Abstract
In actinomycetes, the onset of secondary metabolite biosynthesis is often triggered by the quorum-sensing signal γ-butyrolactones (GBLs) via specific binding to their cognate receptors. However, the presence of multiple putative GBL receptor homologues in the genome suggests the existence of an alternative regulatory mechanism. Here, in the model streptomycete Streptomyces coelicolor, ScbR2 (SCO6286, a homologue of GBL receptor) is shown not to bind the endogenous GBL molecule SCB1, hence designated “pseudo” GBL receptor. Intriguingly, it could bind the endogenous antibiotics actinorhodin and undecylprodigiosin as ligands, leading to the derepression of KasO, an activator of a cryptic type I polyketide synthase gene cluster. Likewise, JadR2 is also a putative GBL receptor homologue in Streptomyces venezuelae, the producer of chloramphenicol and cryptic antibiotic jadomycin. It is shown to coordinate their biosynthesis via direct repression of JadR1, which activates jadomycin biosynthesis while repressing chloramphenicol biosynthesis directly. Like ScbR2, JadR2 could also bind these two disparate antibiotics, and the interactions lead to the derepression of jadR1. The antibiotic responding activities of these pseudo GBL receptors were further demonstrated in vivo using the lux reporter system. Overall, these results suggest that pseudo GBL receptors play a novel role to coordinate antibiotic biosynthesis by binding and responding to antibiotics signals. Such an antibiotic-mediated regulatory mechanism could be a general strategy to coordinate antibiotic biosynthesis in the producing bacteria.
Keywords: Antibiotics, Genetics, Ligand-binding Protein, Metabolic Regulation, Receptors, Antibiotic Biosynthesis, γ-Butyrolactone Receptor, Ligand
Introduction
Actinomycetes are well known for their ability to produce a wide variety of antibiotics and other secondary metabolites (1, 2). Earlier investigations indicated that the antibiotic biosynthesis often involves a complex regulatory network that responds to environmental and nutritional factors; one of the initial steps is known as the quorum sensing process (3, 4). In bacteria, this process is often mediated by small molecule autoinducers and their cognate receptors to convert population density information into cellular responses (5, 6). In the Gram-positive streptomycetes, a typical quorum-sensing mechanism to trigger the onset of secondary metabolism involves the γ-butyrolactones (GBLs)4 autoinducers and their cognate GBL receptors (7, 8).
Several GBLs in actinomycetes have been identified so far, and they share a characteristic 2,3-disubstituted-γ-butyrolactone core but differ in the C2 side chains. In most cases, the GBL synthase gene locates next to the gene of its specific receptor (9). However, genome analysis of sequenced Streptomyces indicated the presence of many GBL receptor paralogues in addition to the identified synthase-receptor pairs, raising the question of whether they also respond to GBLs and participate in the quorum-sensing cascade (7). In fact, only a few identified receptors have been proven to interact with their cognate GBL molecules: ScbR with SCBs in Streptomyces coelicolor, ArpA with A-factor in S. griseus, BarA with virginiae butanolides in Streptomyces virginiae, and FarA with IM-2 in Streptomyces lavendulae (9). There are more examples of putative GBL receptor homologues that do not exhibit any GBLs binding ability (10). For instance, BarB from S. virginiae was found not to bind virginiae butanolides (the GBLs in S. virginiae), thereby designated “pseudo” GBL receptor (11). Recent reports also suggest that putative GBL receptor homologues, FarR2 in S. lavendulae, AlpZ in Streptomyces ambofaciens, and SabR in Streptomyces acidiscabies, may bind ligands other than GBL compounds (10, 12, 13). Furthermore, an earlier phylogenetic analysis of GBL receptor homologues has indicated considerable differences between genuine GBL receptors and BarB-like proteins (including BarB, JadR2, SCO6286, CprB, SAV3702, and Aur1R from different streptomycetes), particularly in the region corresponding to the ligand-binding domain (7). It is suspected that they may have developed a different ligand binding capacity, but so far, such a mechanism has not been elucidated.
In this study, we characterized the functions of two BarB-like proteins, ScbR2 (SCO6286) and JadR2, and demonstrated that their ligands are endogenously produced and chemically distinct antibiotics. We further showed that, by responding to antibiotic signals, these pseudo GBL receptors play a novel role to coordinate antibiotic biosynthesis. ScbR2 locates in a cpk (cryptic polyketide synthase) gene cluster of S. coelicolor A3(2), a model streptomycetes that mainly produces two pigmented antibiotics: a blue polyketide antibiotic actinorhodin (Act) and a red prodiginine antibiotic undecylprodigiosin (Red) (14). JadR2 was previously identified as a repressor in the jadomycin biosynthetic gene cluster of Streptomyces venezuelae ISP5230 (15), the producer of chloramphenicol (Cm) and the angucycline antibiotic jadomycin (Jd). Jd is cryptic, being produced only under stresses such as ethanol toxicity, whereas the production of Cm does not require stress stimulus (16).
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
S. coelicolor M145, scbR2 disruption mutant (scbR2DM), scbR2 overexpression mutant (scbR2OE), and scbR2 complementary mutant (scbR2COM) derivatives were grown on supplemented minimum medium solid agar for detecting Act and Red production (14). Act and Red concentrations in supplemented minimum medium liquid-grown cultures were determined spectrophotometrically (17). The procedures for calcium-dependent antibiotic detection were as follows: the freeze and thaw extracts of the S. coelicolor strains grown on Oxoid nutrient agar for 5 days at 30 °C were spotted onto soft Oxoid nutrient agar plate containing 6 mm CaCl2, using Bacillus subtilis as the indicator strain. Calcium-dependent antibiotic production results in zones of clearing of the lawn of the B. subtilis after incubation overnight at 30 °C. R5 medium was used for culturing WT and the actIII, redL double disruption mutant whose supernatants were applied to luciferase assays (14). S. venezuelae ISP5230 wild type, jadR2 disruption mutant (jadR2DM), jadR1 disruption mutant (jadR1DM), jadI disruption mutant (jadIDM), and cmlB disruption mutant (cmlBDM) were grown on maltose-yeast extract-malt extract. The medium and culture conditions for Jd and Cm production were as described previously (15, 16). JadR2, JadR1, ScbR, and ScbR2 were expressed in Escherichia coli BL21 (DE3) to obtain large amounts of C-terminally His-tagged proteins.
Construction of Mutant Strains
For scbR2DM, a 298-bp PshAI-BclI fragment inside scbR2 was replaced by aphII. For scbR2COM, a 982-bp DNA fragment carrying the promoter and coding region of scbR2 was inserted into the EcoRV-cleaved pSET152 to generate the recombinant plasmid, which was then introduced into scbR2DM to obtain the complemented strain. The disruption was confirmed by PCR. For scbR2OE, a 675-bp DNA fragment carrying scbR2 coding region was digested with KpnI and inserted into the EcoRV-KpnI site of pIMEP (18) to generate the recombinant plasmid, which was introduced into S. coelicolor M145 to obtain the overexpression strain. For jadIDM construction, a 273-bp fragment inside jadI was replaced with aphII. For the redL disruption mutant, a 336-bp fragment inside redL was deleted. S. coelicolor strain B41 was an actIII disruption mutant provided by Maureen Bibb. The Act, Red double disruption mutant was obtained by protoplast fusion of the redL disruption mutant with strain B41 (14) and then confirmed by HPLC for no production of Act or Red.
Chemicals Preparation and HPLC Analysis
Act and Red were extracted from cultures of S. coelicolor M145 and purified by HPLC. Jadomycin B (JdB) and jadomycin A (JdA, the aglycone of JdB) were prepared as described previously (19). The HPLC conditions for the simultaneous detection of Cm, JdA, and JdB were modified from previous reports (16, 19). Purified JdB, JdA, Act, and Red were detected by electrospray ionization-MS. The purified Red was determined to be undecylprodigiosin, and the purified Act was determined to be γ-actinorhodin. Partially purified SCB1 was extracted from culture supernatant of S. coelicolor M145, followed by HPLC purification (20) and electrospray ionization-MS confirmation. All the other antibiotics used in this study were purchased from Sigma.
Overexpression and Purification of JadR1, JadR2, ScbR, and ScbR2
jadR1, jadR2, scbR, and scbR2 were amplified from the genomic DNA of S. venezuelae ISP5230 and S. coelicolor M145. The primers used were as follows: jadR1, 5′-acatatgagcctgacgtccgtagaagtgaag-3′ and 5′-actcgaggccgcggccgaagcggaaac-3′; jadR2, 5′-acatatgaccaaacaagagcgggccac-3′ and 5′-actcgagggcgaccgacgtgtacgccc-3′; scbR, 5′-ggaattccatatggccaagcaggaccgggc-3′ and 5′-aaaaactcgaggtccttcccggtcggtgcc-3′; and scbR2, 5′-ggaattccatatgaccaagcaggagcgggc-3′ and 5′-ccgctcgaggtgcggcgcgtcctgccgctc-3′. The amplified fragments were digested with NdeI and XhoI and then inserted into pET23b to obtain expression plasmids pET23b::jadR1, pET23b::jadR2, pET23b::scbR, and pET23b::scbR2, respectively. The plasmids were introduced into E. coli BL21 (DE3) for protein overexpression. Purification and concentration of proteins were carried out as described previously (19).
Band Shift Assay and DNase I Footprinting
Band shift assays were performed as described previously (19). Purified Act, Red, JdA, and JdB were dissolved in Me2SO, and Cm was dissolved in methanol. Dissolved compounds and solvent controls were added at 5% (v/v) in the reaction mixtures.
DNase I footprinting assays were carried out as described previously (19). The sequence ladder was prepared using an fmolTM DNA cycle sequencing kit (Promega) with the labeled primer. The primer pairs used to generate the DNA probes were as follows: for jadR1, the unlabeled primer 5′-gaagtggtcaagagtgcccgtggtc-3′ and the 5′-end [γ-32P]ATP-labeled primer 5′-cggttccccctagcacctatgtcac-3′; and for cmlJ, the unlabeled primer 5′-cggcggcttcggatttctcgtc-3′ and the labeled primer 5′-ggtccgggtgccggtgatcatg-3′.
RNA Isolation, S1 Nuclease Mapping, and RT-PCR
For S. coelicolor strains, RNA was isolated at 15 h (A450 = 0.4) and 20 h (A450 = 0.8) from cultures grown in supplemented minimum liquid medium, and RT-PCR was conducted as described previously (21). The primers of hrdB from S. coelicolor were 5′-gtgacgctgatggtcagtgccg-3′ and 5′-gctcgccgtcttccttcttgg-3′. For S. venezuelae strains, RNA samples were isolated at various times (12, 24, 48, and 72 h) from cultures grown at 28 °C with or without ethanol stress. Ethanol was added to a final concentration of 3% (v/v). Previously described conditions were used for extracting RNA and S1 nuclease mapping (14, 19). The hrdB probe used as a control was amplified using the unlabeled primer 5′-cgggagtgcggagtcggggg-3′ and the labeled primer 5′-tgcccatcagcctttccccgc-3′. The jadJ probe was prepared using the unlabeled primer 5′-aggcgtgggtttccgcttcggc-3′ and the labeled primer 5′-cacggccacgctgccgataccc-3′. The jadR1 and cmlJ probes were the same as those used for DNase I footprinting assays (19).
Binding Reaction of SCB1 with ScbR and ScbR2
Partially purified SCB1 (400 μl) was added to 400 μl of His tag-purified ScbR or ScbR2 (2 mg/ml). They were then processed as described previously (22).
Construction of Biosensor Strains and Luciferase Assays in Vivo
The kasO promoter was ligated to BamHI-treated and blunt-ended pCS26-Pac to give pOkasOlux. For the kasO promoter, primers 5′-ccgctcgaggacgaggagatcgaccgg-3′ and 5′-cgcggatcccggacaacacctcgagtg-3′ were used. The scbR2 and jadR2 genes were amplified with the introduction of Shine-Dalgarno sequences and downstream recognition sites for BamHI (23). For ScbR2, primers 5′-taagaaggaacggagcacgacatgaccaagcaggagcg-3′ and 5′-tcagtgcggcgcgtcctgccgctccg-3′ were used; for JadR2, primers 5′-taagaaggatactggagccattgatgaccaaacaagagcg-3′ and 5′-cgcggatcccgccggtgtcagggcggcgag-3′ were used. After digestion with BamHI, the fragments were ligated to BamHI-EcoRV-cut pACYC184 to give pScbR2 and pJadR2, respectively. Bioluminescence of E. coli reporter cultures and those supplemented with spent Streptomyces culture supernatant was measured using a 20/20n single tube luminometer (Turner Biosystems) as described previously (23).
RESULTS
ScbR2 Directly Represses the Transcription of kasO
ScbR2 is a GBL receptor homologue located next to the cpk gene cluster whose expression is positively controlled by a SARP type regulator, KasO (also known as CpkO (21)) (Fig. 1A). To identify the possible target gene of ScbR2, purified ScbR2 from E. coli was used in band shift assays with all of the potential promoter regions in the cpk gene cluster. It was found to bind the promoter region of kasO (PkasO) in a concentration-dependent manner (Fig. 1B). This binding was specific because no band shift was observed with the promoter region of hrdB (supplemental Fig. S1). To define the regulatory relationship between ScbR2 and its target PkasO, the scbR2DM was constructed. Transcription of kasO was readily detected in scbR2DM at the mid-exponential phase by RT-PCR, as was that of hrdB, whereas no transcript of kasO could be detected in WT at these two early time points (Fig. 1C), which is in accordance with previous evidence (21). Thus, mutation of scbR2 led to earlier expression of kasO. Because the expression of the cpk gene cluster is dependent on kasO, scbR2DM might have activated the biosynthesis of this cluster prematurely.
FIGURE 1.
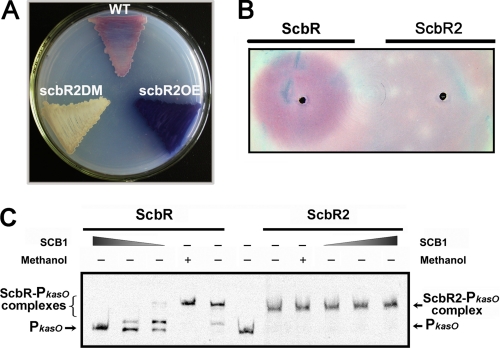
ScbR2 controls the transcription of kasO. A, schematic representation of the relative positions of scbR2, kasO, and scbR in S. coelicolor M145. B, band shift assays of the interaction of PkasO with purified ScbR2 protein. Each lane contains 10 ng of γ-32P-labeled PkasO. Lanes 1–7 contain 0, 1.5, 3, 5, 15, 30, and 50 nm purified protein, respectively. C, transcriptional analysis of kasO. Levels of transcripts in RNA from supplemented minimum medium liquid-grown cultures of WT and ScbR2DM were determined by RT-PCR. The numbers 1 and 2 denote the samples taken at A450 of 0.4 and 0.8, respectively. The hrdB transcription was assayed as a control.
Interestingly, in comparison with the WT, scbR2DM was considerably attenuated in the production of Act, Red (Fig. 2A and supplemental Fig. S2A), and calcium-dependent antibiotic (supplemental Fig. S2B), whereas scbR2OE had an obvious increase of the production of the three antibiotics (Fig. 2A and supplemental Fig. S2B). All these phenotypes were restored to WT in the scbR2COM, which shows that ScbR2 is undoubtedly involved in the control of Act, Red, and calcium-dependent antibiotic biosynthesis (supplemental Fig. S2, C and D). Also, it is noteworthy that scbR2DM produced a yellow compound (supplemental Fig. S2D), whose color and hydrophilicity coincided with the suspected product of the cpk gene cluster (24). Therefore, it is speculated that ScbR2 might act as a transcriptional repressor of the cpk gene cluster by directly repressing kasO. Taken together, these results suggested that ScbR2 has pleiotropic effects on several antibiotic biosynthesis pathways in S. coelicolor.
FIGURE 2.
Effects of ScbR2 on antibiotic production in S. coelicolor. A, pigmented antibiotic (Act and Red) production by S. coelicolor WT, scbR2DM, and scbR2OE on supplemented minimum medium solid agar medium after incubation for 5 days at 30 °C. B, bioassay detection coupled with affinity capture experiment. Molecules harvested by ScbR and ScbR2 were respectively spotted onto confluent lawns of S. coelicolor M145 spores on supplemented minimum medium solid and incubated for 30 h. C, effect of SCB1 on the DNA binding activities of ScbR and ScbR2. Band shift assays of ScbR (2.7 nm) and ScbR2 (38 nm) were carried out in the presence of 2-fold dilution steps of SCB1 captured by ScbR. 50% methanol was used as a solvent control.
ScbR2, a Pseudo GBL Receptor, Binds Antibiotics as Ligands
In previous phylogenetic analyses, ScbR2 was grouped with BarB-like proteins whose GBL receptor designation was questioned. To test the ability of ScbR2 to bind the endogenous γ-butyrolactone SCB1 (the major GBL in S. coelicolor M145), an affinity capture technique previously developed to capture and detect SCB1 via its cognate receptor ScbR was employed (22). Both ScbR and ScbR2 were used to capture SCB1 from a partially purified SCB1 sample (supplemental Fig. S3); the former was included as a positive control here to ensure that the capture works at equal conditions. Based on previous knowledge (25), exogenous addition of SCB1 to agar-grown cultures of S. coelicolor was expected to elicit early Red production. The ScbR-captured molecules could elicit early Red production, but no such effect was observed with the ScbR2-captured material (Fig. 2B). This indicated that purified ScbR2 could not recognize SCB1 under the conditions tested. To further confirm this, molecules captured by ScbR (mostly SCB1) were concentrated, and band shift assays were carried out to assess their effect on the binding of ScbR and ScbR2 to PkasO (Fig. 2C) (ScbR also binds the kasO promoter region (21)). As expected, ScbR-captured molecules, the SCB1 extract dissolved in 50% methanol, inhibited band shifting of PkasO by ScbR in a concentration-dependent manner, although in the case of ScbR2, no effect on the DNA binding activity of ScbR2 was detected with further addition of the SCB1 extract. This clearly indicated that ScbR2 was not able to recognize SCB1 at least in vitro, which further supported the prediction that ScbR2 might be a pseudo GBL receptor.
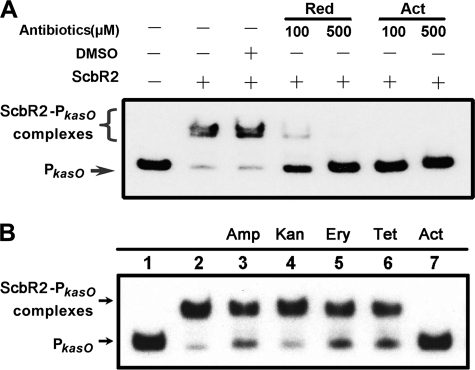
Previously, it was known from the overall structure of CprB (also one of the BarB-like proteins with ligands unidentified) that GBL receptors belong to the TetR family of regulators (26), which bears poor sequence conservation but high global structural similarity in the ligand-binding domain (27). Therefore, the program FUGUE, designed to recognize homologues by sequence-structure comparison, was employed to search for ScbR2 homologues (28). Indeed, all of the top hits calculated by this program are members of the TetR family, and it turned out that ScbR2 shares the greatest similarity with the multi-drug-binding protein TtgR from Pseudomonas putida (29). The richness in helices, which was the structural foundation for binding multiple antibiotics as ligands, was also predicted for ScbR2. Therefore, Act and Red, the endogenously produced metabolites purified from their host, and some other commercially available antibiotics of diverse chemical structures (including ampicillin, kanamycin, erythromycin, and tetracycline) were tested as the candidate ligands in band shifting assays. Indeed, the addition of Act (100 μm) or Red (500 μm) totally inhibited the formation of ScbR2-PkasO complexes (Fig. 3A), whereas other antibiotics did not cause any obvious dissociation (Fig. 3B). These results clearly demonstrated that ScbR2 binds Act and Red specifically.
FIGURE 3.
Effects of ligands on the binding activity of ScbR2. A, effect of Red and Act on the DNA binding activity of ScbR2. Each lane contains 10 ng of labeled PkasO. Band shift assays were performed with 3 nm ScbR2 and a range of ligand concentrations as indicated. Me2SO (DMSO) was used as a solvent control. B, effects of different antibiotics on the DNA binding activity of ScbR2. Each lane contains 10 ng of labeled PkasO. Lanes 2–7 contain 3 nm ScbR2. Lanes 3–7 contain 500 μm of antibiotics with distinct structures.
JadR2 Represses JdB Production by Inhibiting the Transcription of jadR1
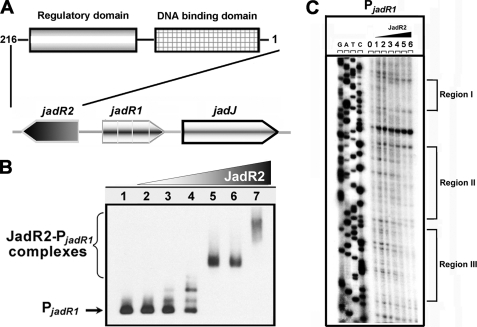
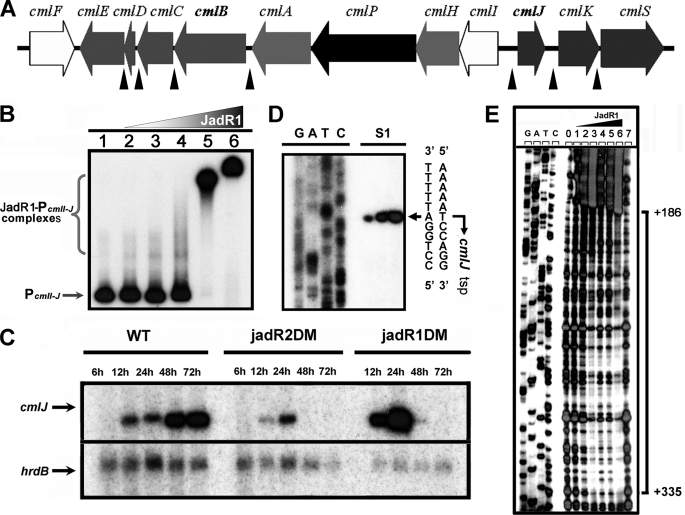
To further define the physiological role of pseudo GBL receptors, another close homologue, JadR2 in S. venezuelae, was studied. Previous works showed that JadR1 is a key activator of the Jd biosynthetic cluster (19), and its encoding gene is adjacent to and divergent from jadR2 (Fig. 4A). Disruption of JadR2 makes production of JdB independent of ethanol stress (15). To examine the molecular mechanism involved, purified JadR2 was used in band shift assays with all of the potential promoter regions in the Jd biosynthetic gene cluster. Protein-DNA complexes were seen only with the jadR1-jadR2 intergenic region in a concentration-dependent manner, indicating its specific binding (Fig. 4B). Negative control hrdB probe showed no obvious band shift (supplemental Fig. S4). In DNase I footprinting, JadR2 protected three regions, at −11 to −38, +11 to +55, and +62 to +83 nucleotides relative to the transcription start point (tsp) of jadR1 (Fig. 4C). The sequences of the target sites in promoters have been determined, and characteristic inverted repeats were found in these binding regions (supplemental Fig. S5). These repeats suggested that JadR2 might impede the access of RNA polymerase to the jadR1 promoter, thereby preventing the JadR1-dependent activation of Jd biosynthesis.
FIGURE 4.
JadR2 binds to the promoter region of jadR1. A, schematic representation of the relative positions of jadR2, jadR1, and jadJ in S. venezuelae. The domain organization of JadR2 is indicated by boxes. B, band shift assays of the interaction of the jadR1 promoter region (PjadR1) with purified JadR2. Each lane contains ∼10 ng of γ-32P-labeled PjadR1 probe. Lanes 1–7 contain 0, 1.5, 4.5, 15, 45, 150, and 450 nm purified JadR2, respectively. C, DNase I footprinting assay of JadR2 on PjadR1. Each lane contains ∼200 ng of labeled DNA probe, and the concentrations of JadR2 from lanes 0–6 are 0, 15, 37.5, 150, 300, 450, and 750 nm, respectively. The brackets denote the regions protected by JadR2.
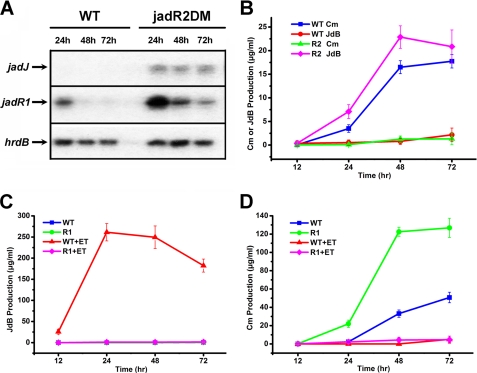
In support of this model, the mRNAs of jadR1 and the Jd biosynthetic structural gene jadJ were markedly more abundant in the jadR2DM than in the WT (Fig. 5A). Indeed, jadJ mRNA was undetectable in WT. Because JadR2 could not bind to the jadJ promoter region, the inhibition of jadJ transcription by JadR2 is an indirect effect of the absence of JadR1, which directly activates jadJ transcription (19).
FIGURE 5.
JadR2 and JadR1 control JdB and Cm biosynthesis. A, the transcriptional level of jadJ and jadR1 in S. venezuelae WT and jadR2DM without ethanol induction was detected by S1 mapping analysis. Total RNA from WT and jadR2DM in the absence of ethanol was hybridized with jadJ and jadR1 probes. The hrdB transcription was assayed as an internal control. B–D, the yields of Cm and JdB in WT, jadR2DM, and jadR1DM with or without ethanol induction. Cm and JdB yields were evaluated by HPLC after organic extraction of the entire culture. The error bars indicate the means ± S.D. with three independent experiments. R2, jadR2DM; R1, jadR1DM; +ET, ethanol added.
JadR1 Activates JdB Biosynthesis while Repressing Cm Biosynthesis Directly
In S. venezuelae, the ethanol stimulus required to initiate Jd biosynthesis was found to abolish Cm biosynthesis, implying that Jd and Cm biosynthesis are cross-regulated in a likely antagonistic mode. Unusually, the Cm biosynthetic gene (cml) cluster has no cluster-situated regulators (30). It thus prompted us to investigate whether JadR2 also regulates Cm biosynthesis. Cm and JdB production were measured by HPLC in WT or jadR2DM cultures. In the absence of ethanol induction, deletion of jadR2 led to JdB production, whereas Cm production was severely reduced (Fig. 5B). In the presence of ethanol, jadR2 deletion resulted in slightly increased JdB production, but no Cm was detected in either strain (supplemental Fig. S6). This suggested that JadR2 might inhibit JdB biosynthesis while positively controlling Cm production at least in the absence of ethanol. However, purified JadR2 failed to bind any of the potential promoter regions in the Cm biosynthetic gene cluster, suggesting that JadR2 might repress Cm biosynthesis indirectly. Because the only known JadR2 target is the promoter of jadR1, we tested whether Cm biosynthesis might be repressed by JadR1 by measuring Cm and JdB yields in WT and jadR1DM (Fig. 5, C and D). In accordance with previous evidence (19), no JdB was detected in WT in the absence of ethanol, although it was relatively abundant in the presence of ethanol, and in jadR1DM, JdB was undetectable under either of the conditions tested (Fig. 5C). In the presence of ethanol, weak Cm production was detected in both strains, but in the absence of ethanol, the Cm yield in jadR1DM grown for 3 days was strikingly higher than in WT, indicating that JadR1 might indeed repress Cm biosynthesis (Fig. 5D).
To examine whether the effect of JadR1 on Cm biosynthesis is direct, JadR1 was used in band shift assays with all of the potential promoter regions from the Cm biosynthetic gene cluster (Fig. 6A). Indeed, it was found to bind to the cmlI-cmlJ intergenic region (Fig. 6B), which is indispensable for Cm biosynthesis as previously shown (16). The result supported the model that JadR1 regulates Cm biosynthesis by direct control of cml expression.
FIGURE 6.
JadR1 inhibits the transcription of cmlJ. A, the gene cluster for Cm biosynthesis in S. venezuelae ISP5230. The intergenic regions containing the potential promoters in this cluster used for band shift assays with JadR1 and JadR2 are indicated by filled triangles. B, band shift assays of JadR1 with the intergenic region of cmlI-cmlJ. Each lane contains ∼10 ng of labeled probe. Lanes 1–6 contain 0, 6, 20, 60, 200, and 600 nm JadR1, respectively. C, the transcriptional analysis of cmlJ. Transcripts from S. venezuelae WT, jadR1DM, and jadR2DM without ethanol induction were determined by S1 mapping analysis. The hrdB transcription was assayed as a control. D, determination of the tsp of cmlJ by S1 mapping of WT RNA (isolated at 48 h after ethanol treatment). E, DNase I footprinting assays of JadR1 on the cmlJ promoter region. Each lane contains ∼300 ng of labeled probe. The concentrations of JadR1 in lanes 1–6 are 25, 50, 100, 500, 750, and 1000 nm. Lanes 0 and 7 are controls without protein. The brackets denote the regions protected by JadR1, and the numbers on the right side indicate the distances relative to the tsp of cmlJ.
To obtain corresponding evidence in vivo, the transcription of cmlJ (which was speculated to be a ketoreductase) in the WT, jadR2DM, and jadR1DM strains was analyzed by S1 mapping. As expected, cmlJ transcripts were much less abundant in mRNA from jadR2DM (i.e. derepressed for jadR1) than from WT (Fig. 6C), whereas they were more abundant in mRNA from jadR1DM than from WT at early time points (12∼24h) (Fig. 6C). An unexpected decrease in cmlJ mRNA abundance in jadR1DM versus WT at later time points (48∼72 h) suggested that some factor(s) in addition to relief from repression by JadR1 was required for cmlJ expression in the later time points under these conditions.
To define the specific sites of interaction between JadR1 and cmlJ promoter, we first determined the transcription start point of cmlJ at 439 nucleotides upstream of the putative cmlJ start codon (Fig. 6D) and then JadR1 binding sites by DNase I footprinting (Fig. 6E). JadR1 protected a large region far downstream of cmlJ tsp (from +186 to +335 nucleotides). This is analogous to the report that four binding sites of AdpA are located far downstream of the tsp of sanG, encoding a pathway-specific regulator for nikkomycin biosynthesis in Streptomyces ansochromogenes (31). Presumably, binding of JadR1 downstream of the tsp inhibits cmlJ transcription either by directly interfering with transcription elongation or by recruiting other repressors. Some alternative sigma factors might be also involved in the presence or absence of ethanol stress. Overall, these results show that JadR1 is not simply “pathway-specific” but a direct regulator of both JdB and Cm biosynthetic pathways.
JadR2 Bind Antibiotics as Ligands
Sharing 46% amino acid identity with ScbR2, JadR2 resembles the multi-drug-binding protein QacR from Staphylococcus aureus (32) in secondary structure, implying that the ligands for JadR2 might also be antibiotics. Indeed, purified JdB and JdA (supplemental Figs. S7 and S8) both inhibited PjadR1 band shifting by JadR2 in a concentration-dependent manner (Fig. 7). It seems that the structurally different Cm could also affect the binding complex of JadR2-PjadR1, suggesting that there might be cross-talk between the Cm and the Jd biosynthesis pathways. Other antibiotics (including ampicillin, kanamycin, erythromycin, and tetracycline) did not cause any obvious dissociation (supplemental Fig. S9). Cm was less effective (which might be due to the solvent involved) than JdA and JdB in relieving the DNA binding of JadR2. This may be due to its relatively small molecule size or coincide with the physiological situation in vivo; in the absence of ethanol stress, the concentration of Cm considerably exceeds that of JdB or JdA. Thus, more sensitive recognition of JdB and JdA by JadR2 might be necessary.
FIGURE 7.
Effects of antibiotics on the binding activity of JadR2. Each lane contains 10 ng of labeled PjadR1. Lanes 2–12 contain 15 nm JadR2. Lanes 3 and 4 contain Me2SO and methanol as solvent controls, respectively. Lanes 5 and 6 contain 50 and 500 μm JdB, respectively. Lanes 7 and 8 contain 50 and 500 μm JdA, respectively. Lanes 9–12 contain Cm 5, 50, 500, and 750 μm, respectively.
ScbR2 and JadR2 Biosensors Are Activated by Endogenous Antibiotics
To further confirm the antibiotic responding activities of ScbR2 and JadR2 in vivo, we employed a bacterial promoter-reporter system that has provided a sensitive tool for high throughput screening for bioactive compounds in microbial supernatants (33). To compare with the WT supernatant, a double mutant lacking actIII and redL (34, 35) structural genes indispensable for the biosynthesis of Act and Red in S. coelicolor was constructed; similarly in the case of S. venezuelae, the jadIDM, producing neither Jd or Cm under ethanol stress (36), was chosen.
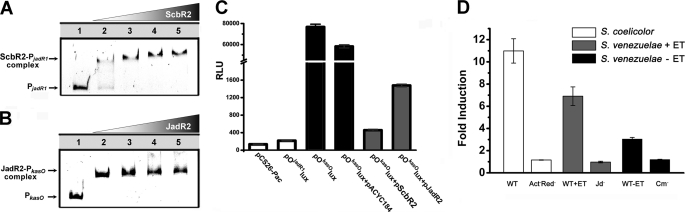
In this promoter-reporter system, the PjadR1 and PkasO promoters used in the band shift assays were introduced upstream of the promoterless lux operon in the vector pCS26-Pac, generating pOjadR1lux and pOkasOlux, respectively, and transformed into E. coli. High levels of luciferase activity were conferred by pOkasOlux, whereas pCS26-Pac and pOjadR1lux conferred only a low background. This suggested that PkasO, unlike PjadR1, could be recognized by RNA polymerase in E. coli. In subsequent band shift assays, JadR2 and ScbR2 were shown to bind to each other's target promoters (Fig. 8, A and B), indicating that the two regulators recognize similar binding sites. This made it possible to evaluate recognition of PkasO by the JadR2 and ScbR2 repressors in vivo. So jadR2 and scbR2 were inserted into pACYC184 to create pJadR2 and pScbR2. They and the vector control pACYC184 were respectively introduced into the E. coli strain bearing pOkasOlux. Both pJadR2 and pScbR2 repressed bioluminescence in the recombinant transformants, although JadR2 repressed less strongly than pScbR2 (Fig. 8C). The plasmid pACYC184 as control had no such effect.
FIGURE 8.
Effects of antibiotics on the DNA binding ability of ScbR2 and JadR2. A, band shift assays of the interaction of the jadR1 promoter region with purified ScbR2. Each lane contains ∼10 ng of probe. Lanes 1–5 contain 0, 5, 15, 30, and 50 nm purified ScbR2, respectively. B, band shift assays of the interaction of the kasO promoter region with purified JadR2. Each lane contains ∼10 ng of probe. Lanes 1–5 contain 0, 4.5, 15, 45, and 150 nm purified JadR2, respectively. C, effects of ScbR2 and JadR2 on various reporter plasmids. The error bars indicate the means ± S.D. with three independent experiments. D, effects of cultural supernatants on ScbR2- and JadR2-mediated repression. The fold induction was compared with the nonsupplemented controls, and the strains whose supernatants were tested are indicated. The error bars indicate the means ± S.D. from three independent experiments. WT, S. coelicolor M145; Act−Red−, actIII and redL double disruption mutant; WT+ET, S. venezuelae WT with ethanol induction; Jd−, jadIDM with ethanol induction; WT-ET, S. venezuelae WT without ethanol induction; Cm−, cmlBDM without ethanol induction.
The recognition of PkasO by JadR2 and ScbR2 in vivo provided a platform for assessing the in vivo effect of culture supernatants on the activities of the two repressors. Culture supernatants from S. coelicolor M145 grown in R5 medium were added to the ScbR2 biosensor strain in a proportion that did not inhibit growth (23). Supernatant isolated before the stationary phase, when only Red was produced, induced no obvious bioluminescence, but supernatants isolated at 72 h and afterward, when Act was abundantly produced, gave strong induction, reaching a maximum at 84 h (supplemental Fig. S10). In similar experiments using culture supernatants of the double mutant lacking actIII and redL (Fig. 8D), induction was much lower than that of WT supernatants. This and the evidence from band shift assays above strongly supported the direct involvement of Act in the endogenous relief of ScbR2-dependent repression of kasO (possibly and/or Red, but this is difficult to determine in vivo because Red accumulates intracellularly (34) and is poorly soluble in culture media).
In a similar experiment, the effects of supernatants of ethanol-induced cultures of S. venezuelae WT and jadIDM (Jd−) and cultures of WT and cmlBDM (Cm−) without ethanol induction were tested on the JadR2 biosensor strain (Fig. 8D). A significant increase of bioluminescence induction (approximately 7-fold compared with Jd−) was observed when the WT + ET culture supernatant was added, whereas that of the WT − ET versus Cm− was ∼2.5-fold. Although less effective than WT + ET versus Jd−, the result is analogous to the case in vitro and consistent with our supposition that Jd and Cm might play a direct role in the endogenous relief of JadR2-dependent repression. Therefore, results in vitro and in vivo all suggested that ScbR2 and JadR2 both utilize an antibiotic responding mechanism to regulate antibiotic biosynthesis.
DISCUSSION
In Gram-negative bacteria, the LuxI-LuxR proteins are commonly involved in synthesis and detection of the quorum-sensing autoinducers. Phylogenetic analyses suggested that the synthase-receptor elements in the LuxI-LuxR systems had evolved concomitantly (37). However, in the GBL signaling cascade of streptomycetes, it is suggested that the GBL receptors had predated GBL synthases and later acquired GBL binding properties (7). The presence of many GBL receptor paralogues in one genome also suggests that their functions have diversified. Here, we demonstrate that pseudo GBL receptors serve a novel role to coordinate antibiotic biosynthesis by responding to antibiotic signals. As briefly summarized in Fig. 9, pseudo GBL receptors (like JadR2 and ScbR2) usually repress the expression of cryptic secondary metabolites (jadomycin and a type I polyketide synthase yellow compound) by directly inhibiting the transcription of cluster-situated activators (JadR1 and KasO). However, unlike genuine GBL receptors, which only bind specific GBL molecules, pseudo GBL receptors act as a signal coordinator to orchestrate antibiotic biosynthesis by binding and responding to disparate antibiotics. In S. venezuelae, JadR2 directly represses the transcription of jadR1, the determinant of an activator of the Jd biosynthetic genes, that is shown to repress Cm biosynthetic genes. These two different antibiotics could bind to JadR2, leading to the dissociation of this repression. Although in S. coelicolor, ScbR2 is the repressor of KasO, a key activator of a cryptic type I polyketide synthase gene cluster. ScbR2 could also indirectly regulate the expression of Red and Act biosynthetic gene clusters. As expected, ScbR2 did not bind the GBL molecule SCB1 but could bind and respond to the endogenous Red and Act. Taken together, their capacity to bind and respond to disparate antibiotics helps them to fulfill a coordinator role in coordinating antibiotic biosynthesis. To our knowledge, this is the first time the ligands for these pseudo GBL receptors are identified as antibiotics.
FIGURE 9.
Cross-coordination of different antibiotic biosynthesis by the pseudo GBL receptors. In S. venezuelae, the pseudo GBL receptor JadR2 directly represses the transcription of jadR1. The cluster-situated regulator JadR1 activates the biosynthesis of Jd by activating the transcription of biosynthetic structural genes. JadR1 also represses the production of Cm by binding to the promoter of the structural genes. Jd and Cm could directly bind to JadR2, leading to its dissociation from the jadR1 promoter. In S. coelicolor, the pseudo GBL receptor ScbR2 directly represses the transcription of kasO. The cluster-situated regulator KasO is an activator in a cryptic type I polyketide synthase (PKSI) gene cluster. ScbR2 could also regulate the production of Red and Act indirectly. ScbR2 did not bind the GBL molecule SCB1 but could bind Red and Act, leading to its dissociation from the kasO promoter. Different antibiotics produced in one strain may affect each other by these pseudo GBL receptors, which bind and respond to antibiotic signals to coordinate their biosynthesis.
Nevertheless, this antibiotic binding capacity is not unexpected. Sequence similarity and phylogenic studies suggested that ScbR2 and JadR2 belong to the TetR superfamily regulators. Because of the sequence diversity of the ligand-binding domain of this family, an extremely wide range of small molecules can serve as the effective ligands. For instance, crystal structures showed that QacR and TtgR are capable of binding and responding to many structurally diverse drugs (29, 32). Investigations of these proteins suggest that this ability is mainly ascribed to the richness of helices and a large cavity in their ligand-binding domains (32). Because ScbR2 and JadR2 were predicted to share significant similarity in secondary structure with them, it is likely that they have also developed this antibiotic binding capacity. Although TetR family regulators comprise a large proportion of prokaryotic transcriptional regulators (27), many of their functions and ligands remain unknown. However, those examples that have been characterized to date include a rich diversity of biologically relevant ligands; some were associated with antibiotic resistance in pathogens. Our results obtained here suggest that some regulators can also mediate secondary metabolism in Streptomyces by responding to antibiotic signals.
Antibiotics are known to serve a general signaling role to induce global changes in gene transcription in a bacterial community (38). However, the mechanisms by which antibiotics trigger specific transcriptional responses are still not elucidated. Our work demonstrates that, just like quorum-sensing molecules, antibiotics can also act as intracellular signals to induce downstream responses, and the key receptors in streptomycetes are pseudo GBL receptors. In the case of ScbR2-KasO and JadR2-JadR1, upon binding to pseudo GBL receptors as ligands, antibiotic signals are transmitted toward cluster-situated regulators to influence specific antibiotic biosynthetic pathways. By dissecting the molecular mechanisms in two model streptomycetes, we speculate that this antibiotic signaling cascade might be widely employed in this genus. Moreover, because GBL receptor homologues are commonly found in actinomycetes, it is suspected that similar mechanisms might also be adopted by closely related actinomycetes or other antibiotic-producing bacteria.
Previously, the understanding of antibiotics-mediated regulation in streptomycetes was limited to feedback autoregulation (19, 39). The ability of ScbR2 and JadR2 to interact with end products of other biosynthetic pathways extends this understanding to antibiotic-mediated communication in which metabolites from a defined biosynthetic pathway indirectly elicit responses in other pathways. Previously, microarray analysis revealed transcription level cross-talk between disparate antibiotic pathways, which is shown to involve cluster-situated regulators (40). In this study, we largely enriched the mechanism by addressing the direct participation of antibiotics and their corresponding receptors. In the regulation network of Jd and Cm biosynthesis, JadR1 is the low level regulator that directly controls their biosynthesis, although JadR2 is the higher level signal coordinator that senses the information from the metabolites and responds by regulating the transcription of jadR1. With the formation of this cross-regulation circuit, Jd and Cm are thus able to cross-regulate the biosynthesis of each other. Knowledge of this antibiotic-induced cross-talk mechanism could prove relevant to rational improvements in the yield of commercially important antibiotics.
Strikingly, the top ten JadR2 homologues in BLAST search are exclusively located in or adjacent to gene clusters for the biosynthesis of antibiotics, most of which are not identified and might well be cryptic. This suggests a possible correlation of pseudo GBL receptors with silent antibiotic clusters, consistent with our findings that inactivation of JadR2 and ScbR2 led to the production of two cryptic metabolites, Jd and a yellow compound. Knowledge of this correlation will likely provide an effective strategy to isolate those novel but cryptic natural products.
Supplementary Material
Acknowledgments
We are grateful to Professor Keith Chater (John Innes Centre, Norwich, UK) for critical reading in preparation of this manuscript. We thank Professor Michael Surette and Dr. Kangming Duan for pCS26-Pac, Maureen Bibb for strain B41, Xia Zhang for samples of jadomycin, and Professor Yiguang Wang for relative biochemical reagent.
This work was supported by Ministry of Science and Technology of China Grant 2009CB118905 and National Natural Science Foundation of China Grants 31030242 and 30870041.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S10.
- GBL
- γ-butyrolactone
- Act
- actinorhodin
- Red
- undecylprodigiosin
- Cm
- chloramphenicol
- Jd
- jadomycin
- JdB
- jadomycin B
- JdA
- jadomycin A
- tsp
- transcription start point
- WT
- wild type
- ET
- ethanol
- SCBs
- Streptomyces coelicolor butyrolactones.
REFERENCES
- 1.Ikeda H., Ishikawa J., Hanamoto A., Shinose M., Kikuchi H., Shiba T., Sakaki Y., Hattori M., Omura S. (2003) Nat. Biotechnol. 21, 526–531 [DOI] [PubMed] [Google Scholar]
- 2.Bentley S. D., Chater K. F., Cerdeño-Tárraga A. M., Challis G. L., Thomson N. R., James K. D., Harris D. E., Quail M. A., Kieser H., Harper D., Bateman A., Brown S., Chandra G., Chen C. W., Collins M., Cronin A., Fraser A., Goble A., Hidalgo J., Hornsby T., Howarth S., Huang C. H., Kieser T., Larke L., Murphy L., Oliver K., O'Neil S., Rabbinowitsch E., Rajandream M. A., Rutherford K., Rutter S., Seeger K., Saunders D., Sharp S., Squares R., Squares S., Taylor K., Warren T., Wietzorrek A., Woodward J., Barrell B. G., Parkhill J., Hopwood D. A. (2002) Nature 417, 141–147 [DOI] [PubMed] [Google Scholar]
- 3.Bibb M. J. (2005) Curr. Opin. Microbiol. 8, 208–215 [DOI] [PubMed] [Google Scholar]
- 4.Camilli A., Bassler B. L. (2006) Science 311, 1113–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng W. L., Bassler B. L. (2009) Annu. Rev. Genet. 43, 197–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lazdunski A. M., Ventre I., Sturgis J. N. (2004) Nat. Rev. Microbiol. 2, 581–592 [DOI] [PubMed] [Google Scholar]
- 7.Nishida H., Ohnishi Y., Beppu T., Horinouchi S. (2007) Environ. Microbiol. 9, 1986–1994 [DOI] [PubMed] [Google Scholar]
- 8.Bassler B. L., Losick R. (2006) Cell 125, 237–246 [DOI] [PubMed] [Google Scholar]
- 9.Takano E. (2006) Curr. Opin. Microbiol. 9, 287–294 [DOI] [PubMed] [Google Scholar]
- 10.Kitani S., Iida A., Izumi T. A., Maeda A., Yamada Y., Nihira T. (2008) Gene 425, 9–16 [DOI] [PubMed] [Google Scholar]
- 11.Matsuno K., Yamada Y., Lee C. K., Nihira T. (2004) Arch. Microbiol. 181, 52–59 [DOI] [PubMed] [Google Scholar]
- 12.Bunet R., Mendes M. V., Rouhier N., Pang X., Hotel L., Leblond P., Aigle B. (2008) J. Bacteriol. 190, 3293–3305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Healy F. G., Eaton K. P., Limsirichai P., Aldrich J. F., Plowman A. K., King R. R. (2009) J. Bacteriol. 191, 4786–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kieser T., Bibb M. J., Buttner M. J., Chater K. F., Hopwood D. A. (2000) Practical Streptomyces Genetics, The John Innes Foundation, Norwich, UK [Google Scholar]
- 15.Yang K., Han L., Vining L. C. (1995) J. Bacteriol. 177, 6111–6117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He J., Magarvey N., Piraee M., Vining L. C. (2001) Microbiology 147, 2817–2829 [DOI] [PubMed] [Google Scholar]
- 17.Strauch E., Takano E., Baylis H. A., Bibb M. J. (1991) Mol. Microbiol. 5, 289–298 [DOI] [PubMed] [Google Scholar]
- 18.Wang S. L., Fan K. Q., Yang X., Lin Z. X., Xu X. P., Yang K. Q. (2008) J. Bacteriol. 190, 4061–4068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L., Tian X., Wang J., Yang H., Fan K., Xu G., Yang K., Tan H. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8617–8622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takano E., Nihira T., Hara Y., Jones J. J., Gershater C. J., Yamada Y., Bibb M. (2000) J. Biol. Chem. 275, 11010–11016 [DOI] [PubMed] [Google Scholar]
- 21.Takano E., Kinoshita H., Mersinias V., Bucca G., Hotchkiss G., Nihira T., Smith C. P., Bibb M., Wohlleben W., Chater K. (2005) Mol. Microbiol. 56, 465–479 [DOI] [PubMed] [Google Scholar]
- 22.Yang Y. H., Joo H. S., Lee K., Liou K. K., Lee H. C., Sohng J. K., Kim B. G. (2005) Appl. Environ. Microbiol. 71, 5050–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tahlan K., Ahn S. K., Sing A., Bodnaruk T. D., Willems A. R., Davidson A. R., Nodwell J. R. (2007) Mol. Microbiol. 63, 951–961 [DOI] [PubMed] [Google Scholar]
- 24.Pawlik K., Kotowska M., Chater K. F., Kuczek K., Takano E. (2007) Arch. Microbiol. 187, 87–99 [DOI] [PubMed] [Google Scholar]
- 25.Takano E., Chakraburtty R., Nihira T., Yamada Y., Bibb M. J. (2001) Mol. Microbiol. 41, 1015–1028 [DOI] [PubMed] [Google Scholar]
- 26.Natsume R., Ohnishi Y., Senda T., Horinouchi S. (2004) J. Mol. Biol. 336, 409–419 [DOI] [PubMed] [Google Scholar]
- 27.Ramos J. L., Martínez-Bueno M., Molina-Henares A. J., Terán W., Watanabe K., Zhang X., Gallegos M. T., Brennan R., Tobes R. (2005) Microbiol. Mol. Biol. Rev. 69, 326–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi J., Blundell T. L., Mizuguchi K. (2001) J. Mol. Biol. 310, 243–257 [DOI] [PubMed] [Google Scholar]
- 29.Alguel Y., Meng C., Terán W., Krell T., Ramos J. L., Gallegos M. T., Zhang X. (2007) J. Mol. Biol. 369, 829–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piraee M., White R. L., Vining L. C. (2004) Microbiology 150, 85–94 [DOI] [PubMed] [Google Scholar]
- 31.Pan Y., Liu G., Yang H., Tian Y., Tan H. (2009) Mol. Microbiol. 72, 710–723 [DOI] [PubMed] [Google Scholar]
- 32.Schumacher M. A., Miller M. C., Grkovic S., Brown M. H., Skurray R. A., Brennan R. G. (2001) Science 294, 2158–2163 [DOI] [PubMed] [Google Scholar]
- 33.Yim G., Wang H. H., Davies J. (2006) Int. J. Med. Microbiol. 296, 163–170 [DOI] [PubMed] [Google Scholar]
- 34.Bystrykh L. V., Fernández-Moreno M. A., Herrema J. K., Malpartida F., Hopwood D. A., Dijkhuizen L. (1996) J. Bacteriol. 178, 2238–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mo S., Sydor P. K., Corre C., Alhamadsheh M. M., Stanley A. E., Haynes S. W., Song L., Reynolds K. A., Challis G. L. (2008) Chem. Biol. 15, 137–148 [DOI] [PubMed] [Google Scholar]
- 36.Wohlert S. E., Wendt-Pienkowski E., Bao W., Hutchinson C. R. (2001) J. Nat. Prod. 64, 1077–1080 [DOI] [PubMed] [Google Scholar]
- 37.Gray K. M., Garey J. R. (2001) Microbiology 147, 2379–2387 [DOI] [PubMed] [Google Scholar]
- 38.Davies J., Spiegelman G. B., Yim G. (2006) Curr. Opin. Microbiol. 9, 445–453 [DOI] [PubMed] [Google Scholar]
- 39.Jiang H., Hutchinson C. R. (2006) Res. Microbiol. 157, 666–674 [DOI] [PubMed] [Google Scholar]
- 40.Huang J., Shi J., Molle V., Sohlberg B., Weaver D., Bibb M. J., Karoonuthaisiri N., Lih C. J., Kao C. M., Buttner M. J., Cohen S. N. (2005) Mol. Microbiol. 58, 1276–1287 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.