ying Su for technical assistance, and the staff at Southeast Regional Collaborative Access Team for help with data collection.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 GM063843 (to K. J. C.).
The atomic coordinates and structure factors (code 3MTR) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1, Figs. 1 and 2, and additional references.
- PST
- ST8Sia IV
- polyST
- polysialyltransferase
- NCAM
- neural cell adhesion molecule
- sNCAM
- soluble NCAM
- FNIII
- fibronectin type III repeat of NCAM
- FN1
- first fibronectin type III repeat of NCAM
- FN2
- second fibronectin type III repeat of NCAM
- PNGaseF
- peptide N-glycosidase F
- Endo H
- endoglycosidase H
- Endo N
- endo-N-acylneuraminidase.
REFERENCES
- 1.Eckhardt M., Mühlenhoff M., Bethe A., Koopman J., Frosch M., Gerardy-Schahn R. (1995) Nature 373, 715–718 [DOI] [PubMed] [Google Scholar]
- 2.Kojima N., Yoshida Y., Tsuji S. (1995) FEBS Lett. 373, 119–122 [DOI] [PubMed] [Google Scholar]
- 3.Nakayama J., Fukuda M. N., Fredette B., Ranscht B., Fukuda M. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 7031–7035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scheidegger E. P., Sternberg L. R., Roth J., Lowe J. B. (1995) J. Biol. Chem. 270, 22685–22688 [DOI] [PubMed] [Google Scholar]
- 5.Rothbard J. B., Brackenbury R., Cunningham B. A., Edelman G. M. (1982) J. Biol. Chem. 257, 11064–11069 [PubMed] [Google Scholar]
- 6.Finne J., Finne U., Deagostini-Bazin H., Goridis C. (1983) Biochem. Biophys. Res. Commun. 112, 482–487 [DOI] [PubMed] [Google Scholar]
- 7.Curreli S., Arany Z., Gerardy-Schahn R., Mann D., Stamatos N. M. (2007) J. Biol. Chem. 282, 30346–30356 [DOI] [PubMed] [Google Scholar]
- 8.Yabe U., Sato C., Matsuda T., Kitajima K. (2003) J. Biol. Chem. 278, 13875–13880 [DOI] [PubMed] [Google Scholar]
- 9.Zuber C., Lackie P. M., Catterall W. A., Roth J. (1992) J. Biol. Chem. 267, 9965–9971 [PubMed] [Google Scholar]
- 10.Mühlenhoff M., Eckhardt M., Bethe A., Frosch M., Gerardy-Schahn R. (1996) EMBO J. 15, 6943–6950 [PMC free article] [PubMed] [Google Scholar]
- 11.Close B. E., Colley K. J. (1998) J. Biol. Chem. 273, 34586–34593 [DOI] [PubMed] [Google Scholar]
- 12.Rutishauser U., Acheson A., Hall A. K., Mann D. M., Sunshine J. (1988) Science 240, 53–57 [DOI] [PubMed] [Google Scholar]
- 13.Fujimoto I., Bruses J. L., Rutishauser U. (2001) J. Biol. Chem. 276, 31745–31751 [DOI] [PubMed] [Google Scholar]
- 14.Johnson C. P., Fujimoto I., Rutishauser U., Leckband D. E. (2005) J. Biol. Chem. 280, 137–145 [DOI] [PubMed] [Google Scholar]
- 15.Angata K., Nakayama J., Fredette B., Chong K., Ranscht B., Fukuda M. (1997) J. Biol. Chem. 272, 7182–7190 [DOI] [PubMed] [Google Scholar]
- 16.Ong E., Nakayama J., Angata K., Reyes L., Katsuyama T., Arai Y., Fukuda M. (1998) Glycobiology 8, 415–424 [DOI] [PubMed] [Google Scholar]
- 17.Cremer H., Lange R., Christoph A., Plomann M., Vopper G., Roes J., Brown R., Baldwin S., Kraemer P., Scheff S., Barthels D., Rajewsky K., Wille W. (1994) Nature 367, 455–459 [DOI] [PubMed] [Google Scholar]
- 18.Cremer H., Chazal G., Goridis C., Represa A. (1997) Mol. Cell. Neurosci. 8, 323–335 [DOI] [PubMed] [Google Scholar]
- 19.Weinhold B., Seidenfaden R., Röckle I., Mühlenhoff M., Schertzinger F., Conzelmann S., Marth J. D., Gerardy-Schahn R., Hildebrandt H. (2005) J. Biol. Chem. 280, 42971–42977 [DOI] [PubMed] [Google Scholar]
- 20.Kiss J. Z., Troncoso E., Djebbara Z., Vutskits L., Muller D. (2001) Brain Res. Rev. 36, 175–184 [DOI] [PubMed] [Google Scholar]
- 21.Durbec P., Cremer H. (2001) Mol. Neurobiol. 24, 53–64 [DOI] [PubMed] [Google Scholar]
- 22.Gascon E., Vutskits L., Kiss J. Z. (2007) Brain Res. Rev. 56, 101–118 [DOI] [PubMed] [Google Scholar]
- 23.Rutishauser U. (2008) Nat. Rev. Neurosci. 9, 26–35 [DOI] [PubMed] [Google Scholar]
- 24.Livingston B. D., Jacobs J. L., Glick M. C., Troy F. A. (1988) J. Biol. Chem. 263, 9443–9448 [PubMed] [Google Scholar]
- 25.Hildebrandt H., Becker C., Glüer S., Rösner H., Gerardy-Schahn R., Rahmann H. (1998) Cancer Res. 58, 779–784 [PubMed] [Google Scholar]
- 26.Suzuki M., Suzuki M., Nakayama J., Suzuki A., Angata K., Chen S., Sakai K., Hagihara K., Yamaguchi Y., Fukuda M. (2005) Glycobiology 15, 887–894 [DOI] [PubMed] [Google Scholar]
- 27.Komminoth P., Roth J., Lackie P. M., Bitter-Suermann D., Heitz P. U. (1991) Am. J. Pathol. 139, 297–304 [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka F., Otake Y., Nakagawa T., Kawano Y., Miyahara R., Li M., Yanagihara K., Nakayama J., Fujimoto I., Ikenaka K., Wada H. (2000) Cancer Res. 60, 3072–3080 [PubMed] [Google Scholar]
- 29.Glüer S., Schelp C., Gerardy-Schahn R., von Schweinitz D. (1998) J. Pediatr. Surg. 33, 1516–1520 [DOI] [PubMed] [Google Scholar]
- 30.Roth J., Zuber C., Wagner P., Taatjes D. J., Weisgerber C., Heitz P. U., Goridis C., Bitter-Suermann D. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 2999–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roth J., Zuber C., Wagner P., Blaha I., Bitter-Suermann D., Heitz P. U. (1988) Am. J. Pathol. 133, 227–240 [PMC free article] [PubMed] [Google Scholar]
- 32.Seidenfaden R., Krauter A., Schertzinger F., Gerardy-Schahn R., Hildebrandt H. (2003) Mol. Cell. Biol. 23, 5908–5918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cunningham B. A., Hemperly J. J., Murray B. A., Prediger E. A., Brackenbury R., Edelman G. M. (1987) Science 236, 799–806 [DOI] [PubMed] [Google Scholar]
- 34.Soroka V., Kolkova K., Kastrup J. S., Diederichs K., Breed J., Kiselyov V. V., Poulsen F. M., Larsen I. K., Welte W., Berezin V., Bock E., Kasper C. (2003) Structure 10, 1291–1301 [DOI] [PubMed] [Google Scholar]
- 35.Nelson R. W., Bates P. A., Rutishauser U. (1995) J. Biol. Chem. 270, 17171–17179 [DOI] [PubMed] [Google Scholar]
- 36.Kojima N., Tachida Y., Yoshida Y., Tsuji S. (1996) J. Biol. Chem. 271, 19457–19463 [DOI] [PubMed] [Google Scholar]
- 37.Angata K., Suzuki M., McAuliffe J., Ding Y., Hindsgaul O., Fukuda M. (2000) J. Biol. Chem. 275, 18594–18601 [DOI] [PubMed] [Google Scholar]
- 38.Close B. E., Mendiratta S. S., Geiger K. M., Broom L. J., Ho L. L., Colley K. J. (2003) J. Biol. Chem. 278, 30796–30805 [DOI] [PubMed] [Google Scholar]
- 39.Mendiratta S. S., Sekulic N., Lavie A., Colley K. J. (2005) J. Biol. Chem. 280, 32340–32348 [DOI] [PubMed] [Google Scholar]
- 40.Mendiratta S. S., Sekulic N., Hernandez-Guzman F. G., Close B. E., Lavie A., Colley K. J. (2006) J. Biol. Chem. 281, 36052–36059 [DOI] [PubMed] [Google Scholar]
- 41.Hall A. K., Rutishauser U. (1987) J. Cell Biol. 104, 1579–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Becker J. W., Erickson H. P., Hoffman S., Cunningham B. A., Edelman G. M. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 1088–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson C. P., Fujimoto I., Perrin-Tricaud C., Rutishauser U., Leckband D. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6963–6968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ranheim T. S., Edelman G. M., Cunningham B. A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 4071–4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carafoli F., Saffell J. L., Hohenester E. (2008) J. Mol. Biol. 377, 524–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 47.Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 48.McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., Read R. J. (2005) Acta Crystallogr. Sect. D Biol. Crystallogr. 61, 458–464 [DOI] [PubMed] [Google Scholar]
- 49.Lamzin V. S., Perrakis A., Wilson K. S. (2001) in International Tables for Crystallography. Vol. F: Crystallography of Biological Macromolecules (Rossmann M. G., Arnold E. eds) pp. 720–722, Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 50.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 51.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 52.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, Palo Alto, CA [Google Scholar]
- 53.Kleywegt G. J. (1996) Acta Crystallogr. D Biol. Crystallogr. 52, 842–857 [DOI] [PubMed] [Google Scholar]
- 54.Pelkonen S., Pelkonen J., Finne J. (1989) J. Virol. 63, 4409–4416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jensen P. H., Soroka V., Thomsen N. K., Ralets I., Berezin V., Bock E., Poulsen F. M. (1999) Nat. Struct. Biol. 6, 486–493 [DOI] [PubMed] [Google Scholar]
- 56.Kasper C., Rasmussen H., Kastrup J. S., Ikemizu S., Jones E. Y., Berezin V., Bock E., Larsen I. K. (2000) Nat. Struct. Biol. 7, 389–393 [DOI] [PubMed] [Google Scholar]
- 57.Atkins A. R., Chung J., Deechongkit S., Little E. B., Edelman G. M., Wright P. E., Cunningham B. A., Dyson H. J. (2001) J. Mol. Biol. 311, 161–172 [DOI] [PubMed] [Google Scholar]
- 58.Harpaz Y., Chothia C. (1994) J. Mol. Biol. 238, 528–539 [DOI] [PubMed] [Google Scholar]
- 59.Liedtke S., Geyer H., Wuhrer M., Geyer R., Frank G., Gerardy-Schahn R., Zähringer U., Schachner M. (2001) Glycobiology 11, 373–384 [DOI] [PubMed] [Google Scholar]
- 60.Wuhrer M., Geyer H., von der Ohe M., Gerardy-Schahn R., Schachner M., Geyer R. (2003) Biochimie 85, 207–218 [DOI] [PubMed] [Google Scholar]
- 61.Albach C., Damoc E., Denzinger T., Schachner M., Przybylski M., Schmitz B. (2004) Anal. Bioanal. Chem. 378, 1129–1135 [DOI] [PubMed] [Google Scholar]
- 62.Kiselyov V. V., Berezin V., Maar T. E., Soroka V., Edvardsen K., Schousboe A., Bock E. (1997) J. Biol. Chem. 272, 10125–10134 [DOI] [PubMed] [Google Scholar]
- 63.Mühlenhoff M., Eckhardt M., Bethe A., Frosch M., Gerardy-Schahn R. (1996) Curr. Biol. 6, 1188–1191 [DOI] [PubMed] [Google Scholar]
- 64.Angata K., Suzuki M., Fukuda M. (1998) J. Biol. Chem. 273, 28524–28532 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.