Abstract
Our previous studies showed that basic fibroblast growth factor 2 (FGF2) null mice display markedly reduced bone mass and bone formation. However, the mechanism by which FGF2 regulates bone mass or bone formation is not fully defined. Activating transcription factor 4 (ATF4), one member of activating transcription factor/cAMP response element binding family, is a transcription factor required for osteoblast terminal differentiation. Here we investigate the ability of FGF2 to increase expression of ATF4 in bone marrow stromal cells (BMSCs) and examine ATF4 expression in Fgf2−/− BMSCs. We found that FGF2 stimulated ATF4 mRNA expression as early as 20 min and increased ATF4 protein expression after three hours of treatment. BMSCs from Fgf2+/+ and Fgf2−/− mice were cultured in osteogenesis medium. We observed reduced alkaline phosphatase staining, decreased mineralized nodules and reduced osteocalcin expression, and reduced expression of ATF4 in Fgf2−/− BMSC cultures compared to Fgf2+/+ BMSCs. This study is the first demonstration that ATF4 expression can be stimulated by FGF2 in osteoblasts and that ATF4 expression is significantly reduced in differentiated Fgf2−/− BMSCs. These results suggest that impaired bone mass and bone formation in Fgf2 null mice may be due in part to reduced ATF4 expression.
Keywords: FGF2, ATF4, osteoblasts
Introduction
Fibroblast growth factor (FGF) 2 is one member of the FGF polypeptide family. It is expressed in osteoblasts and stored in extracellular matrix [1]. In previous studies, we reported that FGF2 positively regulates bone formation and osteoblast differentiation. Disruption of the Fgf2 gene results in significantly decreased bone mass and bone formation revealed by histomorphometry and micro-CT in mice [2]. This phenotype may be due to the defect in osteoblast differentiation as shown by decreased alkaline phosphatase (ALP) and von Kossa staining in cultured Fgf2−/− BMSCs [2]. The decreased colony area can be partially rescued by exogenous FGF2 administration to Fgf2−/− BMSCs in vitro [2, 3]. FGF2 also stimulates bone formation in vivo [4, 5]. These studies demonstrate that FGF2 is a positive regulator of bone formation. However, the specific genes controlled by FGF2 signaling that mediate osteoblast differentiation and bone formation are not clear.
Differentiation of BMSCs into osteoblast is controlled by transcription factors, including Runx2, Osterix, and ATF4 [6]. ATF4 is required for osteoblast terminal differentiation and for maintaining mature osteoblast function including synthesizing collagen, the most abundant extracellular protein of bones [7]. One of the transcriptional targets of ATF4 is osteocalcin (OCN) [7], which is osteoblast specific and a marker for late-stage of osteoblast differentiation. The binding site for ATF4 in the OCN promoter is osteoblast specific element 1 [7]. The transcriptional activity of ATF4 requires phosphorylation. Decreased ATF4 phosphorylation is correlated with decreased bone formation [7]. The transcriptional activity of ATF4 in osteoblasts is further confirmed by ATF4 knock out mice. Atf4−/− mice display a dramatic decrease in OCN expression together with severe reduction of bone mass and bone formation [7]. In contrast, ATF4 overexpression increases OCN expression in osteoblasts [8, 9]. These studies demonstrate that ATF4 is a transcriptional activator required for osteoblast terminal differentiation including OCN expression.
In this report, we provide evidence that exogenous FGF2 is able to simulate ATF4 mRNA and protein expression rapidly in bone marrow stromal cells. Furthermore, we found that ATF4 mRNA and protein expression were greatly reduced in Fgf2−/− BMSCs compared to Fgf2+/+ BMSCs. We also found that FGF2 induces ATF4 expression in Fgf2−/− BMSCs. These studies indicate that impaired bone mass and bone formation in FGF2 null mice may due in part to reduced ATF4 expression.
Materials and Methods
Animals
Development of Fgf2 null mice was previously described [10]. Mice were bred and housed in the transgenic facility in the center for laboratory animal care at the University of Connecticut Health Center. Three or eight months old mice were utilized. Mice were sacrificed by CO2 narcosis and cervical dislocation. The Animal Care and Use Committee of the University of Connecticut Health Center approved animal protocols.
Bone marrow stromal cell cultures
Cells isolation was modified from previous methods [2]. Briefly, femurs and tibiae from male Fgf2+/+ and Fgf2−/− mice were dissected free of adhering tissue. After removing both ends of the bones, they were centrifuged briefly in eppendorf tubes. Then marrow cells were collected with α-MEM (Gibco-BRL, Grand Island, New York, USA). Cells were plated at 10×106 cells/well in six-well plates in α-MEM containing 10% fetal bovine serum, penicillin (100 U/ml) and streptomycin (50 μg/ml). For study of exogenous FGF2 stimulation of ATF4 expression, cells were half medium changed on day 3 and full medium changed on day 6 with α-MEM without FBS, then serum deprived 14 hrs before adding exogenous FGF2. For study of ATF4 expression in cultured BMSCs, cells were fed with α-MEM containing 10% FBS every 3 days in the first 8 days; from day 9, cells were fed with osteogenesis medium (α-MEM, 10% FBS, 8 mM β-glycerophosphate, and 50 μg/ml ascorbic acid) every other day until day 17. Cells were fixed and stained for alkaline phosphatase (ALP) positive colonies using a kit (Sigma Chemical, St. Louis, Missouri, USA). ALP stained dishes were re-stained with silver nitrate using von Kossa method to determine mineralization. Colony area was quantified with NIH Image (version 1.61; NIH, Bethesda, Maryland, USA).
RNA isolation and quantitative real-time PCR analysis
RNA was extracted from cells with TRIzol reagent (Invitrogen, CA, USA) according to the manufacture’s protocol. RNA (3 μg) was reverse transcribed using a commercial kit (Clontech, CA, USA). Quantitative real-time PCR was carried out using the QuantiTect™ SYBR Green PCR kit on a MyiQ™ instrument (Bio-Rad, Laboratories Inc Hercules, CA, USA). β-Actin or GAPDH was used as an internal reference for each sample. Mouse specific primers used were as follows: Atf4, 5′ –GAG CTT CCT GAA CAG CGA AGT G-3′ (forward), 5′-TGG CCA CCT CCA GAT AGT CAT C-3′ (reverse); Ocn, 5′-TAG TGA ACA GAC TCC GGC GCT A-3′ (forward), 5′-TGT AGG CGG TCT TCA AGC CAT-3′ (reverse); Gapdh, 5′-CAG TGC CAG CCTCGT CCC GTA GA-3′ (forward), 5′-CTG CAA ATG GCA GCC CTGGTG AC-3′ (reverse), β – Actin, 5′-ATC TGG CAC CAC CCT TCT ACAA-3′ (forward), 5′-ATG GCT GGG GTG TTG AAG GT-3′ (reverse).
Protein extraction and Western blot analysis
Whole cell extracts were harvested in RIPA buffer (Cell Signaling, Danvers, MA, USA). Cytoplasmic and nuclear extraction was harvested in Buffer A and buffer B seperately. Buffer A includes 10 mM Tris-HCl (pH 8.0), 10 mM KCl, 0.1 mM EGTA, 0.1 mM EDTA, 1X protease inhibitor cock tail; Buffer B includes 20 mM Tris-HCl (pH 8.0), 0.4 M NaCl, 0.1 mM EGTA, 0.1 mM EDTA, 1X protease inhibitor cock tail. Total protein concentration was assayed with BCA protein assay reagent (Pierce, IL, USA). After SDS-PAGE on 15% gels, resolved samples were transferred onto a PVDF membrane (Bio Rad, CA, USA). Membranes were blocked for 1 h with 5% nonfat dry milk, and then incubated overnight at 4°C with anti-ATF4 (Santa Cruz, CA, USA). Membranes were incubated with appropriate secondary antibody (Amersham Bioscience) at room temperature for 45 min. Blots were developed with ECL plus reagents (Amersham Bioscience). Finally, blots were reprobed with β-actin antibody (Sigma, MO, USA) for normalization. Western blot bands were analyzed by NIH Image (version 1.61; NIH, Bethesda, Maryland, USA).
Statistical analysis
Results were expressed as means ± S.E. Differences between groups were analyze using student’s t test, and differences were considered significant at p values of less than 0.05.
Results
FGF2 stimulates ATF4 expression in Fgf2+/+ BMSCs
We examined the ability of FGF2 to increase ATF4 expression in BMSCs. We first assessed the expression of ATF4 in FGF2 (0.1 nM) treated Fgf2+/+ BMSCs. We found that exogenous FGF2 was able to increase ATF4 mRNA expression 2.5 folds as early as 20 min (Fig 1A). Consistent with elevated mRNA expression, ATF4 protein expression was also increased after three hours of treatment with exogenous FGF2 (0.1 nM) (Fig 1B). The increased ATF4 protein expression was observed both in cytosolic and nuclear fractions.
Figure 1.
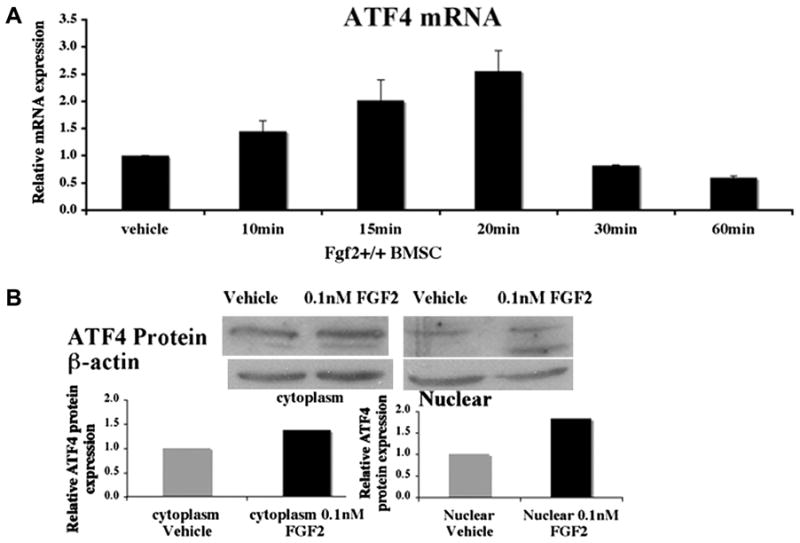
FGF2 stimulates ATF4 expression in Fgf2+/+ BMSCs. Freshly isolated BMSCs were cultured in αMEM 10% FBS for 6 ds, serum deprived 14 hrs, then (A) treated with 0.1 nM FGF2 or vehicle at indicated times, followed by RNA extraction. ATF4 mRNA was quantified by quantitative real time PCR, normalized to GAPDH. (B) BMSCs cultured as above were treated with 0.1 nM FGF2 or vehicle for 3 hrs, followed by extraction of cytosolic and nuclear protein fraction and Western Blots. Western blot Bands were analyzed by NIH Image. Two or three independent experiments were conducted with similar results, and results of representative experiments are shown.
FGF2 deficiency results in decreased ATF4 expression
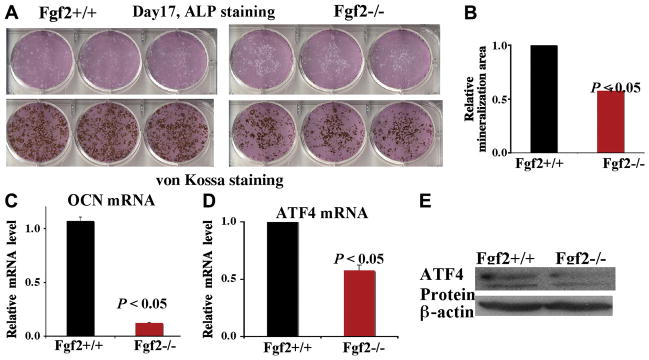
To further study that FGF2 regulates ATF4 expression in BMSCs, we next examined bone nodule formation and the expression level of ATF4 in differentiated BMSCs. BMSCs isolated from Fgf2+/+ and Fgf2−/− mice were cultured in proliferation medium for nine days, then cultured in differentiation medium for another eight days. After 17 days of culture, we assessed the differentiation status of BMSCs by ALP staining and von Kossa staining as well as the expression of genes important in osteoblast terminal differentiation. We found decreased ALP staining and marked reduction of mineralized nodules in Fgf2−/− BMSCs (Fig 2A). Mineralization area in Fgf2−/− cultures was also significantly reduced compared to Fgf2+/+ cultures (Fig 2B), analyzed by NIH image software. The decreased ALP staining and mineralization area suggest impaired differentiation and mineralization in Fgf2−/− BMSC cultures. OCN is a hallmark of osteoblast terminal differentiation. Consistent with decreased mineralization, we observed reduced OCN expression in Fgf2−/− BMSC cultures (Fig 2C). Interestingly, we also found significantly decreased ATF4 mRNA (Fig 2D) as well as reduced ATF4 protein expression (Fig 2E) in Fgf2−/− BMSC cultures compared to Fgf2+/+ BMSC cultures.
Figure 2.
FGF2 deficiency results in decreased ATF4 expression. Freshly isolated BMSCs were cultured in αMEM/10% FBS for 9 ds, then in αMEM/10% FBS with 50μg/ul ascorbic acid and 8 mM β-glycerophosphate for another 8 ds, followed by ALP, Von Kossa staining (A) and quantification of total mineralization area (B) and RNA extraction for gene analysis for OCN (C) and for ATF4 (D) by quantitative real-time PCR, normalized to β –actin, (E) whole cell extracts for ATF4 protein analysis by Western Blots. Three independent experiments were conducted with similar results, and result of a representative experiment is shown.
FGF2 increases ATF4 expression in Fgf2−/− BMSCs
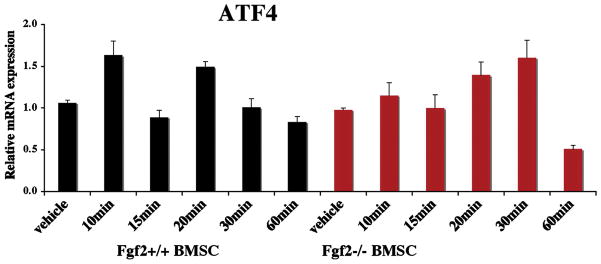
We next assessed whether exogenous FGF2 is able to induce ATF4 expression in Fgf2−/− BMSCs. We observed increased ATF4 mRNA expression as early as 20 min after FGF2 (0.1 nM) treatment (Fig 3).
Figure 3.
Exogenous FGF2 increases ATF4 expression in Fgf2−/− BMSCs. Freshly isolated BMSCs were cultured in MEM 10% FBS for 6 ds, serum deprived 14 hrs, then treated with 0.1 nM FGF2 or vehicle at indicated times, followed by RNA extraction. ATF4 mRNA was quantified by quantitative PCR, normalized to GAPDH. Two independent experiments were conducted with similar results, and result of a representative experiment is shown.
Discussion
In the present study, we found that exogenous FGF2 is able to rapidly stimulate transcription factor ATF4 in Fgf2+/+ BMSCs. Furthermore, FGF2 deficiency resulted in greatly reduced ATF4 expression in cultured BMSCs. We also observed that FGF2 is able to induce ATF4 mRNA expression in Fgf2−/− BMSCs. These results indicate that FGF2 positively regulates ATF4 expression in osteoblasts.
ATF4 is a recently identified transcription factor for osteoblast terminal differentiation. ATF4 positively regulates OCN expression in osteoblasts. ATF4 knock out in mice results in dramatic reduction of OCN expression in bone [7] while ATF4 overexpression increases OCN expression [9]. Atf4−/− mice display greatly decreased bone mass and bone formation [7]. These studies demonstrate that ATF4 is essential for osteoblast differentiation and function.
Our previous studies showed that disruption of Fgf2 gene results in decreased differentiation potential of BMSCs into osteoblasts [2]. The decreased differentiation ability of osteoblasts lead to markedly reduced bone mass as well as bone formation in Fgf2 null mice. The mechanism(s) by which that FGF2 positively regulates bone formation and bone mass is not well understood. We now show that FGF2 could stimulate ATF4 expression rapidly in BMSCs and that Fgf2−/− BMSCs display greatly reduced ATF4 expression. Therefore, we propose that ATF4 is likely one of the novel downstream targets of FGF2 signaling in osteoblasts (Fig 4). The impaired bone mass and bone formation in Fgf2 null mice may be partially due to reduced ATF4 expression.
Figure 4.
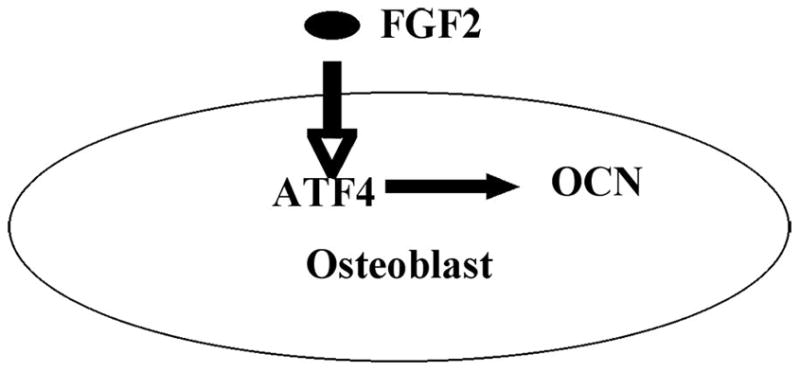
Schematic model – FGF2 elevates ATF4 expression in osteoblasts. FGF2 increases expression of ATF4, which promotes osteoblast terminal differentiation and regulates osteoblast specific gene including OCN expression.
Our finding that FGF2 is able to stimulate ATF4 expression is supported by other studies in non-osteoblastic cells. A recent study showed that FGF2 induced ATF4 expression in vascular smooth muscle cells [11]. However, in the same study FGF2 was not able to induce ATF4 expression in bovine aortic endothelial cells [11]. This phenomenon suggests that either FGF2 signaling response is unique to a particular cell type or regulation of ATF4 expression in various cell types is different. Our results demonstrate for the first time increased ATF4 expression induced by FGF2 in osteoblasts.
ATF4 is required for osteoblast terminal differentiation. Disruption of Atf4 gene did not alter expression of Runx2 or Osterix, two other transcription factors required for differentiation of bone marrow stromal cell into osteoblasts [7]. Consistent with an important role in terminal osteoblast differentiation, disruption of Atf4 gene did not modulate α1(I) Collagen and other genes that are expressed in the early stage of osteoblast differentiation [7]. In contrast, expression of OCN, a gene characteristic of late stage of osteoblast differentiation was dramatically reduced in the absence of Atf4 [7]. In accordance with the function of ATF4 promoting osteoblast terminal differentiation, we observed reduced ATF4 expression and dramatically decreased OCN expression in Fgf2−/− BMSCs.
As previously described, another critical transcription factor for osteoblast differentiation is Runx2 [6]. Previous studies showed that FGF2 was able to induce expression of Runx2 [12–14]. FGF2 was also able to promote Runx2 nuclear localization [15], a requirement for its function as a transcription factor. Here, we report that FGF2 is able to induce expression of ATF4 in BMSCs, another key transcription factor for osteoblast differentiation and function. Moreover, FGF2 has been shown to increase osteoblast specific OCN gene expression in osteoblasts [12–14]. Induction of OCN expression required interaction between ATF4 and Runx2 in the OCN promoter region [9]. We previously reported reduced OCN expression, reduced mineralization, and decreased expression of Runx2 in Fgf2−/− BMSCs compared to Fgf2+/+ BMSCs [2, 3]. Since we also found decreased ATF4 expression in Fgf2−/− BMSCs in the present study, it is highly likely that disruption of Fgf2 gene leads to decreased expression of transcription factors ATF4 and Runx2 and thus attenuated interaction between ATF4 and Runx2, which result in reduced osteoblast differentiation, reduced mineralization and decreased OCN expression.
To our knowledge, this study is the first demonstration that ATF4 expression can be stimulated by FGF2 in osteoblasts and that ATF4 expression is significantly reduced in the absence of endogenous FGF2 in BMSCs cultures. These results suggest that impaired bone mass and bone formation in Fgf2 null mice may be due in part to reduced ATF4 expression.
Acknowledgments
This project was supported by NIH Grant AG02119-05 to MM. Hurley.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Yurong Fei, Email: yfei@student.uchc.edu.
Liping Xiao, Email: xiao@uchc.edu.
Marja M. Hurley, Email: hurley@exchange.uchc.edu.
References
- 1.Hurley MM, Marie PJ, Florkiewics RZ. Fibroblast growth factor and fibroblast growth factor receptor families. In: Bilezikian JP, Raisz LG, Rodan G, editors. Principles of bone biology. Academic Press; San Diego: 2002. pp. 627–645. [Google Scholar]
- 2.Montero A, Okada Y, Tomita M, Ito M, Tsurukami H, Nakamura T, Doetschman T, Coffin JD, Hurley MM. Disruption of the fibroblast growth factor-2 gene results in decreased bone mass and bone formation. J Clin Invest. 2000;105:1085–1093. doi: 10.1172/JCI8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naganawa T, Xiao L, Abogunde E, Sobue T, Kalajzic I, Sabbieti M, Agas D, Hurley MM. In vivo and in vitro comparison of the effects of FGF-2 null and haplo-insufficiency on bone formation in mice. Biochemical and Biophysical Research Communications. 2006;339:490–498. doi: 10.1016/j.bbrc.2005.10.215. [DOI] [PubMed] [Google Scholar]
- 4.Mayahara IT, Nagai H, Miyajima H, Tsukuda R, Taketomi S, Mizoguchi J, Kato K. In vivo stimulation of endosteal bone formation by basic fibroblast growth factor in rats. Growth Factors. 1993;9:73–80. doi: 10.3109/08977199308991583. [DOI] [PubMed] [Google Scholar]
- 5.Nakamura K, Kurokawa T, Aoyama I, Hanada K, Tamura M, Kawaguchi H. Stimulation of bone formation by intraosseous injection of basic fibroblast growth factor in ovariectomised rats. Int Orthop. 1998;22:49–54. doi: 10.1007/s002640050207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karsenty G. Transcriptional control of skeletogenesis. Annu Rev Genomics Hum Genet. 2008;9:183–196. doi: 10.1146/annurev.genom.9.081307.164437. [DOI] [PubMed] [Google Scholar]
- 7.Yang X, Matsuda K, Bialek P, et al. ATF4 Is a Substrate of RSK2 and an Essential Regulator of Osteoblast Biology. Cell. 2004;117:387–398. doi: 10.1016/s0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- 8.Yu S, Franceschi RT, Luo M, et al. Parathyroid Hormone Increases Activating Transcription Factor 4 Expression and Activity in Osteoblasts: Requirement for Osteocalcin Gene Expression. Endocrinology. 2008;149:1960–1968. doi: 10.1210/en.2007-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao G, Jiang D, Ge C, et al. Cooperative Interactions between Activating Transcription Factor 4 and Runx2/Cbfa1 Stimulate Osteoblast-specific Osteocalcin Gene Expression. J Biol Chem. 2005;280:30689–30696. doi: 10.1074/jbc.M500750200. [DOI] [PubMed] [Google Scholar]
- 10.Zhou M, Sutliff RL, Paul PJ, et al. Fibroblast growth factor 2 control of vascular tone. Nat Med. 1998;4:201–207. doi: 10.1038/nm0298-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malabanan KP, Kanellakis P, Bobik A, Khachigian LM. Activation Transcription Factor-4 Induced by Fibroblast Growth Factor-2 Regulates Vascular Endothelial Growth Factor-A Transcription in Vascular Smooth Muscle Cells and Mediates Intimal Thickening in Rat Arteries Following Balloon Injury. Circ Res. 2008;103:378–387. doi: 10.1161/CIRCRESAHA.107.168682. [DOI] [PubMed] [Google Scholar]
- 12.Xiao G, Jiang D, Gopalakrishnan R, Franceschi RT. Fibroblast growth factor 2 induction of the osteocalcin gene requires MAPK activity and phosphorylation of the osteoblast transcription factor, Cbfa1/Runx2. J Biol Chem. 2002;277:36181–36187. doi: 10.1074/jbc.M206057200. [DOI] [PubMed] [Google Scholar]
- 13.Kim HJ, Kim JH, Bae SC, Choi JY, Ryoo HM. The Protein Kinase C Pathway Plays a Central Role in the Fibroblast Growth Factor-stimulated Expression and Transactivation Activity of Runx2. J Biol Chem. 2003;278:319–326. doi: 10.1074/jbc.M203750200. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, Sobue T, Hurley MM. FGF2 increases colony formation, PTH receptor, and IGF-1 mRNA in mouse marrow stromal cells. Biochemical and Biophysical Research Communications. 2002;290:526–531. doi: 10.1006/bbrc.2001.6217. [DOI] [PubMed] [Google Scholar]
- 15.Sabbieti MG, Agas D, Xiao L, Marchetti L, Coffin JD, Doetschman T, Hurley MM. Endogenous FGF-2 is critically important in PTH anabolic effects on bone. J Cell Physiol. 2009;219:143–151. doi: 10.1002/jcp.21661. [DOI] [PMC free article] [PubMed] [Google Scholar]