Abstract
Environmental DNA-derived cosmid clones containing type II polyketide synthase genes were screened for the ability to produce clone-specific metabolites in Streptomyces. One of the Streptomyces recombinants produces erdacin, a novel polyketide with a previously unknown pentacyclic skeleton. Erdacin provides tangible evidence that environmental DNA gene clusters are likely to encode the biosynthesis of molecules that are substantively different from those that have been identified using culture-based strategies.
Keywords: Natural product, polyketides, environmental DNA, metagenomics, biosynthesis
It is now well established that environmental samples contain a significantly more diverse collection of bacteria than that which is readily grown in the laboratory.[1-4] Large insert clone libraries of DNA extracted directly from environmental samples (environmental DNA, eDNA) provide a means to retrieve the natural product biosynthetic gene clusters found in the genomes of these previously inaccessible bacteria. One of the challenges associated with the discovery of new molecules from large eDNA libraries has been the identification of clones containing intact gene clusters that can be used to produce small molecules in heterologous hosts.[5-10] Here we report the recovery of a collection of type II (iterative, aromatic) polyketide synthase (PKS) containing clones from an eDNA cosmid library and the characterization of erdacin, a novel pentacyclic polyketide produced by Streptomyces albus transformed with one of these clones. This study provides one of the clearest indications yet that DNA derived from the large unstudied collection of bacteria present in most environmental samples has the potential to encode the biosynthesis of molecules that are substantively different from known metabolites and not simply derivatives of molecules previously identified in culture-based studies.
Type II PKS gene clusters contain a conserved minimal PKS that is composed of two ketosynthase genes (KSα and KSβ) and an acyl carrier protein (ACP). The minimal PKS is responsible for the iterative condensation of malonyl-CoA into a nascent polyketide chain that can be cyclized, aromatized, reduced, rearranged and functionalized into a vast assortment of structurally unique metabolites.[11-15] Because the minimal PKS is used iteratively, type II PKS gene clusters are smaller than most natural product gene clusters that encode the biosynthesis of comparably complex metabolites. As a result, it is possible to capture functionally complete type II PKS pathways on individual cosmid clones. We hypothesized that eDNA cosmid clones containing type II PKS biosynthetic machinery would be a productive starting point for the discovery of novel secondary metabolites from eDNA cosmid libraries. While both PCR based studies and high throughput sequencing efforts indicate that eDNA samples contain a plethora of novel type II PKS genes, none of these previous studies have focused on the recovery of functionally intact type II PKS gene clusters, and, as a result, no new metabolites have been produced from eDNA-derived type II PKS gene clusters.[9, 16-18]
Degenerate primers based on conserved regions of minimal PKS KSα and ACP genes were used to amplify full length KSβ sequences captured in an eDNA library constructed from desert soil collected in Utah (Fig. 1).[9, 19, 20] Of the 21 unique KSβ sequences amplified from this eDNA library, only one showed greater than 80% identity to a previously reported KSβ gene. The low identity these sequences exhibit to known KSβ sequences suggested they are associated with gene clusters that are functionally distinct from any previously sequenced gene clusters and might, therefore, encode the biosynthesis of novel secondary metabolites. To explore this possibility, eDNA-derived KSβ sequences were used as probes to identify and recover ten cosmid clones containing type II PKS biosynthetic genes from the soil eDNA library. The recovered clones were retrofitted with the genetic elements necessary for conjugation and integration into Streptomyces; and eight of the retrofitted clones were successfully integrated into both S. lividans and S. albus. Based on antibacterial assays, HPLC analysis and color production (Fig. 1), five of the eight clones appear to produce clone-specific metabolites. V167, which produces large quantities (15-20 mg/L) of one major clone-specific metabolite in S. albus, was selected for further analysis.
Figure 1.
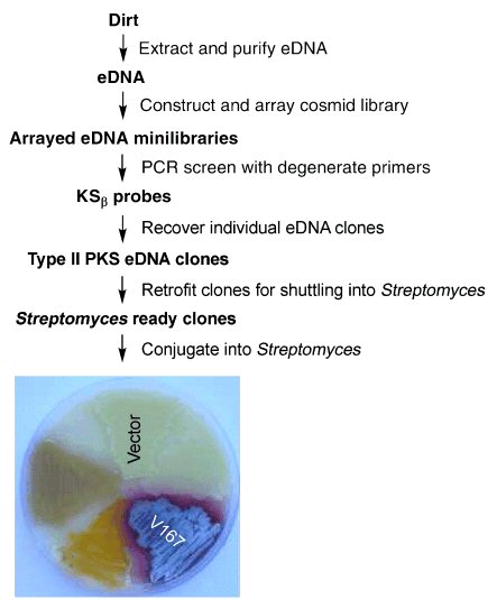
General scheme used for the recovery of Type II PKS eDNA clones and the heterologous expression of erdacin (1) from clone V167. S. albus transformed with a vector control and eDNA-derived clones containing Type II PKS biosynthetic genes are shown. The colored phenotype exhibited by some clones is indicative of the production of clone-specific small molecules.
The major clone-specific metabolite found in ethyl acetate extracts of V167 cultures grown in modified Streptomyces supplemented minimal media was purified by reversed phase HPLC and given the trivial name erdacin (1).[21] Extensive 1- and 2-D NMR analysis of erdacin, obtained from both 13C-labeled and naturally abundant 12C cultures, allowed us to unambiguously assign the position of all but five carbon atoms (C-2, -3, -4, -22 and -23) in the final structure (Fig. 2).[22] The position of the final five carbons could not be assigned with certainty because many of the NMR signals from this region of the structure were either weak or absent.
Figure 2.
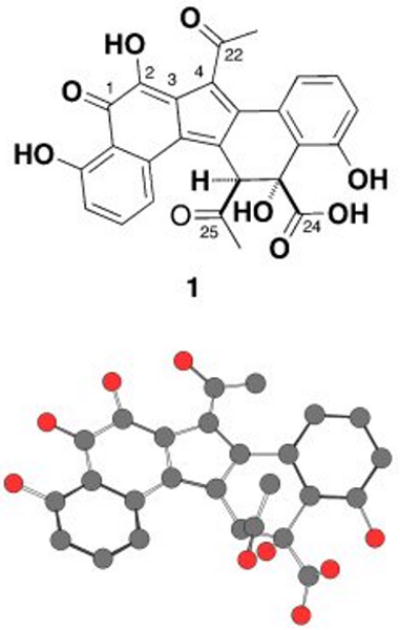
Chemical and computer-generated perspective drawings of erdacin (1). No hydrogens have been included in the computer-generated perspective drawing. The pentacyclic ring system found in erdacin has not been previously reported from culture-dependent natural product studies.
To complete the structural characterization of erdacin, crystals were obtained by slow evaporation from acetonitrile and water and then analyzed by single crystal X-ray diffraction (CDCC deposit number 721152). The X-ray structure confirmed that erdacin is based on a novel pentacyclic (6-6-5-6-6) ring system. As drawn in Figure 2, the upper edge of the central five-membered ring is functionalized with an acetyl group and the bottom edge of the third six-membered ring is functionalized with both an acetyl group and a carboxylate. To the best of our knowledge, erdacin is a novel natural product. Neither the erdacin carbon skeleton nor the pentacyclic ring system appears as substructures in any known secondary metabolites characterized from culture-dependent studies. Pentacylic structures with the same number of five- and six-membered rings have been reported as natural products; however, the organization of the rings in known structures does not match that seen in erdacin.[23]
The weak NMR signals that prevented the unambiguous assignment of the complete erdacin structure by NMR alone likely result from an interaction between the C-2 hydroxyl and the C-22 carbonyl oxygen. Based on the final erdacin structure, a C-2 hydroxyl assisted tautomerization of the C-22 carbonyl could explain both the broad NMR signals in this region of the structure and the rapid disappearance of the C-23 methyl protons that occurs in protic NMR solvents.
A hint as to the biosynthetic origin of this new structure comes from the characterization of juglomycin F (4),[24] a second less abundant clone-specific metabolite found in V167 culture broth extracts. The carbon skeleton of this known 13-carbon metabolite maps onto one half of the erdacin structure suggesting that erdacin might arise from the heterodimerization of 4 (or a close relative of 4) and a second 13-carbon subunit (Fig. 3b). In an attempt to better understand the origin of 1, the V167 insert was sequenced and transposon mutagenized. Figure 3a shows the position of key transposon insertions that result in easily discernable changes in the V167 colony phenotype. All of these transposons are found in a 20 kb region with 22 predicted open reading frames that we have called the Erd gene cluster. Due to the very high GC content (>90%) of some regions of the Erd gene cluster, we were not able to obtain transposon insertions in every predicted Erd open reading frame. The Erd gene cluster contains a minimal PKS, a collection of post PKS enzymes, regulatory proteins, a transporter and a number of hypothetical proteins.
Figure 3.
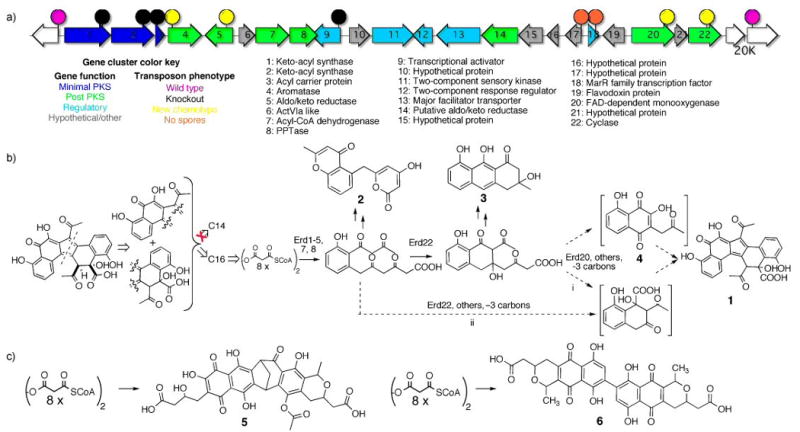
a. The Erd gene cluster (GenBank accession number FJ719113) and the location of key transposon insertions discussed in the text. b. A retrobiosynthetic analysis of erdacin suggests that it might arise from two 13-carbon monomers, however, only 16-carbon compounds accumulate in transposon knockouts of early post miminal PKS genes. A proposed biosynthetic scheme for the formation of erdacin from an octaketide precursor is shown. Compounds 1-4 were isolated from wild type and mutant V167 cultures. Shown in brackets are juglomycin F (4) and a hypothetical 13-carbon polyketide which together have carbon skeletons that accout for the upper and lower halves of erdacin, respectively. c. The Erd minimal PKS genes are related to minimal PKS genes involved in the biosynthesis of the two structurally distinct octaketide dimers naphthocyclinone (5) and actinorhodin (6).
Transposon insertions in early post minimal PKS biosynthetic genes (Erd4, 5, and 22) knock out the production of erdacin and unexpectedly lead to the accumulation known octaketide shunt products instead of the 13-carbon (or 14-carbon) intermediates that are suggested by the structure of erdacin. The known early octaketide shunt products SEK4b, mutactin, and SEK34b (2) were found in extracts from Erd5, -4 and -22 mutants, respectively (Fig. 1 supporting information).[14] Transposon insertions in Erd20, a predicted monooxygenase, lead to the production of apparently reactive intermediates that generate a number of compounds with a range of molecular weights (m/z from 240 to 315). One of the stable components of this mixture was fully characterized and found to be the decarboxylated octaketide prechrysophanol (3).[25] Based on the metabolites identified in extracts from both wild type and mutant V167 cultures, it appears likely that the Erd gene cluster converts an octaketide produced by the minimal PKS into two distinct intermediates that give rise to erdacin (Fig. 3).
The formation of a juglomycin-like structure from an octaketide precursor has been reported previously.[26] While the isolation of juglomycin F (4) supports the existence of one 13-carbon precursor, the origin of the second 13-carbon subunit is not immediately obvious. Two possible routes for the formation of the second 13-carbon monomer with the necessary carbon skeleton are shown in Figure 3b (3bi rearrangement, 3bii alternate cyclization). To distinguish between these two potential routes a [1,2-13C]acetate stable isotope feeding experiment was carried out on cultures of V167. The appearance of the C-24 carboxylate as a doublet in the 13C NMR spectra of erdacin derived from this feeding study indicates that C-24 and C-12 are incorporated as an intact acetate unit and therefore supports 3bii as the likely route for the formation of the lower half of erdacin (Fig. 4).
Figure 4.
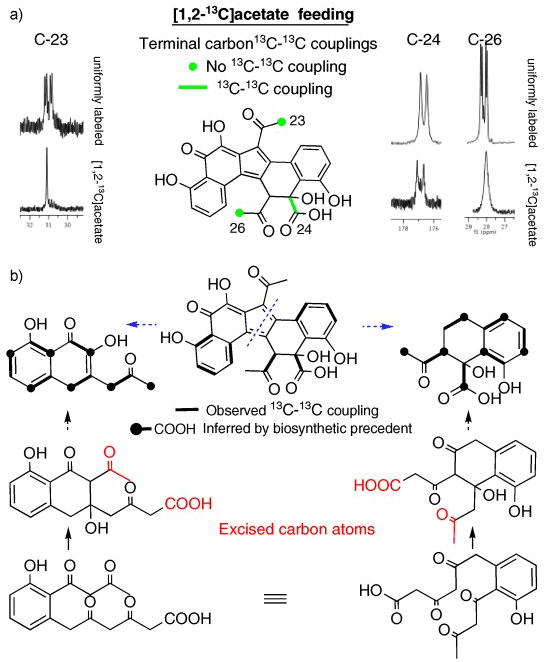
13C-13C couplings (or in some instances the absence of coupling) observed in 13C (a) and 2D-INADEQUATE (b) NMR spectra of [1,2-13C]acetate labelled erdacin indicate that two distinct polyketide cyclization patterns are used in the biosynthesis of erdacin. Couplings between weak or overlapping NMR signals not clearly seen by 2D-INADEQUATE were inferred based on polyketide biosynthetic precedent. The exact structure of the late stage polyketide intermediates used in the biosynthesis of erdacin is not known.
In the biosynthetic scheme shown in Figure 3b both methyl ketones are predicted to arise from the decarboxylation of terminal acetates, instead of intact acetates as might be predicted from glancing at the structure of erdacin. In the 13C NMR spectra of [1,2-13C]acetate labelled erdacin the methyl carbons are not coupled to the adjacent carbonyls confirming that, as predicted, neither methyl ketone arises from the intact incorporation of acetate. Additional 13C-13C couplings observed in 2D-INADEQUATE experiments (Fig. 4b) run on [1,2-13C]acetate labelled erdacin support the cyclization patterns suggested by both the transposon mutagenesis studies and the couplings observed in the 13C NMR spectra of [1,2-13C]acetate labelled erdacin (Fig. 4a).
While the exact nature of the late stage polyketide intermediates used in the biosynthesis of erdacin is not known, it appears that erdacin arises from two distinct 13-carbon substructures that are derived from a common octaketide intermediate. In the proposed biosynthetic scheme a common octaketide precursor would undergo two distinct cyclizations. At some point in the biosynthetic process these octaketide intermediates would loose the same three carbons (terminal carboxylate and terminal methyl ketone) to yield a pair of 13-carbon intermediates that make up the two halves of erdacin (Fig. 4).
The Erd minimal PKS enzymes are most closely related to ACP, KSα and KSβ sequences from gene clusters that encode the biosynthesis of naphthocyclinon, actinorhodin, granaticin and medermycin.[14, 27-29] While granaticin and medermycin are simple octaketide structures, naphthocyclinone (5) and actinorhodin (6) are octaketide dimers that arise from two distinct dimerization strategies (Fig. 3c).[14, 27-29] Erdacin appears to result from a third previously unseen octaketide dimerizatoin strategy. By leveraging biosynthetic information from sequenced gene clusters we have been able to recover an eDNA-derived gene cluster that is sufficiently different from known clusters such that it encodes the biosynthesis of a structurally unprecedented secondary metabolite.
Transposon insertions in either the hypothetical protein Erd17 or the MarR-like transcription factor, Erd18, lead to mutants that no longer sporulate, indicating that the Erd gene cluster may interfere with sporulation.[30] During our exploration of potential bioactivities for erdacin, we found that it exhibits significant antioxidant activity. In a standard copper reduction assay, erdacin exhibits antioxidant activity that is twice that reported for many well-known antioxidants, including ascorbic acid (vitamin C).[31] Studies are currently underway to further explore the bioactivity of erdacin.
The systematic recovery and large-scale investigation of eDNA clones containing minimal PKS gene clusters is likely to be a productive avenue for the culture-independent discovery of structurally unique secondary metabolites. More generally, this work suggests that “compact” natural product biosynthetic pathways that can be captured on individual cosmid sized clones (i.e. Type II PKS, Type III PKS, aminoglycosides) are likely to be rewarding systems to examine in the search for novel metabolites from eDNA libraries. The discovery of erdacin, from the small sample of eDNA-derived clones investigated in this study, provides tangible evidence that the diverse collection of gene clusters predicted to be present in environmental bacteria likely encodes the biosynthesis of metabolites that are, in many cases, distinct from those that have been characterized using traditional cultured-based discovery strategies.
Footnotes
This work was supported by NIH GM077516, The Beckman Foundation, The Searle Foundation and The Hershel Foundation. We thank Emil Lobkovsky for his assistance with our X-ray studies.
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author
References
- 1.Rappe MS, Giovannoni SJ. Annu Rev Microbiol. 2003;57:369. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- 2.Torsvik V, Ovreas L, Thingstad TF. Science. 2002;296:1064. doi: 10.1126/science.1071698. [DOI] [PubMed] [Google Scholar]
- 3.Curtis TP, Sloan WT, Scannell JW. Proc Natl Acad Sci U S A. 2002;99:10494. doi: 10.1073/pnas.142680199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Torsvik V, Salte K, Sorheim R, Goksoyr J. Appl Environ Microbiol. 1990;56:776. doi: 10.1128/aem.56.3.776-781.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piel J, Hui D, Wen G, Butzke D, Platzer M, Fusetani N, Matsunaga S. Proc Natl Acad Sci U S A. 2004;101:16222. doi: 10.1073/pnas.0405976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schirmer A, Gadkari R, Reeves CD, Ibrahim F, DeLong EF, Hutchinson CR. Appl Environ Microbiol. 2005;71:4840. doi: 10.1128/AEM.71.8.4840-4849.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Courtois S, Cappellano CM, Ball M, Francou FX, Normand P, Helynck G, Martinez A, Kolvek SJ, Hopke J, Osburne MS, August PR, Nalin R, Guerineau M, Jeannin P, Simonet P, Pernodet JL. Appl Environ Microbiol. 2003;69:49. doi: 10.1128/AEM.69.1.49-55.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brady SF, Bauer JD, Clarke-Pearson MF, Daniels R. J Am Chem Soc. 2007;129:12102. doi: 10.1021/ja075492v. [DOI] [PubMed] [Google Scholar]
- 9.Seow KT, Meurer G, Gerlitz M, Wendt-Pienkowski E, Hutchinson CR, Davies J. J Bacteriol. 1997;179:7360. doi: 10.1128/jb.179.23.7360-7368.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ginolhac A, Jarrin C, Gillet B, Robe P, Pujic P, Tuphile K, Bertrand H, Vogel TM, Perriere G, Simonet P, Nalin R. Appl Environ Microbiol. 2004;70:5522. doi: 10.1128/AEM.70.9.5522-5527.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hertweck C, Luzhetskyy A, Rebets Y, Bechthold A. Nat Pro Rep. 2007;24:162. doi: 10.1039/b507395m. [DOI] [PubMed] [Google Scholar]
- 12.Shen B. Curr Top Chem. 2000;209:1. [Google Scholar]
- 13.McDaniel R, Ebert-Khosla S, Hopwood DA, Khosla C. Nature. 1995;375:549. doi: 10.1038/375549a0. [DOI] [PubMed] [Google Scholar]
- 14.McDaniel R, Ebert-Khosla S, Hopwood DA, Khosla C. J Am Chem Soc. 1994;116:10855. [Google Scholar]
- 15.Matharu AL, Cox RJ, Crosby J, Byrom KJ, Simpson TJ. Chem Biol. 1998;5:699. doi: 10.1016/s1074-5521(98)90663-9. [DOI] [PubMed] [Google Scholar]
- 16.Wawrik B, Kerkhof L, Zylstra GJ, Kukor JJ. Appl Environ Microbiol. 2005;71:2232. doi: 10.1128/AEM.71.5.2232-2238.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pang M, Tan GA, Abdullah N, Lee C, Ng C. Biotechnology. 2008;7:660. [Google Scholar]
- 18.Wawrik B, Kutliev D, Abdivasievna UA, Kukor JJ, Zylstra GJ, Kerkhof L. Appl Environ Microbiol. 2007;73:2982. doi: 10.1128/AEM.02611-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brady SF. Nature Protocols. 2007;2:1297. doi: 10.1038/nprot.2007.195. [DOI] [PubMed] [Google Scholar]
- 20.Banik JJ, Brady SF. Proc Natl Acad Sci U S A. 2008;105:17273. doi: 10.1073/pnas.0807564105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Four-day-old cultures of V167 grown in modified SMM (30°C at 250 rpm) were acidified to pH 3 and then extracted twice with an equal volume of ethyl acetate. The dried extract was dissolved in methanol, passed through a SPE Previal C-18 plug and then partitioned by reversed phase HPLC (XBridge C18, 10 mm × 150 mm, 5 μm, 2 min of 20:80 acetonitrile:water with 0.1% formic acid followed by a gradient of increasing acetonitrile (20:80 to 50:50) over 24 min, 7 mL/min). Erdacin (1) eluted at 14.9 min. The trivial name erdacin is derived from “erda”, the Old English word for earth or soil.
- 22.Erdacin (1) 13C NMR (150 MHz, CD3OD with 0.1% TFA): δ=205.5 (C-22), 203.5 (C-25), 188.4 (C-1), 176.6 (C24), 165.3 (C-20), 156.7 (C-10), 153.7 (C-2), 143.3 (C-5), 138.8 (C-18), 135.6 (C-16), 134.6 (C-14), 133.4 (C-6), 131.3 (C-8), 131.2 (C-4), 130.6 (C-15), 127.7 (C-3), 123.8 (C-11), 120.6 (C-7), 119.1 (C-17), 118.7 (C-9), 118.4 (C-19), 114.1 (C-21), 75.5 (C-12), 64.6 (C-13), 30.3 (C-23), 27.9 (C-26); 1H NMR (600 MHz, CD3OD with 0.1% TFA): δ=7.40 (m, 2H; H-17,18), 7.28 (t, J=7.7 Hz, 1H; H-8), 7.16 (d, J=7.1 Hz, 1H; H-7), 6.92 (d, J=7.7 Hz, 1H; H-9), 6.73 (d, J=7.7 Hz, 1H; H-19), 4.29 (s, 1H; H-13), 2.53 (s, 3H; H-23), 1.72 (s, 3H; H-26); HR-ESI-MS m/z 475.1037, calcd. for C26H19O9, 475.1029. 1H and 13C spectra for erdacin are included as supporting information.
- 23.Lessmann H, Maskey RP, Fotso S, Lackner H, Laatsch H. Z Naturforsch. 2005;60b:189. [Google Scholar]
- 24.Lessmann H, Krupa J, Lackner H, Jonesz PG. Naturforsch. 1989;44b:353. [Google Scholar]
- 25.Yenesew A, Ogur JA, Duddeck H. Phytochemistry. 1993;34:1442. [Google Scholar]
- 26.Ozawa M, Taguchi T, Itoh T, Ebuizuka Y, Booker-Milburn KI, Stephenson GR, Ichinose K. Tetrahedron. 2003;59:8793. [Google Scholar]
- 27.Brunker P, McKinney K, Sterner O, Minas W, Bailey JE. Gene. 1999;227:125. doi: 10.1016/s0378-1119(98)00618-0. [DOI] [PubMed] [Google Scholar]
- 28.Ichinose K, Bedford DJ, Tornus D, Bechthold A, Bibb MJ, Revill WP, Floss HG, Hopwood DA. Chem Biol. 1998;5:647. doi: 10.1016/s1074-5521(98)90292-7. [DOI] [PubMed] [Google Scholar]
- 29.Ichinose K, Ozawa M, Itou K, Kunieda K, Ebizuka Y. Microbiology. 2003;149:1633. doi: 10.1099/mic.0.26310-0. [DOI] [PubMed] [Google Scholar]
- 30.Wilkinson SP, Grove A. Curr Issues Mol Biol. 2006;8:51. [PubMed] [Google Scholar]
- 31.Prior RL, Wu X, Schaich K. J Agric Food Chem. 2005;53:4290. doi: 10.1021/jf0502698. [DOI] [PubMed] [Google Scholar]


