Abstract
Pathological forms of left ventricular hypertrophy (LVH) often progress to heart failure. Specific transcription factors have been identified that activate the gene program to induce pathological forms of LVH. It is likely that apart from activating transcriptional inducers of LVH, constitutive transcriptional repressors need to be removed during the development of cardiac hypertrophy. Here, we report that the constitutive presence of Krüppel-like factor 15 (KLF15) is lost in pathological hypertrophy and that this loss precedes progression toward heart failure. We show that transforming growth factor-β-mediated activation of p38 MAPK is necessary and sufficient to decrease KLF15 expression. We further show that KLF15 robustly inhibits myocardin, a potent transcriptional activator. Loss of KLF15 during pathological LVH relieves the inhibitory effects on myocardin and stimulates the expression of serum response factor target genes, such as atrial natriuretic factor. This uncovers a novel mechanism where activated p38 MAPK decreases KLF15, an important constitutive transcriptional repressor whose removal seems a vital step to allow the induction of pathological LVH.
Keywords: Gene Transcription, Heart, MAP Kinases (MAPKs), Transcription Factors, Transcription Repressor, Transforming Growth Factor beta (TGFbeta)
Introduction
Pathological forms of LVH3 often progress to heart failure. Many transcription factors have been identified that play a role in the development of pathological LVH (1). Most of these transcription factors are inducers of hypertrophy, for instance GATA4 (2), MEF2, NFAT, and SRF (3), and their function and regulation are increasingly understood.
Myocardin is an extraordinarily potent transcriptional coactivator expressed exclusively in cardiomyocytes and smooth muscle cells (4). Myocardin stimulates transcription from CArG-dependent muscle enhancers but does not bind DNA directly. Instead, myocardin forms a stable ternary complex on CArG-boxes by associating with SRF (4, 5). Recently, Parmacek and co-workers (6) showed that ablation of myocardin in the adult mouse heart leads to rapid onset of heart failure, which was accompanied by dissolution of sarcomeric organization. Moreover, patients with dilated or hypertrophic cardiomyopathy have been reported to harbor causal mutations in myocardin-regulated genes such as ACTN2, MYH7, ACTC, and TPM1 (6–10). Altogether, these studies have shown that myocardin regulates the organization of the contractile unit and adaptive responses of the cardiomyocyte to stress.
In contrast to these activators, repressors of cardiac gene expression and hypertrophy are less well explored (11). One of the best studied repressors of cardiac hypertrophy is the family of histone deacetylases. For instance, HDAC9 null mice display spontaneous cardiac hypertrophy and are hypersensitive to pressure overload (12, 13). In addition, NAB1 has been identified to repress hypertrophy by direct inhibition of Egr-dependent transcription (14). Recently, it was also shown that KLF15 acts a transcriptional repressor of pathological cardiac hypertrophy (15). KLF15 belongs to the family of Krüppel-like factors, which has 17 members (16, 17). KLF15 is widely expressed, but the highest expression levels are found in liver, kidney, pancreas, and cardiac and skeletal muscle (18). The zinc finger transcriptional regulator KLF15 was first identified as a repressor of the kidney-specific chloride channel CLC-K1 (18). KLF15 can also act as an activator of transcription as illustrated by the increased GLUT4 promoter activity and GLUT4 expression in response to KLF15 overexpression (19). Expression of KLF15 in the mouse heart is very low during development but increases after birth to reach high levels in the adult heart (19). Mice lacking KLF15 have higher base-line expression of ANF and BNP. Interestingly, when these mice are exposed to pressure overload, they develop rapid and severe hypertrophy and heart failure, suggesting that KLF15 may repress cardiac hypertrophy. Here, we sought to understand how KLF15 is regulated and how it represses cardiac hypertrophy. Our results reveal that the TGFβ-p38-MAPK axis down-regulates KLF15 and that KLF15 acts as a potent competitive inhibitor of myocardin, thereby preventing transcription of SRF target genes.
EXPERIMENTAL PROCEDURES
Animal Models
Ren-2 rats were obtained from Möllegard Breeding Center, Lille Skensveld, Denmark, and were studied as described previously (20).
Neonatal Rat Cardiomyocyte Isolation and Transfection
1–2-Day-old Lewis neonatal rats were sacrificed by decapitation. Hearts were carefully removed, and left ventricles were cut into small pieces. Cardiac cells were then isolated by enzymatic dissociation using 0.05% collagenase I (Invitrogen, catalog no. 17100-017) and 0.05% pancreatin (Sigma, catalog no. p3292) dissolved in 1× Hanks' balanced salt solution (Sigma, catalog no. H4641). Cells were pre-plated for 1 h in DMEM (Invitrogen, catalog no. 11966) (supplemented with 10% horse serum, 5% newborn calf serum, 0.16% glucose and antibiotics) to separate myocytes from fibroblasts. After 1 h, cardiomyocytes were collected, counted, and plated in plates coated with 1% gelatin (Fluka, via Sigma). Overnight, cells were grown in DMEM supplemented with 10% horse serum, 5% newborn calf serum, 0.16% glucose, and antibiotics. Dharmacon ON-TARGETplus siRNA SMARTpools for non-targeting control (D-001810-10) and KLF15 (L-080131-01) were transfected (Lipofectamine 2000, Invitrogen) to a final concentration of 300 nm and incubated for 5 h. Medium was changed to medium containing 2% BSA, and after 72 h cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100 in PBS, and stained with phalloidin 1:40 (Invitrogen, catalog no. F432) in PBS for actin. Images were taken with a Leica inverted microscope, and cell size (actin-positive area) was analyzed with Scion image.
Stimulation of Cardiomyocytes
Prior to stimulation of cardiomyocytes with TGFβ1 (10 ng/ml) (PeproTech, via Tebu-Bio, the Netherlands), cells were serum-starved for 24 h with DMEM supplemented with 0.16% glucose and antibiotics. Cells were stimulated for 24 h. One hour prior to TGFβ1, cells were treated with two independent p38 MAPK inhibitors, SB202190 or SB203580 (Sigma). Endothelin-1 (100 nm), phenylephrine (50 μm), insulin (100 nm) (Sigma), IGF-I (100 nm), and IGF-II (100 nm) (PeproTech, via Tebu-Bio) were used to stimulate cardiomyocytes as described for TGFβ1.
[3H]Leucine Measurements
TGFβ1- (10 ng/ml) and SB202190 (10 μm)-stimulated cells were exposed to [3H]leucine (1 μCi) for 24 h. After 24 h of TGFβ1 stimulation, cells were scraped in 5% trichloroacetic acid (TCA) and centrifuged, and pellets were dissolved in 0.5 m NaOH. Activity was measured in 4 ml of high ionic count fluid in a scintillation counter.
Adenoviral Infection
Infection of cardiomyocytes with AdMKK6 was performed to phosphorylate p38 MAPK as described previously (21). For infection of cardiomyocytes, 200 μl of viral supernatant (AdMKK6 and AdGFP) was added to 5 × 105 cardiomyocytes that were plated in a gelatinized 6-well plate and cultured as described above. Three days after infection, cells were harvested for protein analyses by scraping the cells in an SDS buffer containing 10% 2-mercaptoethanol and 0.5 mm sodium orthovanadate. Western blotting was performed using phospho-p38 (Thr-180/Tyr-182) (1:1000) and p38 MAPK (1:1000) (Cell Signaling Technology, via Bioké, the Netherlands).
Neonatal Mouse Cardiomyocyte Isolation and Transfection
Mouse neonatal cardiomyocytes were isolated from 1- to 3-day-old FVB mice. Heart tissue was incubated overnight at 4 °C in 0.5 mg/ml trypsin (United States Biochemical, catalog no. 22715), HBSS (Sigma, catalog no. H4641) supplemented with 1 g/liter d-glucose (Merck). Tissue was digested in collagenase buffer (150 units/ml collagenase type II (Worthington 4176) in DMEM 41965 (Invitrogen) for 45 min at 37 °C. Individual cells were obtained by trituration and filtering over a 100-μm filter. Cells were centrifuged and pre-plated for 1.5 h at 37 °C and 5% CO2. Cells were seeded in plating medium (10% FCS, 20 μm l-glutamine, and 10 μm AraC in DMEM) on 1% gelatin (Sigma, catalog no. G1890)-coated culture plates at a final density of 100,000 cells/cm2. After 48 h, medium was changed to DMEM 41965 supplemented with 25 mm l-glutamine, 2% BSA (Sigma, catalog no. A6003), 0.25 milliunits/ml insulin (Sigma, catalog no. I6634), 250 μm l-carnitine (Sigma, catalog no. C0283), and 10 μm AraC (Sigma, catalog no. C6645)). 24 h later, medium was changed to DMEM 41965 without penicillin/streptomycin for transfection. All solutions were supplemented with 100 units/ml penicillin/streptomycin and 10 mm HEPES. Transfection was performed according to the manufacturer's protocol. Dharmacon ON-TARGETplus siRNA SMARTpools for nontargeting control (D-001810-10) and KLF15 (L-059453-01) were added with Lipofectamine 2000 (Invitrogen, catalog no. 11668) as described above.
Quantitative Real Time PCR
For quantitative real time PCR, RNA was isolated from cardiomyocytes or left ventricles using the RNeasy mini kit (Qiagen) or TRIzol (Invitrogen) according to the manufacturer's protocols. cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad). Quantitative real time was performed using iQ SYBR Green Supermix (Bio-Rad), 20 ng of total RNA, and 10 pmol/μl forward and reverse primers (supplemental Table 1a). Quantification was performed using LinRegPCR analysis software (22).
Transfections and Luciferase Assays
Transfection of COS7 cells for luciferase assays were performed as described previously (23). SM22 (505 bp) luciferase reporter was a kind gift of Dr. J. M. Miano (Rochester, NY) (24). The 3×CArG and ANF (638 bp) luciferase reporter and the expression plasmids encoding myocardin (935 amino acids) and SRF were provided by Dr. Eric Olson (Dallas, Tx) (25). A pcDNA3.1-based expression vector encoding FLAG-tagged KLF15 was donated by Dr. M. K. Jain (Cleveland, OH). Transfection of COS7 cells and luciferase assays was performed as described previously (23). In short, 24-well plates with COS7 cells were transfected per well with 30 ng of pcDNA-β-gal, 75 ng of luciferase reporter, and 50 ng of pcDNA-KLF15 and myocardin(935). Transfection of vectors was facilitated using GeneJammer (Stratagene, via Bio-Connect, Huissen, the Netherlands). For titration experiments, we transfected 50 ng of ANF- or Sm22-luciferase, 50 ng of pcDNA-myocardin, and increasing concentrations of KLF15 (1–100 ng) or pcDNA-SRF (0.5–25 ng) as indicated. Luciferase assays were performed using the luciferase assay system (Promega). Measurements were performed in duplicate and repeated at least three times.
GST Pulldown
Glutathione S-transferase (GST)-KLF15 fusion proteins were generated by subcloning the KLF15 open reading frame into the pGEX-KG vector. GST fusion proteins were produced and isolated by standard procedures. Deletion mutants of myocardin were constructed through PCR-based mutagenesis and subcloning of these DNA fragments into FLAG-tagged pcDNA3.1 expression vector. Myocardin proteins were translated in vitro and labeled with [35S]methionine in a coupled transcription-translation T7 reticulocyte lysate system (Promega) and assayed for binding to GST fusion proteins.
Statistical Analyses
Data are shown as mean ± S.E. Unpaired t test was used. p values of ≤0.05 were considered statistically significant.
RESULTS
Loss of KLF15 Expression Is Specific for Pathological LV Hypertrophy
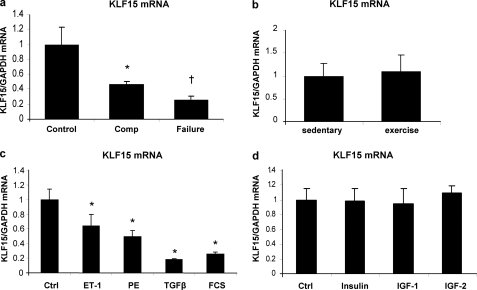
To explore whether KLF15 expression, besides in the TAC model (15), is also regulated in other models of hypertrophy and failure, we studied KLF15 expression in the homozygous TGR(mRen2)27 rat (Ren-2). In this model, KLF15 is down-regulated already in an early stage of LVH. We employed this model to evaluate whether down-regulation of KLF15 could distinguish the hypertrophied hearts that will quickly progress to failure versus those where hypertrophy remains compensated for a prolonged period. We obtained cardiac biopsies at a time when all Ren-2 rats displayed similar cardiac hypertrophy and were well compensated. After cardiac biopsies were taken, we followed each rat to see whether it would progress to failure or not (20). This revealed that KLF15 expression was decreased significantly more in the hypertrophied hearts that later progressed toward failure (Fig. 1a). To evaluate whether KLF15 is also down-regulated in physiological LVH, we assessed KLF15 expression levels in rats with exercise-induced LVH. Heart weights from trained rats were significantly increased compared with those from sedentary rats. Training resulted in hypertrophy of individual cardiomyocytes as shown by an increased length, but not width, of these cells (supplemental Fig. 1, a–d). Despite this pronounced cardiac remodeling, no difference in KLF15 gene expression between the sedentary group and the trained rats was noted (Fig. 1b).
FIGURE 1.
Loss of KLF15 is specific for pathological hypertrophy and is mediated by the TGFβ1-p38 MAPK pathway. a, KLF15 expression is down-regulated in cardiac biopsies of hypertensive Ren-2 rats. KLF15 is decreased more in the hearts of the rats that after the biopsy progressed to heart failure, as compared with the hypertrophied hearts from the rats that remained compensated after the biopsy (n = 4–6 rats per group; *, p < 0.05 compared with control group; †, p < 0.02 compared with the compensated (Comp) hypertrophy group). b, in hypertrophic myocardium from rats that had undergone exercise training for 10 weeks, 5 days per week (n = 5), KLF15 expression was not altered compared with control rat hearts (n = 7). c, KLF15 mRNA expression is decreased in cultured neonatal rat cardiomyocytes in response to several hypertrophic stimuli. Cardiomyocytes were serum-starved overnight and then stimulated with endothelin 1 (ET-1) (100 nm), phenylephrine (PE) (50 μm), TGFβ (10 ng/ml), and 10% fetal calf serum (FCS) for 24 h. Ctrl, control. KLF15 levels were detected using quantitative real time PCR. (*, p < 0.01 compared with control cells, n = 3 per group). d, KLF15 mRNA levels do not decrease on stimulation of cultured neonatal rat cardiomyocytes with IGF-I (100 nm), IGF-II (100 nm), or insulin (100 nm) for 24 h after 24 h of serum starvation (n = 3 per group).
KLF15 Expression Is Regulated by the TGFβ-p38 MAPK Pathway
KLF15 is specifically down-regulated in pathological hypertrophy, which prompted us to search for the stimuli and pathways that repress KLF15. We used cultured neonatal rat cardiomyocytes to study the effect of several hypertrophic stimuli such as phenylephrine, endothelin-1, TGFβ, and FCS on the expression of KLF15. All stimuli were able to decrease KLF15 expression levels (Fig. 1c). In contrast, when cardiomyocyte growth was stimulated with stimuli that are known to induce physiological hypertrophy (insulin, IGF-1, and IGF-2), KLF15 levels did not decrease (Fig. 1d). This further supports the in vivo findings that KLF15 is down-regulated only in pathological hypertrophy. Because TGFβ was the most robust inhibitor of KLF15 expression, we explored this pathway more extensively.
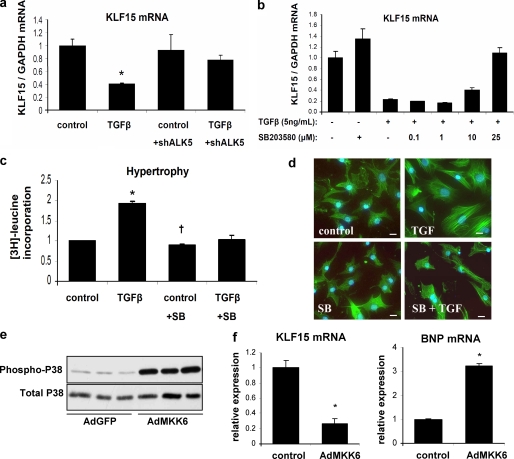
The observed TGFβ-mediated down-regulation of KLF15 was completely abolished in cultured cardiomyocytes after knockdown of TGFβ receptor 1 (TGFBR1 or ALK5) (Fig. 2a and supplemental Fig. 2), demonstrating that TGFβ signaling via ALK5 is vital to regulate KLF15 expression.
FIGURE 2.
In vitro activation of p38 MAPK is both necessary and sufficient to decrease KLF15 levels. a, TGFβ regulates KLF15 expression in vitro. Neonatal rat cardiomyocytes were infected with a lentivirus containing shRNA against ALK5 or with a control lentivirus. TGFβ decreased expression of KLF15 after control virus but had no effect in the cells treated with the shRNA. (n = 3/group; *, p < 0.01 compared with control group; †, p < 0.01 compared with TGFβ-treated cells without shRNA against ALK5. b, specific inhibitor of p38 MAPK, SB203580, abolishes the TGFβ-induced down-regulation of KLF15 in cardiomyocytes in a dose-dependent manner. c, inhibition of p38 MAPK prevents TGFβ-induced hypertrophy, measured by [3H]leucine incorporation (n = 3/group; *, p < 0.05 compared with control group; †, p < 0.05 compared with TGFβ-treated cells without SB203580 (SB) treatment), and d, inhibition of p38 MAPK prevents TGFβ-induced increase in cell size as visualized by phalloidin staining of F-actins. Bars in panels represent 50 μm. e, Western blot analysis shows an increase in phosphorylated p38 MAPK after infection of cardiomyocytes with constitutively active adMKK6, the upstream kinase of p38. f, adenoviral overexpression of MKK6 resulted in decreased KLF15 mRNA levels and increased expression of the hypertrophy marker BNP (n = 3/group; *, p < 0.05 compared with control group).
It is known that during hypertrophy of cardiomyocytes, TGFβ induces p38 MAPK phosphorylation via TAK1 (26), suggesting that TGFβ may decrease KLF15 expression by activation of p38 MAPK. To investigate this hypothesis, we treated cultured cardiomyocytes with two different specific p38 MAPK inhibitors, SB202190 and SB203580. Indeed, TGFβ-induced down-regulation of KLF15 was dose-dependently abolished by either inhibitor (Fig. 2b and supplemental Fig. 3). Strikingly, KLF15 expression was inversely correlated with ANF expression (supplemental Fig. 3). Apart from preventing KLF15 down-regulation, inhibition of p38 phosphorylation blunted the TGFβ-induced hypertrophic response of cardiomyocytes as assessed by [3H]leucine incorporation and phalloidin staining of F-actins (Fig. 2, c and d). From this, we conclude that in cardiomyocytes p38 MAPK signaling is necessary for TGFβ-induced hypertrophy and KLF15 down-regulation. To elucidate whether activation of p38 MAPK is not only necessary but also sufficient to repress KLF15 expression downstream of TGFβ, we activated p38 MAPK by adenoviral overexpression of the upstream kinase of p38 MAPK, MKK6, which induced a robust increase of phosphorylated p38 (Fig. 2e). This resulted in an almost 80% decrease in KLF15 mRNA levels and induced expression of the hypertrophy marker BNP in cultured cardiomyocytes (Fig. 2e). In conclusion, we show that activation of p38 MAPK is necessary and sufficient to decrease KLF15 expression in cardiac myocytes, providing one of the first examples of a cardiac transcription factor that is actually down-regulated by activated p38.
KLF15 Inhibits the Activity of Myocardin, a Transcriptional Coactivator of SRF
Earlier studies have shown that loss of KLF15 aggravates cardiac hypertrophy and dysfunction (15).
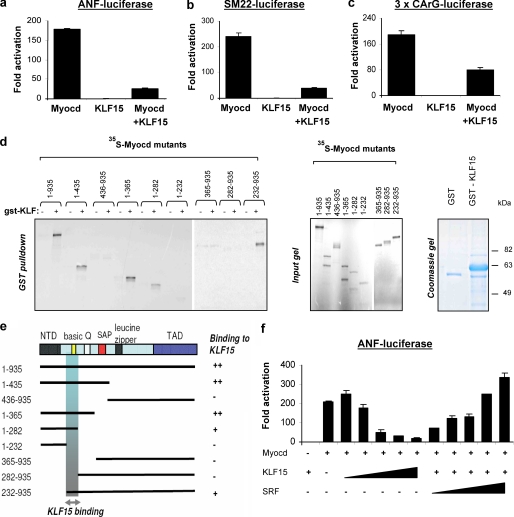
We confirmed these observations, after transfecting cultured neonatal rat and mouse cardiomyocytes with two independent SMARTpool siRNAs against KLF15 or a nontargeting control siRNA. This resulted in a 16–30% increase in cardiomyocyte size and shows that loss of KLF15 alone is sufficient to induce cardiomyocyte hypertrophy and elevate ANF expression (supplemental Fig. 4). The opposite also holds true; overexpression of KLF15 using a lentiviral approach seems to inhibit cardiomyocyte hypertrophy, as demonstrated by decreased mRNA levels of ANF in rat neonatal cardiomyocytes (supplemental Fig. 4). It remains unknown how KLF15 inhibits LVH. It has been shown that KLF15 interacts with different major transcriptional regulators of cardiac hypertrophy, like the MADS box transcription factor MEF2A and the zinc finger transcription factor GATA4, but it remains unclear whether KLF15 displays specificity to certain cardiac transcription factors or whether it represses the general transcriptional machinery through other mechanisms. This prompted us to investigate whether KLF15 could repress SRF, “the other” MADS box transcription factor with an established role in cardiac gene expression. We first performed GST pulldown assays using in vitro translated MEF2A and SRF and a GST-KLF15 fusion protein, but we did not observe a direct interaction of KLF15 to either SRF or MEF2A (supplemental Fig. 5a). This suggested to us that the previously reported repression of MEF2 activity by KLF15 is indirect (15, 19). We subsequently tested if KLF15 could physically interact with myocardin, an extraordinarily potent coactivator of the MADS box transcription factors, which has recently been reported to play a role in cardiomyocyte hypertrophy and cardiac failure (6, 25, 27). Indeed, GST-pulldown assays showed that KLF15 directly binds to myocardin (Fig. 3d). We next tested whether KLF15 could affect the transcriptional activity of myocardin. Indeed, luciferase assays showed that KLF15 virtually abolished myocardin activity on SRF-dependent promoters such as ANF(638), SM22(505), and an artificial promoter that contains three SRF binding domains (3×CArG) in COS7 cells. Repression of these promoters by KLF15 only took place in the presence (Fig. 3, a–c) and not in the absence of myocardin (supplemental Fig. 5b), indicating that KLF15 does not affect the basic transcriptional machinery in our assays but specifically functions through its interaction with myocardin. To map the region of myocardin that is required for the interaction with KLF15, we compared binding of several deletion mutants of myocardin to KLF15 using GST pulldown assays. As shown in Fig. 3, d and e, KLF15 binds to a 50-amino acid region within amino acids 232–282 of myocardin. Interestingly, this region contains the basic domain of myocardin, which is also required for binding to SRF (4). Binding of SRF and KLF15 to the same region within myocardin implicates that KLF15 can trigger the displacement of myocardin from SRF by competition for a common docking site. Indeed, competition experiments in COS7 cells using the ANF(638) reporter show that repression of myocardin activity by KLF15 is relieved in a dose-dependent manner by addition of SRF (Fig. 3f). In conclusion, competition of SRF and KLF15 for myocardin provides a mechanism whereby lower KLF15 levels in response to TGFβ signaling can enhance the expression of SRF-dependent genes during the hypertrophic response of the heart (Fig. 5).
FIGURE 3.
Binding of KLF15 to the basic region of myocardin results in repression of myocardin-responsive reporters. KLF15 inhibits myocardin activity. a–c, myocardin activates the following SRF-dependent reporters: ANF, SM22, and the 3×CArG-luciferase. This activity is blocked by KLF15. COS cells were transfected with the luciferase reporters and expression vectors encoding myocardin-935 and KLF15. Luciferase activity is expressed as fold change over the empty expression vector, pcDNA3.1 (n = 3, mean ± S.D.). d, GST pulldown assays show that KLF15 associates with myocardin. 35S-Labeled myocardin mutant proteins were translated in vitro and incubated with GST-KLF15 fusion protein. Proteins were captured on glutathione-agarose beads and analyzed by SDS-PAGE. The input lanes contain 10% of the amount of 35S-labeled myocardin protein in the pulldown lanes. GST-KLF15 fusion protein is shown on a Coomassie gel on the right. e, schematic representation of full-length myocardin (1–935 amino acids) and myocardin mutants used for GST pulldown (as shown in d) and their binding to KLF15 are indicated to the right. The shaded box represents the region that is necessary for KLF15 binding. NTD, N-terminal domain; Q, glutamine-rich domain; TAD, transactivation domain. f, KLF15 competes with SRF for myocardin interaction. COS cells were transfected with ANF luciferase and expression vectors encoding myocardin-935 and increasing concentrations of KLF15 and SRF as indicated. Luciferase activity expressed as fold change over the empty expression vector, pcDNA3.1, decreases dose dependently with increasing amounts of KLF15. (n = 3, mean ± S.D.)
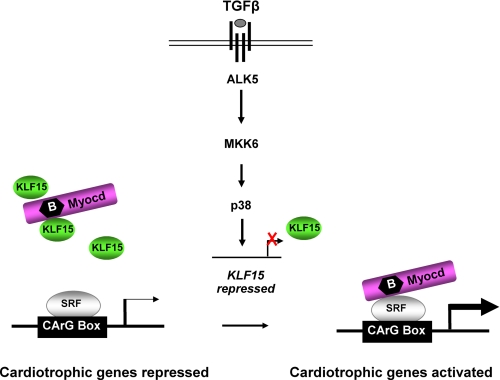
FIGURE 5.
Proposed mechanism of action of TGFβ-p38 MAPK-KLF15 cascade to explain induction of cardiomyocyte hypertrophy. KLF15 competes with SRF for a common docking site within myocardin, thereby limiting myocardin to activate cardiotrophic genes in normal adult myocytes. Growth factors repress KLF15 expression, allowing myocardin to bind SRF and activate cardiotrophic genes.
TGFβ Regulates the Expression of Myocardin Targets in Cardiomyocytes
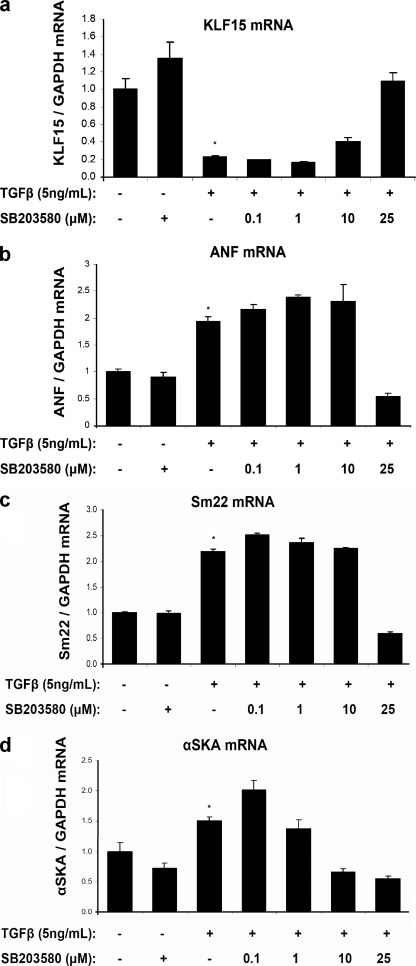
The observations that TGFβ represses KLF15 expression and that KLF15 inhibits myocardin activity predict that TGFβ increases the expression of myocardin target genes. To evaluate this, we stimulated cultured cardiomyocytes under serum-free conditions for 24 h with 5 ng/ml TGFβ and measured the expression of ANF, SM22, and αSKA (i.e. ACTC1), three bona fide myocardin/SRF target genes (4, 6, 27). As shown in Fig. 4, b–d, these SRF targets were significantly up-regulated in response to TGFβ stimulation, whereas myocardin levels were unchanged (data not shown). Regulation of myocardin target genes by TGFβ was fully prevented by titrating in p38 inhibitors (Fig. 4). These results provide further evidence that TGFβ regulates myocardin activity in cardiomyocytes via activation of p38 MAPK as depicted in the model in Fig. 5.
FIGURE 4.
Induction of myocardin/SRF target genes by TGFβ-p38 MAPK signaling. Quantitative real time PCR demonstrates that SRF targets, such as ANF, SM22, and α-skeletal actin are induced in cardiomyocytes in response to TGFβ treatment (24 h, 10 ng/ml serum-free medium), whereas KLF15 mRNA levels are down-regulated (n = 3/group; *, p < 0.05). This increase in SRF targets by TGFβ is counteracted by the p38 inhibitor SB203580.
DISCUSSION
In contrast to transcriptional activators, transcriptional inhibitors of LVH are less well explored. Recently, KLF15 was described as a novel transcriptional inhibitor of LVH (15). This study aimed to elucidate how KLF15 is regulated and how it inhibits pathological LVH. There is increasing recognition of the intrinsic differences between load induced, pathological LVH, and more physiological forms of cardiac hypertrophy as occurs after exercise (28). However, many factors involved in pathological LVH are also involved in adaptive LVH (29). More recently, some transcriptional mechanisms have been identified that specifically inhibit pathological LVH, like class II histone deacetylases and the transcriptional repressor NAB1 (12, 14). Although NAB1 is activated during LVH, class II histone deacetylases have been suggested to constitutively repress the expression of hypertrophy genes like MEF2 (12). Here, we show that KLF15 is a second constitutive transcriptional inhibitor of pathological LVH. The specific role of KLF15 in pathological forms is exemplified by the following findings: (a) KLF15 was not suppressed in exercise-induced LVH, and (b) KLF15 was significantly more suppressed in the hypertrophied Ren-2 hearts that would soon progress to failure. These findings suggest a novel mechanism in pathological LVH, where activated p38 MAPK actually down-regulates a repressive transcription factor. Earlier work showed the relation between loss of KLF15 and LVH in general. KLF15 was found to be expressed less in hypertrophied hearts as compared with the healthy adult myocardium. In addition, KLF15 null mice have higher base-line levels of BNP and ANF and respond to pressure overload with exaggerated expression of these genes, accompanied by higher mortality due to heart failure (15). This underlines the protective role of constitutive expression of KLF15. Because activation of p38 MAPK has been shown to activate numerous mechanisms, but has not yet been shown to repress important transcriptional regulators, we were surprised to find that activation of p38 MAPK was sufficient to decrease KLF15 (30). Although there is some debate about the role of p38 in the development of LVH (30, 31), published findings are in agreement with those we report here. Studies in cultured cardiomyocytes conclude that p38 activation is sufficient to induce myocyte hypertrophy marked by increased cell size and induction of hypertrophy markers like ANF, BNP, and αSKA (32). In vivo studies using transgenic overexpression mice of the p38 MAPK upstream kinases, MKK3 and MKK6, do not show myocyte hypertrophy, but instead they rapidly progress toward heart failure and have increased levels of embryonic genes like ANF, β-MHC, and αSKA (33), which is in line with the loss of repression of myocardin as we suggest here. Finally, it has been shown that activation of p38 MAPK via the TAK1 axis induces hypertrophy, which is in line with our findings (26). Furthermore, very recently it has been shown that KLF15 is not the only Krüppel-like factor that is regulated in cardiomyocytes. In cultured cardiomyocytes, ET-1 up-regulated several Krüppel-like factors like KLF2, KLF4, KLF5, and KLF6 and down-regulated KLF3, KLF11, and KLF15 (34) The possible involvement of p38 MAPK in the regulation of KLF expression was studied, but blockade of p38 by a p38 inhibitor did not have an effect on the ET-1-induced expression of KLF2, KLF4, KLF5, and KLF6.
We explored a possible mechanism by which KLF15 can repress pathological LVH. The role of myocardin in the development of hypertrophy and failure has recently been described (6, 27). Myocardin levels are reported to be increased upon induction of hypertrophy in cultured cardiomyocytes and in patients with LVH (27). Overexpression of myocardin in cardiomyocytes induces hypertrophy and increases ANF, BNP, and β-MHC expression (27). In vivo ablation of myocardin in adult cardiomyocytes in mice results in rapid heart failure, and the expression of myocardin/SRF-regulated sarcomeric genes is extinguished (6). Our study demonstrates that KLF15 is a very potent inhibitor of myocardin activity. Luciferase assays show that KLF15 suppresses myocardin-induced activation of three reporter vectors harboring one or more GArG-boxes (Fig. 2a). In addition, binding studies revealed that KLF15 is able to bind myocardin specifically in a 50-amino acid region where the SRF binding domain (basic domain) is also situated. Competition studies show that KLF15-mediated repression of myocardin activity is abolished by SRF in a dose-dependent manner. This indicates that KLF15 and SRF compete for binding the same region within myocardin. When we studied the effect of TGFβ on the expression of established myocardin targets SM22, ANF, and αSKA, we found these targets to be up-regulated. Moreover, this up-regulation was completely abolished by p38 inhibitors. This is congruent with the notion that TGFβ represses KLF15 expression via p38 MAPK, resulting in enhanced physical association of myocardin to SRF. Unchanged myocardin mRNA levels in response to TGFβ further underscore the interpretation that myocardin activity is not regulated at the level of transcription but rather by interactions at the protein level. Interestingly, a recent study has shown that myocardin mRNA and protein levels are increased in human and mouse failing hearts (27). This indicates that in failing hearts not only myocardin activity is enhanced by a decrease in KLF15 but also by enhancing protein levels of myocardin itself.
Taken together, our data suggest a novel pathway in pathological cardiac hypertrophy. We show here that down-regulation of KLF15 is a vital step in the development of hypertrophy and possibly its progression toward heart failure. We propose that in the healthy post-natal heart, where KLF15 levels are high, KLF15 inhibits the activity of myocardin, a potent transcriptional activator of numerous genes involved in cardiomyocyte hypertrophy like Anf, Sm22, and αska. During cardiac hypertrophy, when levels of TGFβ increase, p38 MAPK signaling is activated, which subsequently reduces KLF15 and releases the endogenous inhibition that KLF15 exerts on myocardin (Fig. 4).
In conclusion, our studies show that molecular mechanisms counteracting cardiomyocyte growth are dysregulated in pathological hypertrophy of the heart. The fact that KLF15 counteracts hypertrophy and is significantly down-regulated in pathological LVH suggests that therapeutic interventions aimed at preventing the decrease of KLF15 levels could be beneficial in the prevention of heart failure.
Supplementary Material
Acknowledgment
All animal experiments using Ren2 rats and neonatal rats were approved by the Animal Care and Use Committee of the University Maastricht and the University of Amsterdam and were performed according to the official rules formulated in the Dutch law on care and use of experimental animals.
This work was supported by Netherlands Heart Foundation Grants 2003T302 (to Y. M. P.) and 2007B077 (to E. E. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5 and Table 1.
- LVH
- left ventricular hypertrophy
- TGFβ
- transforming growth factor-β
- SRF
- serum response factor
- ANF
- atrial natriuretic factor
- BNP
- brain natriuretic peptide.
REFERENCES
- 1.Oka T., Xu J., Molkentin J. D. (2007) Semin. Cell Dev. Biol. 18, 117–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oka T., Maillet M., Watt A. J., Schwartz R. J., Aronow B. J., Duncan S. A., Molkentin J. D. (2006) Circ. Res. 98, 837–845 [DOI] [PubMed] [Google Scholar]
- 3.Zhang X., Azhar G., Chai J., Sheridan P., Nagano K., Brown T., Yang J., Khrapko K., Borras A. M., Lawitts J., Misra R. P., Wei J. Y. (2001) Am. J. Physiol. Heart Circ. Physiol. 280, H1782–H1792 [DOI] [PubMed] [Google Scholar]
- 4.Wang D., Chang P. S., Wang Z., Sutherland L., Richardson J. A., Small E., Krieg P. A., Olson E. N. (2001) Cell 105, 851–862 [DOI] [PubMed] [Google Scholar]
- 5.Pipes G. C., Creemers E. E., Olson E. N. (2006) Genes Dev. 20, 1545–1556 [DOI] [PubMed] [Google Scholar]
- 6.Huang J., Min Lu M., Cheng L., Yuan L. J., Zhu X., Stout A. L., Chen M., Li J., Parmacek M. S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 18734–18739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiu C., Bagnall R. D., Ingles J., Yeates L., Kennerson M., Donald J. A., Jormakka M., Lind J. M., Semsarian C. (2010) J. Am. Coll. Cardiol. 55, 1127–1135 [DOI] [PubMed] [Google Scholar]
- 8.Carballo S., Robinson P., Otway R., Fatkin D., Jongbloed J. D., de Jonge N., Blair E., van Tintelen J. P., Redwood C., Watkins H. (2009) Circ. Res. 105, 375–382 [DOI] [PubMed] [Google Scholar]
- 9.Mogensen J., Klausen I. C., Pedersen A. K., Egeblad H., Bross P., Kruse T. A., Gregersen N., Hansen P. S., Baandrup U., Borglum A. D. (1999) J. Clin. Invest. 103, R39–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Driest S. L., Jaeger M. A., Ommen S. R., Will M. L., Gersh B. J., Tajik A. J., Ackerman M. J. (2004) J. Am. Coll. Cardiol. 44, 602–610 [DOI] [PubMed] [Google Scholar]
- 11.Hardt S. E., Sadoshima J. (2004) Cardiovasc. Res. 63, 500–509 [DOI] [PubMed] [Google Scholar]
- 12.Zhang C. L., McKinsey T. A., Chang S., Antos C. L., Hill J. A., Olson E. N. (2002) Cell 110, 479–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong Y., Tannous P., Lu G., Berenji K., Rothermel B. A., Olson E. N., Hill J. A. (2006) Circulation 113, 2579–2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buitrago M., Lorenz K., Maass A. H., Oberdorf-Maass S., Keller U., Schmitteckert E. M., Ivashchenko Y., Lohse M. J., Engelhardt S. (2005) Nat. Med. 11, 837–844 [DOI] [PubMed] [Google Scholar]
- 15.Fisch S., Gray S., Heymans S., Haldar S. M., Wang B., Pfister O., Cui L., Kumar A., Lin Z., Sen-Banerjee S., Das H., Petersen C. A., Mende U., Burleigh B. A., Zhu Y., Pinto Y. M., Pinto Y., Liao R., Jain M. K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7074–7079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feinberg M. W., Lin Z., Fisch S., Jain M. K. (2004) Trends Cardiovasc. Med. 14, 241–246 [DOI] [PubMed] [Google Scholar]
- 17.van Vliet J., Crofts L. A., Quinlan K. G., Czolij R., Perkins A. C., Crossley M. (2006) Genomics 87, 474–482 [DOI] [PubMed] [Google Scholar]
- 18.Uchida S., Tanaka Y., Ito H., Saitoh-Ohara F., Inazawa J., Yokoyama K. K., Sasaki S., Marumo F. (2000) Mol. Cell. Biol. 20, 7319–7331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray S., Feinberg M. W., Hull S., Kuo C. T., Watanabe M., Sen-Banerjee S., DePina A., Haspel R., Jain M. K. (2002) J. Biol. Chem. 277, 34322–34328 [DOI] [PubMed] [Google Scholar]
- 20.Schroen B., Leenders J. J., van Erk A., Bertrand A. T., van Loon M., van Leeuwen R. E., Kubben N., Duisters R. F., Schellings M. W., Janssen B. J., Debets J. J., Schwake M., Høydal M. A., Heymans S., Saftig P., Pinto Y. M. (2007) J. Exp. Med. 204, 1227–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaiser R. A., Bueno O. F., Lips D. J., Doevendans P. A., Jones F., Kimball T. F., Molkentin J. D. (2004) J. Biol. Chem. 279, 15524–15530 [DOI] [PubMed] [Google Scholar]
- 22.Ruijter J. M., Ramakers C., Hoogaars W. M., Karlen Y., Bakker O., van den Hoff M. J., Moorman A. F. (2009) Nucleic Acids Res. 37, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuwahara K., Barrientos T., Pipes G. C., Li S., Olson E. N. (2005) Mol. Cell. Biol. 25, 3173–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L., Miano J. M., Mercer B., Olson E. N. (1996) J. Cell Biol. 132, 849–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Creemers E. E., Sutherland L. B., Oh J., Barbosa A. C., Olson E. N. (2006) Mol. Cell 23, 83–96 [DOI] [PubMed] [Google Scholar]
- 26.Zhang D., Gaussin V., Taffet G. E., Belaguli N. S., Yamada M., Schwartz R. J., Michael L. H., Overbeek P. A., Schneider M. D. (2000) Nat. Med. 6, 556–563 [DOI] [PubMed] [Google Scholar]
- 27.Xing W., Zhang T. C., Cao D., Wang Z., Antos C. L., Li S., Wang Y., Olson E. N., Wang D. Z. (2006) Circ. Res. 98, 1089–1097 [DOI] [PubMed] [Google Scholar]
- 28.McMullen J. R., Jennings G. L. (2007) Clin. Exp. Pharmacol. Physiol. 34, 255–262 [DOI] [PubMed] [Google Scholar]
- 29.Strøm C. C., Aplin M., Ploug T., Christoffersen T. E., Langfort J., Viese M., Galbo H., Haunsø S., Sheikh S. P. (2005) FEBS J. 272, 2684–2695 [DOI] [PubMed] [Google Scholar]
- 30.Liang Q., Molkentin J. D. (2003) J. Mol. Cell. Cardiol. 35, 1385–1394 [DOI] [PubMed] [Google Scholar]
- 31.Wang Y. (2007) Circulation 116, 1413–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zechner D., Thuerauf D. J., Hanford D. S., McDonough P. M., Glembotski C. C. (1997) J. Cell Biol. 139, 115–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao P., Georgakopoulos D., Kovacs A., Zheng M., Lerner D., Pu H., Saffitz J., Chien K., Xiao R. P., Kass D. A., Wang Y. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12283–12288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cullingford T. E., Butler M. J., Marshall A. K., Tham el L., Sugden P. H., Clerk A. (2008) Biochim. Biophys. Acta 1783, 1229–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.