Abstract
Balanced protein synthesis and degradation are crucial for proper cellular function. Protein synthesis is tightly coupled to energy status and nutrient levels by the mammalian target of rapamycin complex 1 (mTORC1). Quality of newly synthesized polypeptides is maintained by the molecular chaperone and ubiquitin-proteasome systems. Little is known about how cells integrate information about the quantity and quality of translational products simultaneously. We demonstrate that cells distinguish moderate reductions in protein quality from severe protein misfolding using molecular chaperones to differentially regulate mTORC1 signaling. Moderate reduction of chaperone availability enhances mTORC1 signaling, whereas stress-induced complete depletion of chaperoning capacity suppresses mTORC1 signaling. Molecular chaperones regulate mTORC1 assembly in coordination with nutrient availability. This mechanism enables mTORC1 to rapidly detect and respond to environmental cues while also sensing intracellular protein misfolding. The tight linkage between protein quality and quantity control provides a plausible mechanism coupling protein misfolding with metabolic dyshomeostasis.
Keywords: Metabolism, mTOR, mTOR Complex (mTORC), Protein Degradation, Protein Synthesis, Molecular Chaperone System, Nutrients, Protein Quality Control, Stress
Introduction
The eukaryotic translational initiation machinery is a tightly controlled system that regulates protein synthesis based on many factors including the availability of growth factors, nutrients, and glucose (1). Accordingly, when there are shortages of these factors, protein synthesis stalls. The molecular pathway responsible for linking the environmental cues and translational control is the mammalian target of rapamycin (mTOR)3 signaling pathway (2, 3). mTOR is an evolutionarily conserved serine/threonine kinase that links environmental status with a host of other cellular processes such as cellular growth, proliferation, metabolism, autophagy, and translational control (4). mTOR is present in two functionally and structurally distinct multiprotein complexes termed mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2) (5). mTORC1 consists of mTOR, Raptor, PRAS40, and mLST8 and is sensitive to rapamycin (6, 7). mTORC2 contains mTOR, Rictor, mSIN1, and mLST8 and is not directly inhibited by rapamycin (8). The pivotal role of mTOR in cellular and organismal homeostasis is reflected in the fact that dysregulation of this pathway results in unrestrained signaling activity in mammals and is associated with the occurrence of disease states including inflammation, cancer, and diabetes (9).
A common theme underlying a variety of stress conditions is the accumulation of misfolded proteins in cells. In response to stress, or heat shock, cells increase the expression of molecular chaperones. Molecular chaperones are essential for protecting nascent polypeptide chains from misfolding, facilitating co- and post-translational folding, assisting in assembly and disassembly of macromolecular complexes, and regulating translocation (10, 11). The concentration of molecular chaperones is titrated closely to the folding requirements within a specific cell type (12), and it has long been suggested that there is a close correlation between stress response and nutrient signaling pathways (13). Adverse environmental and metabolic conditions (including nutrient limitation, hypoxia, and DNA damage) result in a decrease in mTORC1 activity (14). In addition to conserving cellular energy, the reduction in translation that accompanies the decreased mTORC1 activity also prevents the synthesis of unwanted proteins that could interfere with the cellular stress response. However, the effect of stress conditions on mTORC1 signaling appears to be ambiguous. Depending on exposure length, dose/concentration of the stressor, and time between stress stimulus and assay, stress conditions may have distinct effects on mTOR-dependent cellular events. Several stress conditions, such as UV light exposure, H2O2 addition, heat shock, and fluid sheer stress, have been shown to cause an initial increase of mTOR-dependent S6K phosphorylation, with decreases in mTORC1 activity occurring after prolonged or severe exposure (15–17). The importance of this initial up-regulation of mTORC1 activity upon imposition of stressors is not readily apparent. Similarly, it remains unclear how seemingly disparate stress conditions can elicit a similar response of mTORC1 signaling.
The interface between chaperone-mediated stress response and mTORC1-mediated nutrient signaling can be viewed as a central homeostatic mechanism. Despite the obvious importance of this interface, however, the interrelationships between metabolic and cell stress signaling remain poorly understood, and currently, little is known about the role of cytosolic molecular chaperones in the regulation of nutrient signaling pathways. Survival of multicellular organisms depends on the ability to protect against a variety of stressors and the ability to adjust protein synthesis for developmental need. However, the molecular connection between chaperone-mediated stress response and the mTORC1-mediated nutrient signaling pathway remains poorly understood. In particular, it is completely unclear how mTORC1 signaling responds to the accumulation of misfolded proteins in cells. We now demonstrate that accumulation of misfolded proteins in cells, under both acute and chronic conditions, triggers mTORC1 signaling and transiently increases protein synthesis. This is in sharp contrast to other adverse conditions that suppress mTORC1 signaling and may represent an unrecognized cellular adaptation in response to protein quality fluctuations. Importantly, we provide evidence that molecular chaperones coordinate with nutrients to regulate mTORC1 complex assembly. Our results indicate that mTORC1, by sensing chaperone availability in cells, maintains protein homeostasis by linking protein quality and quantity control in cells.
EXPERIMENTAL PROCEDURES
Cell Lines and Reagents
HEK293 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) and cultured at 37 °C. HSF1+/+ and HSF1−/− mouse embryonic fibroblasts were kindly provided by I. J. Benjamin (University of Utah). l-Azetidine-2-carboxylic acid (AZC), l-canavanine, proline, and geldanamycin were purchased from Sigma; [35S]methionine was from Amersham Biosciences; β-galactosidase (β-gal) and β-actin monoclonal antibodies were from Sigma; anti-Hsp70 (SPA810) and anti-Hsp90α (SPA840) were from StressGen; antibodies for phosphorylated and total eIF2α (antibodies 9721 and 9722), S6 (antibodies 2215 and 2217), p70 S6 kinase 1 (S6K1) (antibodies 9234 and 9202), 4EBP1 (antibodies 9459 and 9452), Raptor, (antibody 4978), and Rictor (antibody 2140) were from Cell Signaling. β-Gal activity was measured by the Galacto-Star system (Applied Biosystems). Mammalian expression vectors for β-gal and a truncated version of bovine serum albumin (cBSA) were described previously (26). Constructs containing Myc-mTOR, Myc-Raptor, and Myc-Rictor were obtained from the D. M. Sabatini laboratory via Addgene Inc.
Immunoblotting
Cells were lysed on ice in TBS buffer (50 mm Tris-HCl, 150 mm NaCl, 1 mm EDTA) containing a protease inhibitor mixture tablet, 1% Triton X-100, and 2 units/ml DNase. After incubating on ice for 30 min, the lysates were heated for 10 min in SDS-PAGE sample buffer (50 mm Tris-HCl (pH 6.8), 100 mm dithiothreitol, 2% SDS, 0.1% bromphenol blue, 10% glycerol). Proteins were resolved on SDS-PAGE and transferred to Immobilon-P membranes (Millipore). Membranes were blocked for 1 h in TBS containing 5% BSA followed by incubation with primary antibodies. After incubation with horseradish peroxidase-coupled secondary antibodies, immunoblots were developed using enhanced chemiluminescence (Amersham Biosciences). All experiments were performed at least three times.
Metabolic Labeling
Cells were radiolabeled with [35S]Met (100 μCi/ml) in complete medium for the indicated times. After washing with PBS containing excess cold Met (6.7 mm), cells were frozen in dry ice. Whole cell lysates were made as described in the above section. After resolving on SDS-PAGE, the gel was fixed and dried. Both the image and the quantitation were obtained using a PhosphorImager (Amersham Biosciences). All experiments were performed at least three times.
Immunoprecipitation
Cells were lysed on ice in TBS buffer (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1 mm EDTA, protease inhibitor mixture tablet) containing 1% Triton X-100, 1% Nonidet P-40, or 0.3% CHAPS as indicated. Cell lysates were cleared by centrifugation at 5,000 × g for 5 min. Supernatants were incubated with anti-Myc agarose beads at 4 °C for 60 min. Immunoprecipitates were washed four times with lysis buffer and eluted using 1× SDS-PAGE sample buffer. Samples were resolved on SDS-PAGE followed by immunoblotting. All experiments were performed at least three times.
Fluorescence Imaging
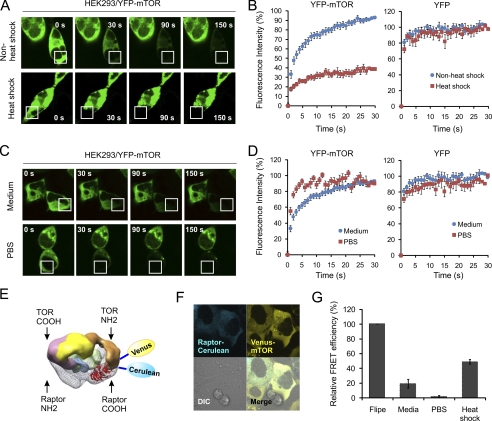
HEK293 cells expressing YFP-mTOR were cultured in glass-bottomed dishes at 37 °C in 5% CO2 in DMEM containing 10% fetal bovine serum. Fluorescence recovery after photobleaching (FRAP) was performed at room temperature on a confocal microscope (LSM 510; Carl Zeiss, Jena, Germany) using a ×40 oil objective and scan zoom = 3. A rectangular region of interest was bleached with 60 iterations and 100% laser power (514-nm argon laser). One image was captured before bleaching. An image was taken every second (514-nm argon laser at 5% power) after bleaching over a 50-s period. For each time point, the fluorescence intensity of the photobleached region of interest was determined using LSM 510 software. Fluorescence loss in photobleaching (FLIP) experiments were also performed on the LSM 510 laser scanning confocal system. Cells were repeatedly bleached at the same defined region, and the whole cell was imaged at intervals of 15 s.
For Föster resonance energy transfer (FRET) experiments, living HEK293 cells expressing Venus-mTOR and Raptor-Cerulean were imaged using the Zeiss LSM510 microscope system. FRET was detected by the method of acceptor photobleaching. Venus and Cerulean were excited by laser light at a 514- or 458-nm wavelength, respectively, by using the multitrack function of the LSM 510 system. To perform multitrack acquisition, two “track” configurations were preset: track 1, acceptor (Venus-mTOR) filter sets (excitation with 514 nm laser; emission passed through an NFT 515 dichroic splitter and a 530–600-nm bandpass filter to PMT 3 (photomultiplier tube 3)); track 2, donor (Raptor-Cerulean) filter set (excitation with 458 nm laser; emission reflected off 515 dichroic splitter and directed through a 470–500-nm bandpass filter to PMT 2). Images from each track were displayed in separate image channels. Cells were then bleached with the laser set at 514 nm and at maximum power for 300 iterations (which corresponds to >80% bleaching), after which postbleaching images were acquired. The fluorescence intensity of donor and acceptor was determined by LSM 510 software before and after photobleaching. After background subtraction, the apparent FRET efficiency was calculated as: apparent FRET efficiency = ((CFPafter − CFPbefore) × YFPbefore) × ((CFPafter × YFPbefore) − (CFPbefore × YFPafter)) − 1, in which the relative CFP increase due to YFP bleaching is corrected for the fraction of YFP bleached. The apparent FRET efficiency was finally expressed relative to control measurements in cells expressing the CFP-YFP fusion protein (FLIPE). All experiments were performed at least three times.
RESULTS
Protein Misfolding Affects Protein Translation
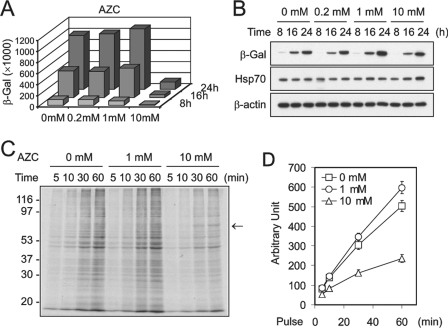
To assess the intracellular adaptation to protein quality variances, we used amino acid analogs to induce protein misfolding. The competitive incorporation between normal amino acids and their analogs enables relatively precise manipulation of the extent of global protein misfolding (18, 19). We followed the fate of β-galactosidase (β-gal) synthesized in the presence of the proline analog AZC. The quality and quantity of β-gal in transfected HEK293 cells were determined respectively by measuring enzyme activity and by immunoblotting with an antibody specific for β-gal. 10 mm AZC, a sublethal but growth-inhibiting concentration, dramatically reduced β-gal activity but caused only a slight decrease in the total amount of synthesized β-gal in cells (Fig. 1, A and B). This discordance is likely due to accumulation of misfolded, enzymatically inactive β-gal in cells (19). To minimize the global disturbance of cellular functions by misfolded proteins, we performed similar experiments using lower concentrations of AZC. Surprisingly, in the presence of 1 mm AZC, we found an increase in both the activity and the total amount of β-gal (Fig. 1, A and B). Although the β-gal activity was increased ∼12.5%, the total amount of β-gal was elevated ∼30%. It is likely that not all of the increased β-gal protein is functional in the presence of AZC.
FIGURE 1.
Effects of amino acid analogs on protein translation. A, HEK293 cells were transfected with plasmids encoding β-gal. 2 h after transfection, AZC was added into the medium with the final concentration as indicated. Cell aliquots were harvested at 8, 16, and 24 h after transfection. The β-gal activity in the whole cell lysates was determined by chemiluminescent assay. B, the total amount of β-gal in the same sample as in A was determined by immunoblotting using a β-gal monoclonal antibody. β-Actin was used as a loading control, and Hsp70 was used as a measure of stress response. C, HEK293 cells were pretreated with AZC for 30 min before labeling with [35S]methionine. The global translation was followed by resolving newly synthesized proteins on a 10% SDS-PAGE. Arrow, approximate size of Hsp70. D, the incorporation of 35S was quantitated by a PhosphorImager. Values are mean ± S.D.
We considered the possibility that protein translation is increased in cells in the presence of certain concentrations of AZC. To test this possibility, we measured the global translation rate by pulsing cells with [35S]methionine in the presence of AZC. 10 mm AZC markedly reduced 35S incorporation as compared with cells treated with the same amount of vehicle (>50% at 60 min labeling; Fig. 1C). In contrast, the presence of 1 mm AZC resulted in ∼20% increase in translation at 60 min of labeling (Fig. 1C). The pattern of translational products as determined by gel fractionation was largely the same in the presence or absence of AZC except that high doses of AZC induced the appearance of several additional bands (likely Hsp70, Fig. 1C, arrow). Taken together, these data suggest that the presence of AZC induces cellular stress and dynamically alters global protein translation.
The incorporation of AZC into synthesizing polypeptides is thought to reduce thermal stability and induce misfolding in a dose-dependent manner. It was therefore surprising to find that translational regulation under these circumstances exhibits a biphasic pattern rather than a gradient of suppression. To confirm that these effects are due to the disturbance of quality of nascent proteins and not to other physiologic responses to AZC, we examined the effects of the arginine analog l-canavanine. Treatment of HEK293 cells with l-canavanine resulted in a similar increase in β-gal levels and [35S]methionine incorporation, although at a higher concentration than that required for AZC (data not shown). The differential dose response could be due either to the fact that AZC is more potent than l-canavanine in altering structural conformations of proteins (19) or to different levels of analog incorporation into nascent proteins (20). In either event, it is likely that the quality of translational products, rather than the concentration of applied amino acid analogs, determines the status of translational regulation.
Regulation of Protein Synthesis in Response to Protein Misfolding Involves mTORC1
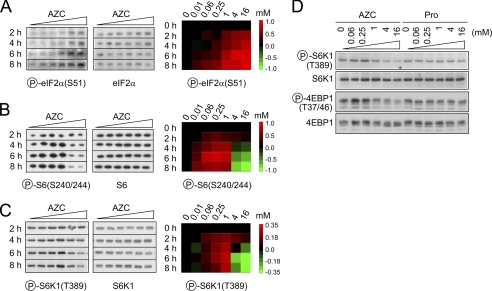
Extensive studies have been devoted to the identification of the mechanisms involved in translational repression under various stress conditions (1). For example, many different types of stress trigger the phosphorylation of eukaryotic initiation factor-2α (eIF2α) at residue Ser-51, resulting in a decrease of translation initiation (21). We thus examined the phosphorylation status of eIF2α in cells treated with AZC at increasing doses and for increasing times. Consistent with an AZC-induced stress response in cells, higher doses and longer treatments of AZC caused a gradient increase of eIF2α phosphorylation in cells (Fig. 2A). No biphasic pattern of eIF2α phosphorylation was detected over a dose range of more than 1,000-fold (from 10 μm to 16 mm) during the 8-h time course. These observations suggest that regulatory nodes independent of eIF2α are responsible for enhanced protein synthesis in the presence of low concentrations of the amino acid analogs.
FIGURE 2.
Modulation of mTORC1 signaling pathway by amino acid analog-induced protein misfolding. A–C, HEK293 cells were treated with AZC at concentrations from 0 to 16 mm. Cell aliquots were harvested every 2 h up to 8 h. Whole cell lysates were immunoblotted using antibodies against total or phosphorylated (circled P) eIF2α (A), S6 (B), and S6K1 (C). After quantitation by densitometry, the fraction of phosphorylated eIF2α was adjusted by total eIF2α. The phosphorylation status was normalized relative to the no treatment control (omitted for clarity) and converted into a heat map by TreeView (right). D, HEK293 cells were treated with AZC or proline at concentrations from 0 to 16 mm for 4 h. See also supplemental Figs. 1 and 2.
We next examined ribosomal protein S6, the phosphorylation of which is closely correlated to translation initiation (22). Interestingly, S6 phosphorylation exhibited a remarkable biphasic pattern in the presence of AZC; higher doses and longer treatments suppressed S6 phosphorylation, whereas lower doses triggered its phosphorylation (Fig. 2B). Similar results were obtained using l-canavanine (supplemental Fig. 1). The phosphorylation of S6 is mediated by p70 S6K1, one of the best characterized targets of mTORC1 (3, 4, 23). Consistent with the phosphorylation status of S6, the presence of AZC caused biphasic phosphorylation of S6K1 at Thr-389, the target residue of mTORC1 (Fig. 2, C and D). Thus, mild reduction in the quality of nascent proteins triggers mTORC1 kinase activity, whereas severe protein misfolding suppresses it.
Amino acids are positive regulators of mTORC1 signaling, although the nature of the sensor remains elusive (24). Several lines of evidence ruled out the possibility that enhanced mTORC1 signaling under low concentrations of AZC is due to altered distribution of normal amino acids in the cytosol. First, our studies were performed in nutrient-rich medium. Second, adding equivalent amounts of proline into the medium did not reproduce the effects of AZC on the phosphorylation status of mTORC1 targets (Fig. 2D). Third, we observed no detectable changes in overall protein degradation in the presence of 1 mm AZC (supplemental Fig. 2). Considering that protein synthesis is increased under these conditions (Fig. 1C), it is unlikely that there are concomitant increases of intracellular amino acid levels under these conditions.
Reduced Chaperone Availability Enhances mTORC1 Signaling
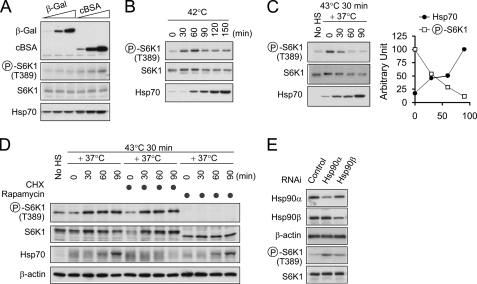
Having shown that mild protein misfolding induced by amino acid analogs enhanced mTORC1 signaling, we were interested in assessing the effect of other forms of protein misfolding on mTORC1 signaling. We first ectopically expressed cBSA, a protein incapable of acquiring its normal conformation in the reducing cytosolic environment (25, 26). Chronic accumulation of cBSA in the cytoplasm triggered the phosphorylation of S6K1 in a dose-dependent manner, whereas equivalent expression of β-gal had negligible effects (Fig. 3A). This result indicates that misfolding of a single protein is able to modulate mTORC1 signaling.
FIGURE 3.
Reduced chaperone availability enhances mTORC1 signaling. A, HEK293 cells were transfected with increasing amounts of β-gal (0.02, 0.1, and 0.5 μg/well in six-well plates) or cBSA (0.1, 0.5, 2.5 μg/well). Whole cell lysates were immunoblotted using antibodies as indicated. Circled P indicates phosphorylation. B, mouse fibroblasts were incubated at 42 °C for the indicated times. Whole cell lysates were immunoblotted using antibodies as indicated. C, mouse fibroblasts were incubated at 43 °C for 30 min followed by recovery at the 37 °C for the indicated times. Whole cell lysates were immunoblotted using antibodies as indicated. No HS, no heat shock. D, mouse fibroblasts were incubated at 43 °C for 30 min followed by recovery at the 37 °C for the indicated times in the absence or presence of 100 μm cycloheximide (CHX). For rapamycin treatment, 20 nm rapamycin was added into the medium prior to heat shock treatment. E, HEK293 cells were transfected with small interference RNA against Hsp90α or Hsp90β. After a 72-h transfection, whole cell lysates were immunoblotted using antibodies as indicated.
We next examined the effects of heat shock, another cellular stressor, on mTORC1 signaling using mouse fibroblasts. Shortly after heat shock treatment (42 °C), we observed a rapid increase of S6K1 phosphorylation (within 30 min; Fig. 3B). The S6K1 phosphorylation was maintained at levels above baseline during heat shock (a slight decrease in S6K1 phosphorylation after 60 min was possibly due to the degradation of total S6K1 molecules as the percentage of S6K1 phosphorylated remained the same). Notably, the surge of S6K1 phosphorylation occurred much earlier than the heat shock-induced Hsp70 expression (<30 min versus >60 min; Fig. 3B). Therefore, it is unlikely that increased Hsp70 expression per se triggered S6K1 phosphorylation.
In contrast to prolonged heat shock, during recovery at 37 °C, S6K1 phosphorylation was gradually reduced to basal levels, although a similar amount of Hsp70 was induced as prolonged heat shock (Fig. 3C). This result led us to hypothesize that chaperone molecules like Hsp70 negatively regulate mTORC1 signaling, at least transiently. Based on this hypothesis, we predicted that mTORC1 should remain responsive to misfolded proteins in the absence of chaperone gene expression. Consistent with this model, treating cells with the protein synthesis inhibitor cycloheximide (CHX) shortly after heat shock did not prevent S6K1 phosphorylation but did completely block the chaperone induction (Fig. 3D). In contrast, rapamycin treatment to block mTOR completely inhibited the S6K1 phosphorylation, indicating that the stress-induced S6K1 phosphorylation occurred through a mTORC1-mediated pathway.
To substantiate the observation that reduced chaperone availability enhances mTORC1 signaling, we used RNAi-mediated knockdown to directly manipulate chaperone levels in cells. Because the basal expression level of Hsp70 is very low under normal conditions, we chose Hsp90 as the knockdown target in these experiments. Due to the abundance of Hsp90 in cells, only ∼50% reduction was obtained for Hsp90α and Hsp90β after a 48-h transfection of small interference RNA (Fig. 3E). However, this moderate reduction of Hsp90 levels was still sufficient to induce a significant increase in S6K1 phosphorylation. Therefore, chronic reduction of chaperone availability, without acute stress exposure, is able to trigger mTORC1 signaling.
Complete Depletion of Chaperoning Capacity Suppresses mTORC1 Signaling
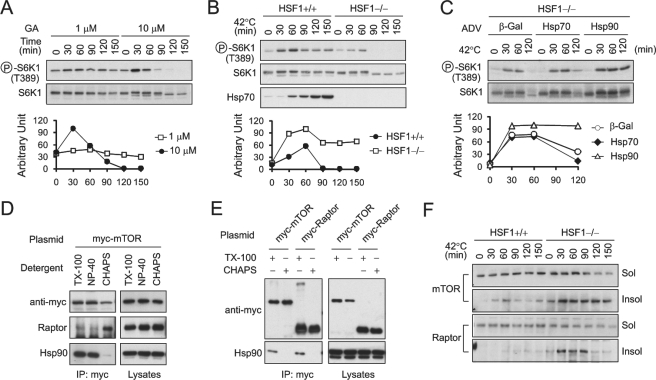
Our discovery that reducing Hsp90 expression enhances mTORC1 signaling is in sharp contrast to the recent report that Hsp90 positively regulates S6K1 phosphorylation in mammalian cells (27). Ohji et al. (27) demonstrated that geldanamycin (GA), a potent and specific Hsp90 inhibitor, suppresses binding of Raptor to Hsp90 and decreases S6K1 phosphorylation. To resolve this apparent contradiction in observations, we re-examined the effects of GA on mTORC1 signaling by performing a time course experiment using different doses of GA. We found that only prolonged GA treatment (>2 h) and high concentrations (10 μm) caused the suppression of S6K1 phosphorylation (Fig. 4A). Remarkably, there was an initial rapid surge of S6K1 phosphorylation shortly after GA treatment (within 30 min) followed by a continual decrease in phosphorylation, similar to the enhanced mTORC1 signaling observed after heat shock treatment (Fig. 3B). This triggering effect was still noticeable even with GA treatment at low concentrations (1 μm; Fig. 4A). Notably, the amount of total S6K1 protein was reduced in the presence of GA, suggesting that S6K1 is one of the clients of Hsp90.
FIGURE 4.
Depleted chaperone availability suppresses mTORC1 signaling. A, mouse fibroblasts were treated with 1 or 10 μm GA for the indicated times. Whole cell lysates were immunoblotted using antibodies as indicated. Circled P indicates phosphorylation. B, HSF1+/+ and HSF1−/− cell lines were incubated at 42 °C for the indicated times. Whole cell lysates were immunoblotted using antibodies as indicated. C, HSF1−/− cells were infected with 20 multiplicity of infection recombinant adenovirus expressing the indicated genes for 24 h. Infected cells were then incubated at 42 °C for the indicated times. Whole cell lysates were immunoblotted using antibodies as indicated. D, HEK293 cells were transfected with Myc-mTOR followed by extraction with 1% Triton X-100 (TX-100), 1% Nonidet P-40 (NP-40), or 0.3% CHAPS. The mTOR complexes were precipitated using anti-Myc antibodies. Both lysates and immunoprecipitates were immunoblotted using antibodies as indicated. E, HEK293 cells were transfected with plasmids encoding Myc-mTOR or Myc-Raptor. Similar experiments as in D were performed to determine the chaperone binding affinity. F, HSF1+/+ and HSF1−/− cell lines were incubated at 42 °C for the indicated times. Whole cell lysates were fractionated using 1% Triton X-100 into soluble and insoluble fractions. Immunoblotting was performed using antibodies as indicated. See also supplemental Fig. 3.
The biphasic pattern of S6K1 phosphorylation after GA treatment is reminiscent of our observations of S6K1 phosphorylation in the presence of the amino acid analog AZC (Fig. 2). Both treatments share a common feature: a potent heat shock response followed by deficient chaperone functionality, presumably because newly synthesized chaperone molecules are nonfunctional in the presence of AZC or GA. We reasoned that both GA and AZC treatments eventually depleted chaperone availability by permanently blocking the functionality of chaperone molecules. Therefore, the two phases in mTORC1 signaling observed in our previous experiments might represent a switch of chaperone availability from modest reduction to total depletion.
To test this hypothesis, we used an immortalized fibroblast cell line derived from HSF1−/− mice (20, 28). HSF1−/− cells cannot induce a heat shock response under stress conditions and therefore experience diminished chaperone availability under prolonged stress conditions. Heat shock (42 °C) induced efficient Hsp70 expression in the HSF1+/+ cells but not the HSF1−/− cells (Fig. 4B). Both cell groups exhibited a rapid increase in S6K1 phosphorylation shortly after heat shock, further confirming the notion that reduced chaperone availability enhances mTORC1 signaling. In contrast to HSF1+/+ cells, in which the S6K1 phosphorylation was sustained at high levels during heat shock, HSF1−/− cells demonstrated a sudden depletion of S6K1 phosphorylation after the initial increase (Fig. 4B). This pattern is strikingly similar to that of GA treatment, where an increase in mTORC1 signaling occurred within 60 min followed by a drastic reduction around 90 min after treatment (Fig. 4, A and B).
In an effort to prove the essential role of molecular chaperones in mTORC1 signaling, we introduced chaperones back into HSF1−/− cells using recombinant adenoviruses. After prolonged heat shock treatment, only the viruses expressing Hsp90 and Hsp70, but not the control virus expressing β-gal, were able to rescue the S6K1 phosphorylation (Fig. 4C). Taken together, our results indicate that complete depletion of chaperone capacity suppresses mTORC1 signaling.
Molecular Chaperones Are Required for Appropriate mTORC1 Assembly
To determine how changing chaperone availability affects mTORC1 signaling, we first examined the integrity of mTORC1 following treatment with either AZC or heat shock. Despite considerable effort, we failed to detect any difference in the amount of Raptor coimmunoprecipitated with mTOR under these conditions (supplemental Fig. 3). We suspect that this is a reflection of the fact that protein-protein interactions detected within the cell lysates may not faithfully represent the mTORC1 integrity in living cells. It is noteworthy that many commonly used detergents readily break down mTOR complexes in lysates (7). It has been suggested that hydrophobicity of the residues at the interface of mTOR-Raptor plays a role in mTORC1 assembly. Consistent with this idea, the interaction of these proteins can be preserved in zwitterionic detergents such as CHAPS (7). Surprisingly, we observed that Hsp90 was associated with mTOR only after extraction using Triton X-100 or Nonidet P-40 (Fig. 4D). 0.3% CHAPS, which maintained mTORC1 integrity, largely abolished Hsp90 binding. Raptor exhibited the same detergent-dependent, mutually exclusive pattern of Hsp90 versus mTOR binding (Fig. 4E). These observations indicate that Hsp90 interacts with isolated components of mTORC1, but not with the assembled complex itself. These characteristics fit with the adopted molecular chaperone definition, a protein that has a functional effect on another protein-protein complex without becoming part of the final operative structure.
Hsp90 is among the most abundant proteins in cells, occupying ∼1–2% of total cellular proteins. The major role of Hsp90 is to suppress protein aggregation by retaining target proteins within the cytosol in the soluble form (29). Recent biochemical analysis indicates that mTORC1 may oligomerize to form complexes of higher order, although the significance of this oligomerization remains to be determined (3). The aggregation-prone property of mTOR complexes prompted us to test whether Hsp90 plays a role in preventing mTORC1 from aggregation and subsequent inactivation. We fractionated lysates of heat-shocked HSF1+/+ and HSF1−/− cells into soluble and insoluble fractions. CHAPS detergent was used to maintain the mTORC1 integrity. The majority of mTOR and Raptor molecules were recovered in the soluble faction of HSF1+/+ cells (Fig. 4F). In contrast, heat shock treatment resulted in progressive accumulation of mTOR and Raptor in the insoluble fraction of HSF1−/− cells. Therefore, diminished chaperone availability due to thermal stress causes relocation of mTOR and its regulatory components into the insoluble fraction, presumably by aggregation.
mTORC1 Undergoes Dynamic Regulation in Living Cells
Following heat shock treatment, a portion of mTOR became insoluble in HSF1+/+ cells (Fig. 4F). Interestingly, the time course of changing solubility of mTOR mirrored the enhanced mTORC1 signaling in these cells (Fig. 4B). It is intuitively appealing to assume that reduced chaperone availability results in more mTORC1 complex formation and thereby enhances mTORC1 signaling, whereas complete chaperone depletion leads to severe mTORC1 aggregation and consequently suppresses mTORC1 signaling (Fig. 4F). Therefore, to elucidate the pattern of mTORC1 signal regulation, we analyzed mTORC1 dynamics in living cells.
We employed live cell imaging to visualize the behavior of mTORC1 in response to changing chaperone availability. After attempting multiple strategies to create functional mTOR fusion proteins, we validated a successful solution by fusing one copy of YFP to the NH2 terminus of mTOR and including a flexible linker between them. The recombinant YFP-mTOR was able to bind endogenous Raptor and endogenous Rictor (supplemental Fig. 4). These observations indicated that the YFP-mTOR fusion protein maintains the functionality of endogenous mTOR by forming complexes with other regulatory components.
We investigated the dynamics of mTOR under three different conditions. First, we established the mobility of YFP-mTOR under normal growth conditions. These experiments provide basic information about whether the majority of mTOR molecules in cells are in the free form or within the complex. Second, we asked how mTOR dynamics change upon reduction of chaperone availability, such as after heat shock treatment. Third, we investigated the effects of nutrients on mTOR dynamics to gain insight into the regulatory mechanisms of mTORC1 signaling.
To analyze the mobility of YFP-mTOR under normal conditions, we first performed fluorescence loss in photobleaching (FLIP). For FLIP, a small region of the cell YFP-mTOR fluorescence was repeatedly photobleached. Over time, YFP-mTOR fluorescence in the entire cell was steadily and uniformly lost, whereas fluorescence of adjacent cells was unaffected (Fig. 5A). These results indicate that YFP-mTOR has access to the region of photobleaching, without any significant immobilized population. We then performed FRAP to quantitate the mobility rate of YFP-mTOR. Under normal growth conditions, the t½ of YFP-mTOR recovery is ∼4.5 s (Fig. 5B). Parameters that directly influence t½ include viscosity (or crowdedness of the environment), size of the protein complex, protein-protein interactions, or combinations of these variables (30). Thus, even modest changes in t½ can be due to biologically significant changes.
FIGURE 5.
mTORC1 dynamics in living cells. A, YFP-mTOR-transfected HEK293 cells were heat-shocked by incubating at 42 °C for 1 h. FLIP analysis was performed to measure the mobility of mTOR. B, YFP-N1- or YFP-mTOR-transfected HEK293 cells were heat-shocked by incubating at 42 °C for 1 h. FRAP analysis was performed to measure the mobility of mTOR. C, YFP-mTOR-transfected HEK293 cells were incubated in PBS for 30 min before FLIP analysis. D, YFP-N1 or YFP-mTOR-transfected HEK293 cells were incubated in PBS for 30 min before FRAP analysis. DIC, differential interference contrast. E, diagram of interaction between Venus-mTOR and Raptor-Cerulean (modified from Ref. 33). F, HEK293 cells were co-transfected with plasmids encoding Venus-mTOR and Raptor-Cerulean. Co-localization of both proteins was examined using blue channel for Raptor and yellow channel for mTOR. G, HEK293 cells were co-transfected with plasmids encoding Venus-mTOR and Raptor-Cerulean. FRET efficiency was calculated as the percentage of positive control FLIP. See also supplemental Figs. 4 and 5. Error bars indicate S.E.
We investigated the consequence of thermal stress on the mobility of YFP-mTOR. Incubation of cells at 42 °C for 60 min resulted in a significant reduction of YFP-mTOR mobility (Fig. 5, A and B) with a resulting t½ > 30 s. This was not due to the increased crowdedness of cytoplasm after heat shock because native YFP demonstrated little change in mobility under the same conditions (Fig. 5B). These data support the notion that heat shock treatment results in mTOR protein oligomerization or complex formation that in turn decreases the diffusibility of mTOR within the cell.
Given the fact that mTORC1 complex formation is a regulated process, we predicted that the presence of nutrients should facilitate mTORC1 complex formation, whereas withdrawal of nutrients should enhance the mTOR mobility because of less complex formation. To test this possibility, we incubated cells in PBS for 10 min before FLIP and FRAP analysis. Remarkably, the absence of nutrients led to much higher mobility of YFP-mTOR in cells (Fig. 5, C and D) with t½ < 1 s. Once again, this was not due to the changing viscosity of cytosol in the absence of nutrients because the mobility of YFP protein demonstrated little change in the absence of nutrients. Therefore, both nutrients and chaperone availability affect mTOR mobility, possibly by regulating mTOR complex formation. Further supporting this notion, recent studies report that nutrient signaling relies on Rag GTPase-mediated mTORC1 recruitment into lysosome-associated Rheb, forming an active super mTORC1 complex (31–33).
Both Nutrients and Chaperones Regulate the mTOR-Raptor Interaction
Having found that both nutrients and chaperones control YFP-mTOR mobility in living cells, we asked whether this mobility change represented mTORC1 complex formation. To this end, we employed FRET to examine the mTOR-Raptor interaction (31). Because Cerulean and Venus are a better combination for FRET analysis, we chose this pair of GFP variants as a donor and an acceptor (32). Studies from cryo-EM have indicated that TOR and KOG1 (the yeast homolog of Raptor) form a head-to-tail ring structure (33). We therefore fused Cerulean to the carboxyl terminus of Raptor (Raptor-Cerulean) and fused Venus to the amino terminus of mTOR (Venus-mTOR) so that both CFP and YFP can be in close proximity when mTOR and Raptor form a complex (Fig. 5E). Raptor-Cerulean functions as endogenous Raptor, as indicated by its capability to coimmunoprecipitate with endogenous mTOR (supplemental Fig. 5).
We first examined the co-localization of mTOR and Raptor by co-transfecting HEK293 cells with plasmids encoding Venus-mTOR and Raptor-Cerulean. As shown in Fig. 5F, both mTOR and Raptor had an almost exclusive cytosolic localization. Because of the rapid mobility of mTOR in cells, co-localization with Raptor does not necessarily mean the formation of an mTORC1 complex. We therefore used the acceptor photobleaching method to measure the FRET efficiency between mTOR and Raptor. In comparison with a positive FRET control (FLIPE) in which both Venus and Cerulean were fused in a single polypeptide (33), we observed ∼20% of FRET signal for mTOR-Raptor under normal growth conditions (Fig. 5G). In complete agreement with the altered mobility of mTOR, nutrient withdrawal dramatically suppressed FRET efficiency, whereas heat shock treatment almost doubled the FRET efficiency (Fig. 5G). These results indicate that both nutrients and chaperone availability control mTORC1 complex formation.
Molecular Chaperones Coordinate with Nutrient Signals in Regulating mTORC1 Signaling
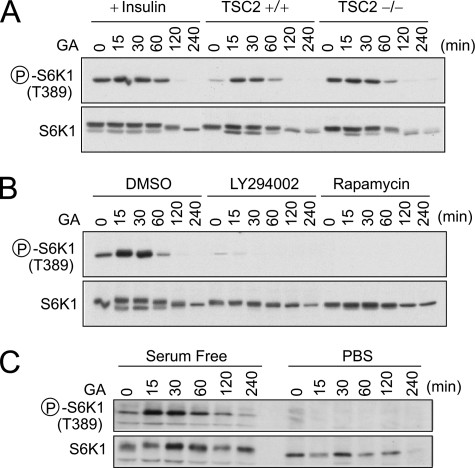
The dynamic assembly of mTORC1 complexes implies that the formation of mTORC1 relies on recycling of existing but not newly synthesized components. Supporting this notion, mTOR demonstrated a very low turnover rate (t½ > 8 h) in cells under normal growth conditions (supplemental Fig. 6). These data are consistent with a model in which nutrients trigger mTORC1 complex formation, whereas molecular chaperones provide a continuous supply of responsive mTOR components. This dynamic remodeling mechanism indicates a strong coordination between nutrients and chaperones in controlling mTORC1 signaling. We have repeatedly demonstrated the indispensable role of chaperones in nutrient-mediated mTORC1 signaling (Fig. 4, A and B). To test whether hyperactive mTORC1 signaling is able to circumvent the requirement of chaperones, we treated cells with high concentrations of insulin (200 nm). After prolonged GA treatment (to inhibit Hsp90), cells were totally resistant to insulin stimulation (Fig. 6A). Long term GA treatment inevitably caused S6K1 degradation, but the phosphorylation ablation was far more rapid than the diminishing total levels of S6K1. Similar results were obtained using TSC2−/− mouse embryonic fibroblasts in which mTORC1 activity is constitutively active (34), indicating that chaperones are required for mTORC1 activity even under conditions of maximal upstream activation of mTORC1.
FIGURE 6.
Coordination between molecular chaperone and nutrients in regulating mTORC1 signaling. A, TSC2+/+ and TSC2−/− cells were treated with 10 μm GA treatment followed by immunoblotting using antibodies as indicated. For insulin stimulation, TSC2+/+ cells were preincubated in the presence of 200 nm insulin prior to GA treatment. Circled P indicates phosphorylation. B, mouse fibroblasts were treated with 50 μm LY294002, 20 nm rapamycin, or an equal amount of dimethyl sulfoxide (DMSO) for 10 min prior to 10 μm GA treatment. Whole cell lysates were immunoblotted using antibodies as indicated. C, mouse fibroblasts were incubated in serum-free medium overnight or in PBS for 30 min prior to 10 μm GA treatment. See also supplemental Fig. 6.
We next examined whether reducing chaperone availability alone is able to trigger mTORC1 signaling. We blocked mTORC1 upstream signals by applying a PI3K inhibitor LY294002 before GA treatment. As shown in Fig. 6B, PI3K inhibition completely prevented the initial surge of S6K1 phosphorylation shortly after GA treatment. Thus, reducing chaperone availability alone does not automatically lead to enhanced mTORC1 signaling. Instead, it relies on the presence of upstream signals such as PI3K-Akt. Consistent with heat shock treatment, GA-induced increase of S6K1 phosphorylation was also rapamycin-sensitive (Fig. 6B).
To define the minimal requirement of upstream signals for GA-induced mTORC1 signaling, we examined S6K1 phosphorylation under either serum depletion or nutrient withdrawal. Interestingly, serum depletion did not prevent mTORC1 from activation after GA treatment (Fig. 6C). Only nutrient withdrawal after a 10-min PBS incubation completely suppressed GA treatment-induced mTORC1 signaling. Taken together, our results indicate that nutrients and chaperones rely on each other in controlling mTORC1 signaling.
Enhanced mTORC1 Signaling in Mice with Accumulation of Misfolded Proteins
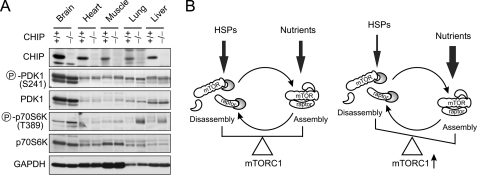
The chaperone-mediated dynamic remodeling of mTORC1 prompted us to examine whether the chronic accumulation of misfolded proteins in mice also enhances mTORC1 signaling. CHIP (carboxyl terminus of Hsp70/Hsp90-interacting protein) has been identified as an important quality control ubiquitin ligase (35, 36). In the absence of CHIP, the global chaperone network is challenged by the accumulation of misfolded proteins (26, 37). Mice lacking CHIP have a markedly reduced life span, along with accelerated age-related pathophysiological phenotypes (38). We examined mTORC1 signaling in various tissues isolated from CHIP−/− mice and wild-type littermates. We observed that the phosphorylation of S6K1 on Thr-389 was markedly increased in CHIP−/− mice (Fig. 7A). This is quite specific because the phosphorylation of PDK1 remained unchanged. These results indicate that the accumulation of misfolded proteins in mice also induces mTORC1 signaling.
FIGURE 7.
Accumulation of misfolded proteins triggers mTORC1 signaling in mice. A, immunoblotting analysis of tissue samples from CHIP+/+ and CHIP−/− mice at 3 months of age. Homogenates were prepared from brain, heart, muscle, lung, and liver. Protein extracts were resolved by SDS-PAGE and blotted for antibodies as indicated. Circled P indicates phosphorylation. B, proposed model for the coordination between molecular chaperones and nutrients in regulating mTORC1 signaling. Both chaperones and nutrients control the balance between mTORC1 assembly and disassembly. Under conditions of excess nutrition or reduced chaperone availability, the increased mTORC1 signaling provides an explanation about how chronic stress fuels metabolic dyshomeostasis.
DISCUSSION
The results presented in this report point to a new paradigm in the regulation of mTORC1 signaling in which chaperone availability directly affects mTORC1 signaling (Fig. 7B). Under normal growth conditions, mTORC1 activity relies on the coordination between molecular chaperones and nutrients. However, in the absence of chaperone availability, mTORC1 is unresponsive to nutrient stimulation. Conversely, higher levels of nutrients or reduced chaperone availability result in increased mTORC1 singling. Thus, mTORC1 perceives proteotoxic stress based on an order-of-severity sensing mechanism.
mTORC1 Links Protein Quality and Quantity Control
The initial increase of mTORC1 signaling upon exposure to adverse conditions was first reported over a decade ago. Work by the Blenis group (16) demonstrated that heat shock treatment unexpectedly increased phosphorylation of S6K1 in mouse fibroblasts, a finding corroborated by Lin et al. (39) in 1997. Likewise, multiple studies have demonstrated that H2O2 treatment of cells regulates S6K activity in a dose- and time-dependent manner (40, 41), similar to our results using amino acid analog treatment. The initial up-regulation of mTORC1 signaling upon imposition of stressors should not be viewed as the cell misinterpreting a life-threatening insult. Rather, this feature enables the cell to distinguish physiological fluctuations in the quality of translational products from the devastating accumulation of misfolded proteins. It is generally accepted that global protein translation is suppressed in response to severe stress conditions, but little is known about how cells respond to fluctuations in the quality of translational products. Like the variation of nutrient availability in the environment, there is a wide fluctuation in the quality of synthesized proteins during cell growth and organismal development. Based on our observations, we propose that once the buffering capacity of chaperones is exceeded, mTORC1 kinase activity is quickly attenuated. Thus, the positive regulation of mTORC1 signaling by misfolded proteins represents an unrecognized cellular adaptation in response to reduced protein quality.
mTORC1 Senses Chaperone Availability
Consistent with other reports (14, 27), we demonstrate here that severe stress conditions suppress mTORC1 signaling. Interestingly, the transition from up-regulation to down-regulation occurs rapidly (Fig. 4, A and B). We reason that there is a threshold of chaperone availability for the mTORC1-mediated sensing mechanism. It is likely that folding status of mTOR determines the threshold. mTOR and other regulator components like Raptor are chaperone clients, as evidenced by the selective binding to Hsp90 when not forming the complex. In agreement with the role of Hsp90 in the ordered assembly and disassembly of large multisubunit complexes (42), this binding shields the hydrophobic region of interacting components that maintains their solubility. Lack of chaperone availability may result in mTOR aggregation. Indeed, we observed a progressive change of mTOR solubility in heat-shocked HSF1−/− cells (Fig. 4D). This provides an explanation for the unresponsiveness of mTORC1 to high insulin levels under the depletion of chaperone availability.
Dynamic Remodeling of mTORC1
mTOR forms a huge complex (up to ∼2 MDa) with other subunits to achieve signal transmission (4, 43). Such multicomponent complexes present special challenges for modulatory signaling pathways, which must sense both increasing and decreasing signal intensities and must also terminate and reinitiate signaling efficiently. A dynamic mode of mTORC1 complexes would fulfill this regulatory requirement. According to this scheme, termination of mTORC1 signaling would not require a separate mechanism but rather would be an intrinsic consequence of continuous disassembly of mTORC1. Indeed, we observed a high degree of mobility of mTOR molecules within the crowded cell interior, suggesting that mTOR molecules undergo rapid exchange between the complexes and free subunits (Fig. 5). Importantly, mTOR mobility is directly affected by both nutrients and chaperone availability. FRET analysis further confirmed that the changing mobility of mTOR is due to mTOR-Raptor interaction. Although it is challenging to recapitulate the mTORC1 dynamics in cell lysates using a biochemical approach (due to possible altered chaperone functionality in solution, detergent-affected protein-protein interactions, and a lack of lipid hydrophobicity), we conclude from our findings that mTORC1 complex assembly is a highly dynamic process inside cells, which consists of an efficient “on demand” assembly by nutrients and a continuous disassembly by molecular chaperones. Analogous to chaperone-regulated HSF1 functionality, the dynamic remodeling of mTORC1 signaling not only ensures tight control, dynamic range, and a rapidly reversible response in sensing upstream signals but provides an elegant mechanism linking protein quality and quantity control in cells. The interconnection of different signaling pathways that feed back through a shared pool of chaperones provides the recurrent framework for adaptation and survival of cells in a changing environment.
Coordination between Nutrients and Chaperones in Regulating mTORC1 Signaling
As illustrated in Fig. 7B, chaperone-mediated mTORC1 disassembly coordinates with nutrient signaling-triggered assembly. This mechanism allows continuous sensing of extracellular nutrients and intracellular protein homeostasis. It remains to be determined how chaperone molecules actively remodel the mTORC1 complex. HSPs may simply “disaggregate” the mTORC1 complex while maintaining a continuous supply of signaling components. An important implication of the model of mTORC1 regulation presented in this study is the synergistic effect of nutrient overload and chaperone stress in metabolic dyshomeostasis. Unrestrained mTOR activity in mammals is associated with the occurrence of disease states including inflammation, cancer, and diabetes (23). By contrast, decreased mTOR signaling by genetic or pharmaceutical approaches (such as rapamycin) has been shown to extend lifespan in a variety of organisms (44–47). Interestingly, a robust stress response is also required for life span extension in these organisms (48, 49). Therefore, there is a close correlation between stress response and nutrient signaling in regulating metabolic homeostasis.
Under physiological conditions, depletion of chaperone availability is rare. In contrast, chronic reduction of chaperone functionality often occurs during aging processes (12). The up-regulation of mTORC1 signaling after reduction of chaperone availability could be beneficial at the initial stage to maintain protein homeostasis. However, prolonged mTORC1 activation might cause metabolic dysregulation. Consistent with this notion, CHIP−/− mice exhibited accelerated aging phenotypes as well as increased mTORC1 signaling (38). It is possible that accumulation of misfolded proteins might fuel aging processes by modulating the mTORC1 signaling pathway.
In summary, our results unveiled a molecular linkage between protein quality and quantity control in cells. Further understanding of the interaction between stress response and the mTOR signaling pathway may provide opportunities for therapeutic avenues to aging and age-related diseases.
Acknowledgments
We thank Dr. I. J. Benjamin (University of Utah) for providing HSF1+/+ and HSF1−/− mouse embryonic fibroblasts and Dr. D. J. Kwiatkowski (Harvard Medical School) for TSC2+/+ and TSC2−/− mouse embryonic fibroblasts.
This work was supported, in whole or in part, by the Intramural Research Program of the National Institutes of Health through NIAID (to J. R. B. and J. W. Y.) and National Institutes of Health Grants GM61728, AG024282, and DK056350 (to C. P.). This work was also supported by Cornell Startup Research Funds and the Ellison Medical Foundation (to S. B. Q.).
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–6.
- mTOR
- mammalian target of rapamycin
- mTORC1
- mTOR complex 1
- mTORC2
- mTOR complex 2
- AZC
- l-azetidine-2-carboxylic acid
- cBSA
- truncated version of bovine serum albumin
- CHIP
- carboxyl terminus of Hsp70/Hsp90-interacting protein
- FLIP
- fluorescence loss in photobleaching
- FRAP
- fluorescence recovery after photobleaching
- GA
- geldanamycin
- HSF1
- heat shock factor 1
- Hsp
- heat shock protein
- PDK1
- pyruvate dehydrogenase kinase isozyme 1
- S6K1
- p70 S6 kinase 1.
REFERENCES
- 1.Holcik M., Sonenberg N. (2005) Nat. Rev. Mol. Cell Biol. 6, 318–327 [DOI] [PubMed] [Google Scholar]
- 2.Gingras A. C., Raught B., Sonenberg N. (2004) Curr. Top. Microbiol. Immunol. 279, 169–197 [DOI] [PubMed] [Google Scholar]
- 3.Wullschleger S., Loewith R., Hall M. N. (2006) Cell 124, 471–484 [DOI] [PubMed] [Google Scholar]
- 4.Sarbassov D. D., Ali S. M., Sabatini D. M. (2005) Curr. Opin. Cell Biol. 17, 596–603 [DOI] [PubMed] [Google Scholar]
- 5.Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J. L., Bonenfant D., Oppliger W., Jenoe P., Hall M. N. (2002) Mol. Cell 10, 457–468 [DOI] [PubMed] [Google Scholar]
- 6.Hara K., Maruki Y., Long X., Yoshino K., Oshiro N., Hidayat S., Tokunaga C., Avruch J., Yonezawa K. (2002) Cell 110, 177–189 [DOI] [PubMed] [Google Scholar]
- 7.Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2002) Cell 110, 163–175 [DOI] [PubMed] [Google Scholar]
- 8.Sarbassov D. D., Ali S. M., Kim D. H., Guertin D. A., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2004) Curr. Biol. 14, 1296–1302 [DOI] [PubMed] [Google Scholar]
- 9.Inoki K., Li Y., Zhu T., Wu J., Guan K. L. (2002) Nat. Cell Biol. 4, 648–657 [DOI] [PubMed] [Google Scholar]
- 10.Bukau B., Weissman J., Horwich A. (2006) Cell 125, 443–451 [DOI] [PubMed] [Google Scholar]
- 11.Frydman J., Nimmesgern E., Ohtsuka K., Hartl F. U. (1994) Nature 370, 111–117 [DOI] [PubMed] [Google Scholar]
- 12.Morimoto R. I. (2008) Genes Dev. 22, 1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neel J. V. (1999) Nutr. Rev. 57, S2–9 [DOI] [PubMed] [Google Scholar]
- 14.Reiling J. H., Sabatini D. M. (2006) Oncogene 25, 6373–6383 [DOI] [PubMed] [Google Scholar]
- 15.Ding M., Li J., Leonard S. S., Shi X., Costa M., Castranova V., Vallyathan V., Huang C. (2002) Mol. Cell. Biochem. 234–235, 81–90 [PubMed] [Google Scholar]
- 16.Jurivich D. A., Chung J., Blenis J. (1991) J. Cell. Physiol. 148, 252–259 [DOI] [PubMed] [Google Scholar]
- 17.Kraiss L. W., Ennis T. M., Alto N. M. (2001) J. Surg Res. 97, 20–26 [DOI] [PubMed] [Google Scholar]
- 18.Goldberg A. L., Dice J. F. (1974) Annu. Rev. Biochem. 43, 835–869 [DOI] [PubMed] [Google Scholar]
- 19.Trotter E. W., Kao C. M., Berenfeld L., Botstein D., Petsko G. A., Gray J. V. (2002) J. Biol. Chem. 277, 44817–44825 [DOI] [PubMed] [Google Scholar]
- 20.Qian S. B., Reits E., Neefjes J., Deslich J. M., Bennink J. R., Yewdell J. W. (2006) J. Immunol. 177, 227–233 [DOI] [PubMed] [Google Scholar]
- 21.Ron D. (2002) J. Clin. Invest. 110, 1383–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruvinsky I., Meyuhas O. (2006) Trends Biochem. Sci. 31, 342–348 [DOI] [PubMed] [Google Scholar]
- 23.Inoki K., Corradetti M. N., Guan K. L. (2005) Nat. Genet. 37, 19–24 [DOI] [PubMed] [Google Scholar]
- 24.Proud C. G. (2004) Biochem. Biophys. Res. Commun. 313, 429–436 [DOI] [PubMed] [Google Scholar]
- 25.Guo Y., Guettouche T., Fenna M., Boellmann F., Pratt W. B., Toft D. O., Smith D. F., Voellmy R. (2001) J. Biol. Chem. 276, 45791–45799 [DOI] [PubMed] [Google Scholar]
- 26.Qian S. B., McDonough H., Boellmann F., Cyr D. M., Patterson C. (2006) Nature 440, 551–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohji G., Hidayat S., Nakashima A., Tokunaga C., Oshiro N., Yoshino K., Yokono K., Kikkawa U., Yonezawa K. (2006) J. Biochem. 139, 129–135 [DOI] [PubMed] [Google Scholar]
- 28.McMillan D. R., Xiao X., Shao L., Graves K., Benjamin I. J. (1998) J. Biol. Chem. 273, 7523–7528 [DOI] [PubMed] [Google Scholar]
- 29.Whitesell L., Lindquist S. L. (2005) Nat. Rev. Cancer 5, 761–772 [DOI] [PubMed] [Google Scholar]
- 30.Lippincott-Schwartz J., Snapp E., Kenworthy A. (2001) Nat. Rev. Mol. Cell Biol. 2, 444–456 [DOI] [PubMed] [Google Scholar]
- 31.Piston D. W., Kremers G. J. (2007) Trends Biochem. Sci. 32, 407–414 [DOI] [PubMed] [Google Scholar]
- 32.Rizzo M. A., Springer G., Segawa K., Zipfel W. R., Piston D. W. (2006) Microsc. Microanal. 12, 238–254 [DOI] [PubMed] [Google Scholar]
- 33.Adami A., García-Alvarez B., Arias-Palomo E., Barford D., Llorca O. (2007) Mol. Cell 27, 509–516 [DOI] [PubMed] [Google Scholar]
- 34.Kwiatkowski D. J., Manning B. D. (2005) Hum. Mol. Genet 14, Spec. No. 2, R251–R258 [DOI] [PubMed] [Google Scholar]
- 35.Cyr D. M., Höhfeld J., Patterson C. (2002) Trends Biochem. Sci. 27, 368–375 [DOI] [PubMed] [Google Scholar]
- 36.McDonough H., Patterson C. (2003) Cell Stress Chaperones 8, 303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dai Q., Zhang C., Wu Y., McDonough H., Whaley R. A., Godfrey V., Li H. H., Madamanchi N., Xu W., Neckers L., Cyr D., Patterson C. (2003) EMBO J. 22, 5446–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Min J. N., Whaley R. A., Sharpless N. E., Lockyer P., Portbury A. L., Patterson C. (2008) Mol. Cell. Biol. 28, 4018–4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin R. Z., Hu Z. W., Chin J. H., Hoffman B. B. (1997) J. Biol. Chem. 272, 31196–31202 [DOI] [PubMed] [Google Scholar]
- 40.Bae G. U., Seo D. W., Kwon H. K., Lee H. Y., Hong S., Lee Z. W., Ha K. S., Lee H. W., Han J. W. (1999) J. Biol. Chem. 274, 32596–32602 [DOI] [PubMed] [Google Scholar]
- 41.Huang C., Li J., Ke Q., Leonard S. S., Jiang B. H., Zhong X. S., Costa M., Castranova V., Shi X. (2002) Cancer Res. 62, 5689–5697 [PubMed] [Google Scholar]
- 42.Frydman J. (2001) Annu. Rev. Biochem. 70, 603–647 [DOI] [PubMed] [Google Scholar]
- 43.Bentzinger C. F., Romanino K., Cloëtta D., Lin S., Mascarenhas J. B., Oliveri F., Xia J., Casanova E., Costa C. F., Brink M., Zorzato F., Hall M. N., Rüegg M. A. (2008) Cell Metab. 8, 411–424 [DOI] [PubMed] [Google Scholar]
- 44.Vellai T., Takacs-Vellai K., Zhang Y., Kovacs A. L., Orosz L., Muller F. (2003) Nature 426, 620. [DOI] [PubMed] [Google Scholar]
- 45.Kapahi P., Zid B. M., Harper T., Koslover D., Sapin V., Benzer S. (2004) Curr. Biol. 14, 885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaeberlein M., Powers R. W., 3rd, Steffen K. K., Westman E. A., Hu D., Dang N., Kerr E. O., Kirkland K. T., Fields S., Kennedy B. K. (2005) Science 310, 1193–1196 [DOI] [PubMed] [Google Scholar]
- 47.Harrison D. E., Strong R., Sharp Z. D., Nelson J. F., Astle C. M., Flurkey K., Nadon N. L., Wilkinson J. E., Frenkel K., Carter C. S., Pahor M., Javors M. A., Fernandez E., Miller R. A. (2009) Nature 460, 392–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hsu A. L., Murphy C. T., Kenyon C. (2003) Science 300, 1142–1145 [DOI] [PubMed] [Google Scholar]
- 49.Morley J. F., Morimoto R. I. (2004) Mol. Biol. Cell 15, 657–664 [DOI] [PMC free article] [PubMed] [Google Scholar]