Abstract
High-throughput genomic data for both lung development and lung cancer continue to accumulate. Significant molecular intersection between these two processes has been hypothesized due to overlap in phenotypes and genomic variation. Examining the network biology of both cancer and development of the lung may shed functional light on the individual signaling modules involved. Stem cell biology may explain a portion of this network intersection and consequently studying lung organogenesis may have relevance for understanding lung cancer. This review summarizes our understanding of the potential overlapping mechanisms involved in lung development and lung tumorigenesis.
Keywords: gene expression profile, lung development, lung cancer, signaling pathway, stem cell, network biology
Introduction
Lung cancer continues to be the leading cause of cancer-related mortality in both women and men in the world with an estimated 160,390 deaths in the United States alone in 2007.1,2 Lung cancer comprises two primary histological subtypes: small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), which account for approximately 13% and 86% of lung cancers respectively. NSCLC is further subdivided into at least three histologic subtypes: adenocarcinoma (ADC), squamous cell carcinoma (SCC) and large cell carcinoma (LCC).3 SCLC has a more aggressive biology than NSCLC and is characterized by early metastatic spread. Despite initial sensitivity to chemotherapy and radiotherapy, overall five-year survival in SCLC patients is less than 5%.4 Outcomes for NSCLC are significantly better for stage I and II disease if successfully treated with surgical resection, but similarly dismal for more advanced disease stages.
Genomic profiling directed at characterizing gene expression, SNP genotypes, methylation status and microRNA profiles has provided a wealth of information at the molecular level for lung cancer. Many of these studies have provided new insight into potential molecular classifiers of patient outcome. While originally thought to be illuminating in terms of biologic understanding, this unfortunately has not turned out to be true to the degree originally suspected. SNP (single nucleotide polymorphism) genotyping has provided information regarding CNV (copy number variation) regions that may harbor alterations relevant for NSCLC, but few if any have yet to be functionally demonstrated as important for disease initiation or progression. Gene expression profiling has given us many genes up or down in disease states, but again few have been shown to be clearly functionally relevant to the tumorigenic process. Micro RNA alterations, particularly those related to the let 7-ras story,5 have potential functional relevance and there are likely many more waiting to be discovered. Projects such as the Cancer Genome Atlas, jointly funded by the NCI and NHGRI (http://cancergenome.nih.gov), promise to provide an ongoing wealth of genomic data for lung cancer that will take many years to fully interpret in a functional context.
One of the interesting findings to date from lung cancer genomic data is the overlap between developmental gene networks and those that appear to be altered in cancer. One hypothesis from this observation is that many of the biological networks utilized during lung organogenesis are those that also go awry in the initiation and progression of lung cancer. This would suggest direct relevance for understanding cancer through organogenesis and vice versa.6 Recent observations suggesting the presence of adult stem cells in solid organs and the possibility that cancers may arise from these cell types adds further relevance to potential onco/devo links in the lung.7
This review summarizes our understanding of the potential overlapping mechanisms involved in lung development and lung tumorigenesis. We explore a number of angles relevant to the issue, including individual signaling pathways, biological networks, and the role of stem cells in development and cancer.
Genomic Alterations in Lung Development and Cancer
Hypotheses abound regarding the possible role for embryonic developmental programs in tumorigenesis. Gene expression profiling tools have recently been used to address the extent of overlap between lung developmental programs and lung cancer circuitry.8 Borczuk et al. identified gene marker sets associated with lung tumor histology and determined the expression of murine orthologues of these genes in normal mouse lung development.9 They demonstrated that adenocarcinoma markers appeared to be expressed late in mouse lung development and were associated with differentiation and glandular formation. Large-cell gene markers were expressed earlier in mouse lung development and were predominantly associated with proliferation and cell cycle processes. Bonner et al.10 investigated changes in gene expression profiles during lung development and identified four patterns of expression and mapped the developmental genes to three primary regulatory pathways involved in lung tumorigenesis: wnt/β-catenin signaling, cell cycle and apoptosis. In another study, Bonner et al.11 performed gene profiling analysis on various lung tissues. They identified potential oncogenes and tumor suppressors based on the comparison of the expression patterns with the gene profiles of lung development and suggested an association at the molecular level between murine lung tumorigenesis and lung development.
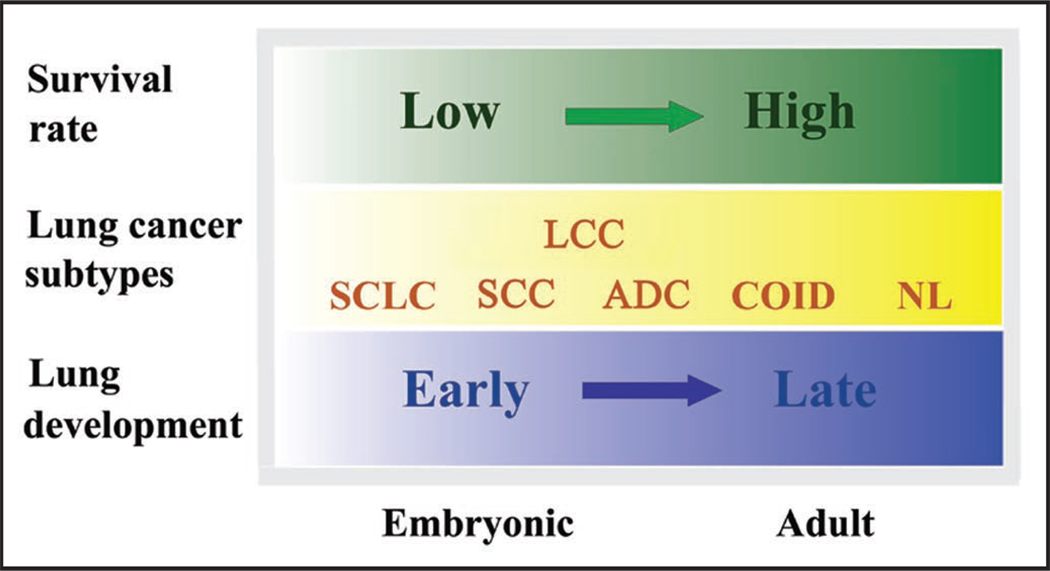
Kho et al.6 have provided compelling data to support common mechanisms underlying lung development and primary tumorigenesis through cross-organ analyses. Adopting a similar principal component analysis method, Liu et al.12 compared gene expression profiles from 186 patients representing four different lung cancer subtypes with the gene expression signatures collected from mouse lung development models. First, they described a temporal analysis of the significantly different lung cancer genes in murine lung development and demonstrated that murine orthologs of genes that are upregulated in human lung cancers tend to have an expression profile that is decreasing with time during mouse lung development. Conversely, genes that are largely downregulated in lung cancers tend to have a monotonically increasing expression profile during mouse lung development. This trend is consistent with that observed in other studies9,10 and has been further validated by a recent study of Kpantzev et al.13 who directly compared gene expression profiles in human non-small cell lung carcinomas with those from human fetal lung development. Secondly, genomic level associations between human lung cancer subtypes and mouse lung development stages were established through projection of normal lung and lung cancer genomic profiles onto a genomic mouse lung developmental framework. This framework was constructed from a 596 gene signature derived from a subset of 3,590 unique genes whose expression varied significantly both in lung tumors and during lung development. As expected, normal human samples were most similar to late mouse lung development stages. In placing samples from three human lung cancer subtypes along this timeline, the most aggressive malignancy, SCLC, appeared the earliest, followed by squamous carcinoma and then adenocarcinoma, reflecting their five-year survival rates. Specifically, the earlier the genomic association between a human tumor profile and the mouse lung development sequence, the poorer the patient’s prognosis. Thirdly, the most enriched molecular pathways in the top 100 most heavily weighted genes of the lung onco/devo signature (596 genes) were identified as being involved in pyrimidine metabolism and the cell cycle. While the pathways represent general biological processes for both lung cancer and lung development, it will be relevant to explore the projection of the signature genes to those pathways that are important to cell fate decisions in the developing lung and their contribution to malignancies. These findings are intriguing as they provide further supportive evidence for lung cancer-development associations that suggest a potential developmental basis for lung cancer classification and prognosis. Figure 1 illustrates an integrated view of our understanding of the associations between lung development stages, lung cancer subtypes and survival rate.
Figure 1.
An integrated view of the associations between lung developmental stages, lung cancer subtypes, and survival rate for small cell lung cancer (SCLC), squamous cell carcinoma (SCC), adenocarcinoma (ADC) large cell carcinoma (LCC), carcinoid (COID) and normal lung (NL) respectively.
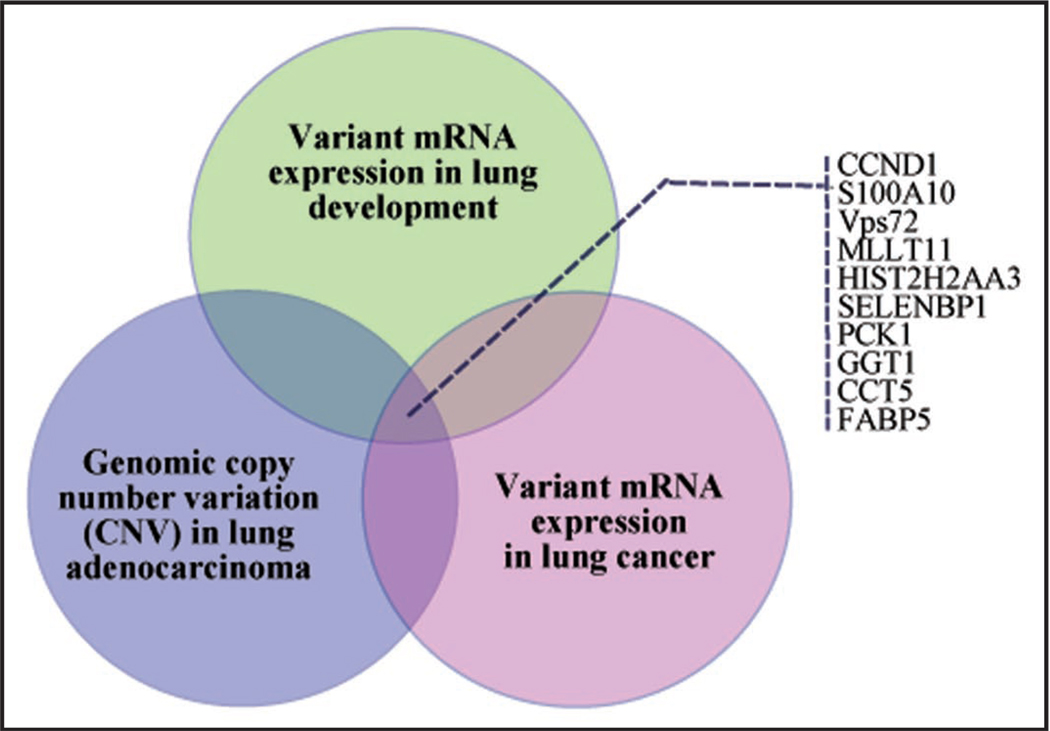
Recently described genomic datasets derived from other technology platforms have provided further opportunity to explore links between lung development and cancer. A recently published study by Weir et al.14 examining CNV in lung adenocarcinomas is the largest dataset to date utilizing SNP genotyping for studying a lung cancer genome. In a preliminary analysis of the developmental overlap with this data, we have extracted the genes identified in the richest regions of amplification and deletion and mapped these to genes known to be variant in expression levels from both lung development and cancer based on the Liu dataset.12 This approach yielded a number of genes with clear copy number variation in lung ADC and correlating alterations in gene expression that are reflected in genes varying over the time course of lung development. Figure 2 shows a schematic of this overlap. Integrated genomic analyses such as this have the promise of pointing toward true functional alterations in disease states with a higher degree of confidence than from data derived on a single genomic platform alone.
Figure 2.
A schematic diagram of the intersection between mRNA expression and copy number variation for lung development and lung adenocarcinoma. Panel insert lists ten genes within this overlap.
MicroRNAs (miRNAs) are small noncoding RNAs thought to be involved in physiologic and developmental processes by negatively regulating the expression of target genes. MicroRNAs regulate gene expression by resulting in direct cleavage of targeted mRNAs or inhibiting translation through complementary sequences to targeted mRNAs at the 3' untranslated regions of their respective targets. Recently, evidence is emerging for specific miRNAs with involvement in both lung development and lung cancer. The miR-17-92 cluster, which comprises seven miRNAs in intron 3 of the C13orf25 gene at 13q31.3, is markedly overexpressed in lung cancers, particularly SCLC.15 Expression of the miR-17-92 cluster is high at early stages of lung development, but declines as development proceeds. Conditional overexpression of the miR-17-92 cluster targeted to the lung in mice results in abnormal lung development with hyperproliferation and diminished differentiation of distal epithelial cell lineages.16 Mice harboring a targeted deletion of miR-17-92 die shortly after birth with lung hypoplasia and cardiac anomalies.17 As the volume of miRNA data for both lung development and cancer continues to grow, further insights with onco/devo overlap are likely to be discovered.
Stem Cells in Development and Cancer
Stem cells are vital to all stages of life, yet the specific roles that stem cells play during different stages of ontogeny change considerably. During early embryogenesis, pluripotent stem cells differentiate to give rise to the three germ layers that establish the basic vertebrate body plan.18 As development proceeds, distinct subsets of stem cells emerge to orchestrate the construction of tissues and organs. These processes are often incomplete at birth and carry over into postnatal life. Once tissues are fully established, stem cells undergo a fundamental change as their role turns from one of tissue building to one of tissue maintenance and repair, which persists throughout adult life. Although best exemplified in the hematopoietic system,19 the existence of hierarchical relationships between stem and progenitor cells is emerging as a common feature of other tissue-specific stem cell compartments.
Cells with long life spans are intuitively more likely to accrue the requisite number and variety of genetic alterations necessary to acquire full tumorigenic capacity.20 The cancer stem cell (CSC) hypothesis has arisen from observations that neoplastic clones are maintained exclusively by rare fractions of cells with stem cell properties. Not only is the existence of CSCs in human leukemia well established,21 the identification of CD133+ human colon CSCs22 and brain tumor CSCs23 provide further evidence for the potential existence of CSCs in solid tumors. A tumor can be viewed as an aberrant organ initiated by a CSC that acquired the capacity for indefinite proliferation through accumulated mutations.21 Both normal stem cells and tumorigenic cells have extensive proliferative potential and the ability to give rise to new (normal or abnormal) tissues. Both tumors and normal tissues are composed of interacting heterogeneous combinations of cells, with different phenotypic characteristics and different proliferative potentials. This suggests that CSC undergo processes that are analogous to the self-renewal and differentiation of normal stem cells. Thus, the principles of normal stem cell biology may be applicable to understand better how tumors develop.24
Recent observations by Kim et al. have ignited interest in the potential role of adult stem cells in lung development, injury and repair, and as a potential cell of origin for tumor initiation.25 A rare population of bronchioalveolar stem cells (BASCs) was described at the bronchoalveolar duct junction in adult mice. BASCs harbor both the alveolar epithelial type II (AT2) cell marker, surfactant protein C (SP-C), and the Clara cell marker, Clara cell secretary protein (CCSP or CC10). They were identified and purified by FACS based on being positive for Sca-1 and CD34, and negative for CD45 and CD31. This population retained the double positive staining characteristics for SP-C and CC10 with serial passage in culture. BASCs demonstrated resistance to naphthalene-induced lung injury and increased in number during a period of bronchiolar epithelial repair. Interestingly, BASC populations demonstrated early expansion in mice harboring an oncogenic K-ras mutation. Viable BASCs were maintained through multiple passages in vitro under distinct culture conditions, and retained the ability to differentiate into Clara cell and AT2 lineages with culture on Matrigel.25 The observation that almost all cells in the epithelium of the smaller distal airway during early embryonic lung development (E13–E15) express markers of AT2, Clara and neuroendocrine (NE) cells26 supports the potential existence of BASCs that are maintained into adulthood once development is complete.
Identification of BASCs makes it possible to begin to map out the pathways that are required for stem cell function in lung morphogenesis and lung tumorigenesis. Using p38a conditional lung knockout mice, Ventura et al.27 found that inactivation of p38a leads to an immature and hyperproliferative lung epithelium that is highly sensitized to K-RasG12V-induced tumorigenesis. Coincident expansion of the BASC population was observed. This suggests that p38a has a key role in the regulation of lung cell renewal and tumorigenesis by coordinating proliferation and differentiation signals in lung stem and progenitor cells. Dovey et al.28 demonstrated that loss of Bmi1 decreases the number and progression of lung tumors at a very early point in a K-ras-initiated mouse model of lung cancer. This correlates with a defect in the ability of Bmi1-deficient BASCs to proliferate in response to the oncogenic stimulus. Yanagi et al.29 generated PTEN knockout mice under control of the SP-C promoter where 90% of mice that received doxycycline in utero died of hypoxia soon after birth. Postnatal deletion of PTEN resulted in spontaneous lung adenocarcinomas with increased BASC numbers. Expression of the oncogenes Spry2, c-Myc and Shh were elevated in the lungs of mutant mice. Zhang et al.30 show that deletion of Gata6 resulted in the expansion of the BASC population and loss of epithelial differentiation with pronounced activation of the canonical Wnt signaling pathway. These molecular mechanisms regulating the balance between BASC expansion and epithelial differentiation and regeneration strongly imply that the process of lung tumorigenesis may share critical common pathways with lung organogenesis.
Early descriptions of putative CD133+ cancer stem cell populations from NSCLC point to further onco/devo links in the lung. Recently, Eramo et al.31 and Chen et al.32 have reported isolation of CD133+ cells from tumor samples of human NSCLC. These CD133+ cells displayed higher Oct-4 expression with the ability to self-renew and may represent a reservoir with proliferative potential for generating further lung cancer cells during tumor propagation and metastasis. Oct-4, a member of the family of POU-domain transcription factors, is expressed in pluripotent embryonic stem and germ cells. Oct-4 was identified as a stem cell biomarker and as one of four essential genes for reprogramming fibroblasts into a pluripotent embryonic stem cell-like state.33 The injection of CD133+ cells either subcutaneously or through the tail vein into immunocompromised mice generated tumor xenografts phenotypically identical to the original tumor. Knock-down of Oct-4 expression in CD133+ cells significantly inhibited tumor invasion and colony formation.
Ling et al. have reported a serum-free culture system for primary neonatal pulmonary cells that can support the growth of Oct-4+ epithelial colonies. In addition to Oct-4, these cells also express other stem cell markers such as stage-specific embryonic antigen 1 (SSEA-1), stem cell antigen 1 (Sca-1), and CCSP. These cells can be maintained for weeks in primary cultures and undergo terminal differentiation to AT1 and AT2-like pneumocytes. They demonstrated these Oct-4+ cells were located at the bronchoalveolar junction of neonatal lung.34
These findings overall provide further supportive evidence to the hypothesis that stem cell biology is a key interface between lung development and lung cancer.
Developmental Signaling Pathways in Lung Cancer
Development of the murine lung is initiated at embryonic day 9.5 (E9.5), followed by the morphologically characterized pseudoglandular (E10.5–16.5), canalicular (E16.5–17.5), saccular (E17.5-P5) and alveolar stages (P5–36).35 The early stages of lung development are classic examples of branching morphogenesis mediated by epithelial-mesenchymal interactions. The primitive airways begin as a ventral outpouching of foregut epithelium, with almost immediate branching to form the two mainstem bronchi. Interactions between the surrounding mesenchyme and the developing airway epithelium function to promote further branching morphogenesis through the pseudoglandular and canalicular stages up to E17.5.36 Alveolarization begins in the saccules of the lung in parallel with development of the alveolar capillary bed, and proceeds up to completion at approximately one month of age. During alveolarization, the gas exchange surface is further enlarged by the derivation of new septa from pre-existing septa. The newly formed septa increase in height and subdivide the existing airspaces into smaller units to form alveoli.37
Growing evidence suggests that cancer may arise from the aberrant activation of normally regulated developmental pathways. Understanding the role of these developmental pathways as they relate to tumor development and progression may lead to new insights for lung cancer biology.
FGF
Fibroblast growth factor (FGF) signaling is thought to play a critical role in branching morphogenesis of the lung. In Drosophila, branching of the tracheal airway is initiated and controlled by the branchless gene, the closest mammalian equivalent of which is the gene that encodes the signaling protein FGF-2. Also, the receptor for the protein product of branchless is breathless, whose mammalian counterparts are FGF receptors.38 Sprouty2 is an inducible downstream inhibitor of FGF-receptor signaling that is evolutionarily conserved from flies to mice. The Fgf10 gene is expressed in mesenchymal tissue, which overlies the epithelial-cell layer lining the emerging branch tip. The FGF receptor, FGFR2, is expressed throughout the epithelium, and Sprouty2 is expressed locally at the branch tips. Mutations in the Fgf10 or Fgfr2b genes that prevent their expression completely abrogate lung branching, and either decreased FGF10 expression or enhanced expression of Sprouty2 produces a small, poorly branched lung. Thus, in both flies and mice—and probably in humans—Sprouty2 mediates fine regulation of FGF signaling at the correct time, place and dose to induce and control orderly airway branching.39–41 Overexpression of FGF-10 in the fetal lung causes adenomatous malformations, perturbed branching morphogenesis and respiratory failure at birth. When expressed after birth, FGF-10 causes multifocal pulmonary tumors. FGF-10-induced tumors appear as highly differentiated papillary and lepidic pulmonary adenomas. Epithelial cells lining the tumors stain intensely for SP-C but not CCSP, indicating that FGF-10 enhanced the differentiation of cells to a peripheral AT2 cell phenotype. 42 Bmp4 is induced and activated in the epithelium of distal buds to limit Fgf10-mediated bud outgrowth.43
Hedgehog
The Hedgehog pathway has been shown to be essential for normal mammalian development. Signaling via this pathway is initiated by the binding of the secreted morphogen (Shh, Ihh and Dhh) to its receptor, Patched (Ptch), relieving the repression of Smoothened (Smo) and ultimately manifesting in the regulation of the Gli transcription factors (Gli1, Gli2 and Gli3) and downstream target genes. During lung embryogenesis, the presence of Shh expressed in the budding airway epithelium is required for lung development and branching morphogenesis, and is a key mediator for epithelial-mesenchymal interactions. Shh null mouse embryos fail to demonstrate separation of the trachea and esophagus, resulting in the formation of a rudimentary sac lacking branching and lung growth.44–46 Conversely, the overexpression of Shh under control of an SP-C promoter results in an absence of functional alveoli, and a hyperproliferation of both epithelial and mesenchymal cells.47 Reactivation of Shh signaling in airway epithelial repair is thought to lead to malignant change by repeatedly expanding the airway progenitor pool. Persistent Shh pathway activation is seen in SCLC, manifested by a high level of expression of Shh, Ptch and Gli1.48 Treatment of SCLC cell lines with cyclopamine (a specific inhibitor of the Shh pathway) produced tumour growth arrest both in vitro and in tumor xenografts. Cell lines were protected from cyclopamine inhibition by constitutive overexpression of Gli1.38 Together, these data suggested that SCLC may represent a malignancy arising from an airway epithelial progenitor that retains both Hh signaling and primitive features of pulmonary NE differentiation.49
Wnt/β-catenin
The wnt/β-catenin pathway plays crucial roles in normal cell growth, motility and differentiation during embryonic development. The pathway is also prominently involved in cancer development and progression. Several wnt ligands, frizzled receptors and β-catenin are present in the developing lung. Disruption of canonical wnt signaling by targeted deletion of β-catenin prevents distal lung buds from forming and markedly interferes with branching morphogenesis. The defect appears to result, at least in part, from failure to induce proper levels of Fgfr2b in the distal lung epithelium where wnt/β-catenin signaling is inhibited.50,51 Activation of β-catenin caused ectopic differentiation of AT2-like cells in conducting airways, goblet cell hyperplasia, and air space enlargement, demonstrating a critical role for the Wnt/β-catenin signal transduction pathway in the differentiation of the respiratory epithelium in the postnatal lung. Solid pulmonary tumors of epithelial cell origin were observed consisting of sheets of epithelial cells that lacked staining for both SP-C and CCSP, but expressed TTF-1 and Foxa2.51 Wnt1 and Wnt2 are increased in NSCLC primary tumors and cell lines. Inhibition of Wnt1 or Wnt2 by siRNA results in apoptosis of NSCLC cell lines.52,53 Clinically, several groups have reported that elevated levels of β-catenin expression correlate with an increased survival advantage for patients with NSCLC.54–56
N-Myc
N-Myc belongs to the three-member Myc oncogene family. It is frequently activated in SCLC.57 N-Myc is required for lung development. The highest Myc expression is in the early embryonic lung (E11.5), tapering to low levels in the adult lung.58 Mice with homozygous deletions of N-Myc die at birth with lungs half of the normal size.57 The conditional deletion of N-myc severely limits proliferation and epithelial differentiation in both lung epithelium and mesenchyme.59 Specific overexpression of N-myc in the pulmonary epithelium reduces the likelihood of progenitor cells differentiating into proximal or distal cell types.59 The role of N-Myc is lung organogenesis appears to be centered on maintaining a population of undifferentiated, proliferating progenitor cells in the developing lung tissue.
Network Biology of the Lung
Information about the molecular networks that define cellular functions, and hence life, is exponentially increasing. One such network is the aggregate collection of experimentally derived protein-protein interactions (PPIs), the volume of which has dramatically increased in a relatively short time period. This volume of PPI data has presented the opportunity to analyze systematically the topology of such networks for functional information using graph theory-based approaches. This information can then be used to construct models for predicting essentiality, genetic interactions, function, protein complexes and cellular pathways. One analogy is to think of a cellular PPI network as a complete representation of the system of a cell. Connections indicate the roadmap for signaling pathways, highly connected members form protein complexes and mutagenic disruptions can be represented as blockages in network flow. It also provides the opportunity to represent how signaling modules might be regulated by dynamically interacting components.
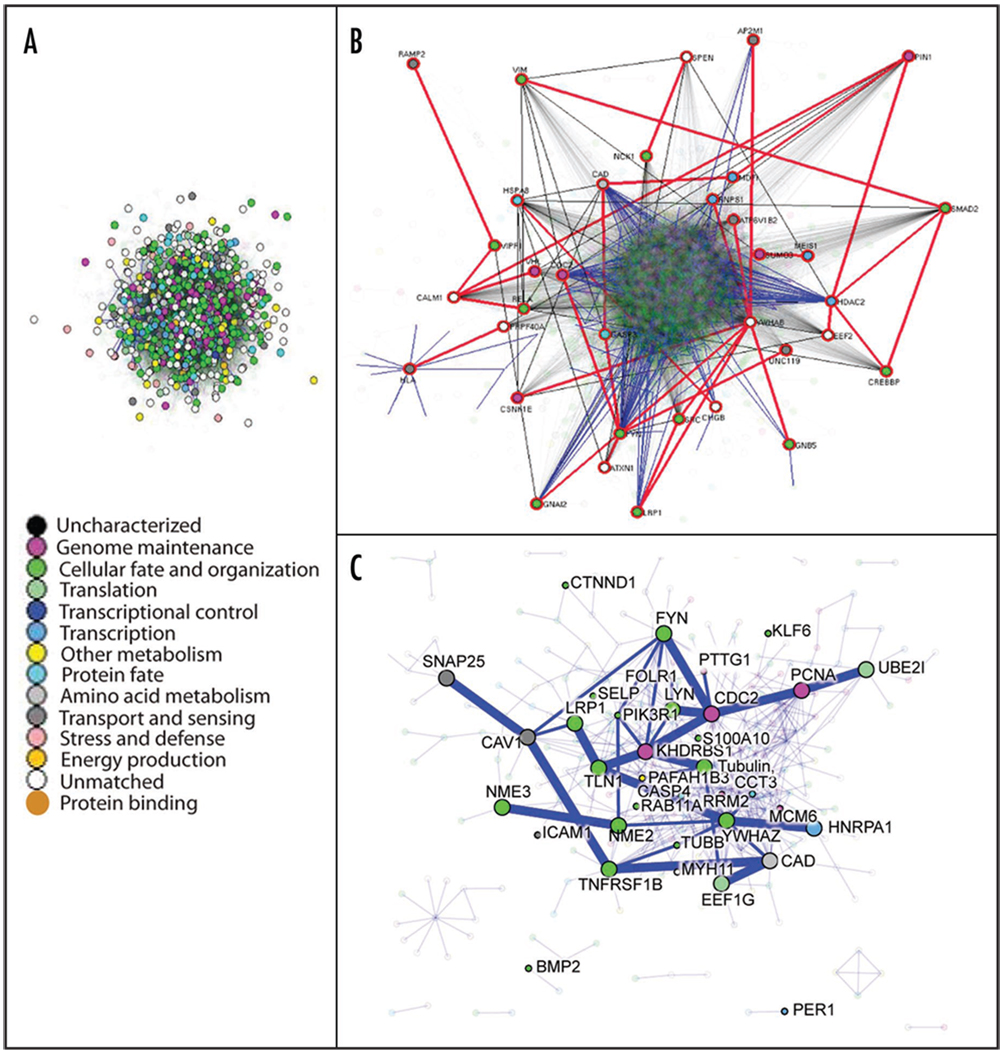
Although organ-specific networks for the lung have yet to be constructed and analyzed, the tools of network biology can play a powerful role in interpreting high-throughput genomic and proteomic data. To demonstrate this, the lung onco/devo gene expression list (containing 596 target genes) from Liu et al.12 was mapped to SwissProt IDs, resulting in 2,487 SPIDs. This target list was then used to query the Interologous Interaction protein-protein interaction database I2D ver.1.71 (http://ophid.utoronto.ca/i2d).60 This produced a dense network with 5,666 proteins and 65,101 interactions (Fig. 3A), including 721 proteins from the target list. Protein color corresponds to GO biological function (see legend in Fig. 3A).61 To aid interpretation, we have analyzed the network in NAViGaTOR ver 2.08 (http://ophid.utoronto.ca/navigator) to determine the central subnetwork, which comprises 36 proteins connected by 60 interactions (Fig. 3B). Inducing a graph on all directly connected target proteins, we generate a subgraph with 380 proteins and 681 interactions (Fig. 3C). We highlight the central subnetwork with thicker blue edges and larger nodes.
Figure 3.
(A) The full protein interaction network generated on 721 target proteins. Node color represents GO biological function as per legend. (B) The central subnetwork comprising 36 proteins connected by 60 interactions. Nodes with red highlight circle are central proteins, and thick red edges are central edges, as determined by frequency counting all pair shortest path algorithm in NAViGaTOR. Thin blue edges are direct interactions among targets, while thin black edges are direct interactions among frequent proteins. The rest of the nodes and edges are partially translucent. (C) Subgraph showing all directly connected target proteins, comprising 380 proteins and 681 interactions. The central subnetwork is highlighted with thicker blue edges and larger nodes. Direct interaction with targets from lung cancer studies with small gene signatures are highlighted with thin blue edges (overlap shown with 158 and five gene signatures,63 50 gene SCC signature,64 lung ADC signature65 and 20 gene signature66). The rest of the network is partially translucent.
To determine biological meaning within the network, the following permutation method was employed. Chosen at random were 721 proteins from a pool of all known human proteins in UniProt (build 8.2). The random proteins (seeds) were then used to search the I2D database ver. 1.7,60 to generate 1,000 random networks. We then compared network properties and network overlap with signaling and cancer-related KEGG pathways62 and to multiple small gene sets in lung cancer. The student’s t-test was then used to compare the properties of the experimentally determined network against the distributions of random networks. The most significant enrichment in the lung cancer small gene set signature lists includes the 158 gene list from Lau et al.63 (p = 0.000248), and the 50 squamous cell carcinoma gene list from Raponi et al.64 (p = 0.000658). The most significant enrichment in KEGG pathways62 includes Hematopoietic_cell_lineage (p = 0.0000108), Asthma (p = 0.0000227), Base_excision_repair (p = 0.000103) and Regulation_of_actin_cytoskeleton (p = 0.000108).
Summary and Future Directions
In this review, we have summarized potential points of overlap between lung development and lung cancer. Common themes relevant to both processes are emerging from the study of stem cell biology and biological networks. Accumulating evidence suggests relevance for onco/devo links to both the study of organogenesis and to cancer biology. We believe further insight remains to be gained from the intersecting study of both development and cancer.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA: A cancer journal for clinicians. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, et al. Cancer statistics, 2008. CA: A cancer journal for clinicians. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 3.Potti A, Mukherjee S, Petersen R, Dressman HK, Bild A, Koontz J, et al. A genomic strategy to refine prognosis in early-stage non-small-cell lung cancer. New Engl J Med. 2006;355:570–580. doi: 10.1056/NEJMoa060467. [DOI] [PubMed] [Google Scholar]
- 4.Kikuchi T, Daigo Y, Katagiri T, Tsunoda T, Okada K, Kakiuchi S, et al. Expression profiles of non-small cell lung cancers on cDNA microarrays: Identification of genes for prediction of lymph-node metastasis and sensitivity to anti-cancer drugs. Oncogene. 2003;22:2192–2205. doi: 10.1038/sj.onc.1206288. [DOI] [PubMed] [Google Scholar]
- 5.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kho AT, Zhao Q, Cai Z, Butte AJ, Kim JY, Pomeroy SL, et al. Conserved mechanisms across development and tumorigenesis revealed by a mouse development perspective of human cancers. Genes Dev. 2004;18:629–640. doi: 10.1101/gad.1182504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tenen DG. Disruption of differentiation in human cancer: AML shows the way. Nat Rev. 2003;3:89–101. doi: 10.1038/nrc989. [DOI] [PubMed] [Google Scholar]
- 8.Powers S, Mu D. Genetic similarities between organogenesis and tumorigenesis of the lung. Cell Cycle. 2008;7:200–204. doi: 10.4161/cc.7.2.5284. [DOI] [PubMed] [Google Scholar]
- 9.Borczuk AC, Gorenstein L, Walter KL, Assaad AA, Wang L, Powell CA. Non-small-cell lung cancer molecular signatures recapitulate lung developmental pathways. Am J Pathol. 2003;163:1949–1960. doi: 10.1016/S0002-9440(10)63553-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonner AE, Lemon WJ, You M. Gene expression signatures identify novel regulatory pathways during murine lung development: Implications for lung tumorigenesis. J Med Genet. 2003;40:408–417. doi: 10.1136/jmg.40.6.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonner AE, Lemon WJ, Devereux TR, Lubet RA, You M. Molecular profiling of mouse lung tumors: Association with tumor progression, lung development and human lung adenocarcinomas. Oncogene. 2004;23:1166–1176. doi: 10.1038/sj.onc.1207234. [DOI] [PubMed] [Google Scholar]
- 12.Liu H, Kho AT, Kohane IS, Sun Y. Predicting survival within the lung cancer histopathological hierarchy using a multi-scale genomic model of development. PLoS Med. 2006;3:232. doi: 10.1371/journal.pmed.0030232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kopantzev EP, Monastyrskaya GS, Vinogradova TV, Zinovyeva MV, Kostina MB, Filyukova OB, et al. Differences in gene expression levels between early and later stages of human lung development are opposite to those between normal lung tissue and non-small lung cell carcinoma. Lung Cancer. 2008;1:23–34. doi: 10.1016/j.lungcan.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 14.Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 16.Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic overexpression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310:442–453. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: Lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 19.Orkin SH, Zon LI. Hematopoiesis: An evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peacock CD, Watkins DN. Cancer stem cells and the ontogeny of lung cancer. J Clin Oncol. 2008;26:2883–2889. doi: 10.1200/JCO.2007.15.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 22.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 23.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 24.Weissman IL. Translating stem and progenitor cell biology to the clinic: Barriers and opportunities. Science (New York, NY) 2000;287:1442–1446. doi: 10.1126/science.287.5457.1442. [DOI] [PubMed] [Google Scholar]
- 25.Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 26.Wuenschell CW, Sunday ME, Singh G, Minoo P, Slavkin HC, Warburton D. Embryonic mouse lung epithelial progenitor cells co-express immunohistochemical markers of diverse mature cell lineages. J Histochem Cytochem. 1996;44:113–123. doi: 10.1177/44.2.8609367. [DOI] [PubMed] [Google Scholar]
- 27.Ventura JJ, Tenbaum S, Perdiguero E, Huth M, Guerra C, Barbacid M, et al. p38alpha MAP kinase is essential in lung stem and progenitor cell proliferation and differentiation. Nat Genet. 2007;39:750–758. doi: 10.1038/ng2037. [DOI] [PubMed] [Google Scholar]
- 28.Dovey JS, Zacharek SJ, Kim CF, Lees JA. Bmi1 is critical for lung tumorigenesis and bronchioalveolar stem cell expansion. Proc Natl Acad Sci USA. 2008;105:11857–11862. doi: 10.1073/pnas.0803574105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yanagi S, Kishimoto H, Kawahara K, Sasaki T, Sasaki M, Nishio M, et al. Pten controls lung morphogenesis, bronchioalveolar stem cells, and onset of lung adenocarcinomas in mice. J Clin Invest. 2007;117:2929–2940. doi: 10.1172/JCI31854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Goss AM, Cohen ED, Kadzik R, Lepore JJ, Muthukumaraswamy K, et al. A Gata6-Wnt pathway required for epithelial stem cell development and airway regeneration. Nat Genet. 2008;40:862–870. doi: 10.1038/ng.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eramo A, Lotti F, Sette G, Pilozzi E, Biffoni M, Di Virgilio A, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Diff. 2008;15:504–514. doi: 10.1038/sj.cdd.4402283. [DOI] [PubMed] [Google Scholar]
- 32.Chen YC, Hsu HS, Chen YW, Tsai TH, How CK, Wang CY, et al. Oct-4 expression maintained cancer stem-like properties in lung cancer-derived CD133-positive cells. PLoS ONE. 2008;3:2637. doi: 10.1371/journal.pone.0002637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 34.Ling TY, Kuo MD, Li CL, Yu AL, Huang YH, Wu TJ, et al. Identification of pulmonary Oct-4+ stem/progenitor cells and demonstration of their susceptibility to SARS coronavirus (SARS-CoV) infection in vitro. Proc Natl Acad Sci USA. 2006;103:9530–9535. doi: 10.1073/pnas.0510232103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maeda Y, Dave V, Whitsett JA. Transcriptional control of lung morphogenesis. Physiol Rev. 2007;87:219–244. doi: 10.1152/physrev.00028.2006. [DOI] [PubMed] [Google Scholar]
- 36.Weaver M, Batts L, Hogan BL. Tissue interactions pattern the mesenchyme of the embryonic mouse lung. Devel Biol. 2003;258:169–184. doi: 10.1016/s0012-1606(03)00117-9. [DOI] [PubMed] [Google Scholar]
- 37.Mund SI, Stampanoni M, Schittny JC. Developmental alveolarization of the mouse lung. Dev Dyn. 2008;237:2108–2116. doi: 10.1002/dvdy.21633. [DOI] [PubMed] [Google Scholar]
- 38.Cooper MK, Porter JA, Young KE, Beachy PA. Teratogen-mediated inhibition of target tissue response to Shh signaling. Science (New York, NY) 1998;280:1603–1607. doi: 10.1126/science.280.5369.1603. [DOI] [PubMed] [Google Scholar]
- 39.Warburton D. Developmental biology: Order in the lung. Nature. 2008;453:733–735. doi: 10.1038/453733a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bellusci S, Grindley J, Emoto H, Itoh N, Hogan BL. Fibroblast growth factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development (Cambridge, England) 1997;124:4867–4878. doi: 10.1242/dev.124.23.4867. [DOI] [PubMed] [Google Scholar]
- 41.Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, et al. Fgf10 is essential for limb and lung formation. Nat Genet. 1999;21:138–141. doi: 10.1038/5096. [DOI] [PubMed] [Google Scholar]
- 42.Clark JC, Tichelaar JW, Wert SE, Itoh N, Perl AK, Stahlman MT, Whitsett JA. FGF-10 disrupts lung morphogenesis and causes pulmonary adenomas in vivo. Am J Physiol. 2001;280:705–715. doi: 10.1152/ajplung.2001.280.4.L705. [DOI] [PubMed] [Google Scholar]
- 43.Bellusci S, Henderson R, Winnier G, Oikawa T, Hogan BL. Evidence from normal expression and targeted misexpression that bone morphogenetic protein (Bmp-4) plays a role in mouse embryonic lung morphogenesis. Development (Cambridge, England) 1996;122:1693–1702. doi: 10.1242/dev.122.6.1693. [DOI] [PubMed] [Google Scholar]
- 44.Pepicelli CV, Lewis PM, McMahon AP. Sonic hedgehog regulates branching morphogenesis in the mammalian lung. Curr Biol. 1998;8:1083–1086. doi: 10.1016/s0960-9822(98)70446-4. [DOI] [PubMed] [Google Scholar]
- 45.Litingtung Y, Lei L, Westphal H, Chiang C. Sonic hedgehog is essential to foregut development. Nat Genet. 1998;20:58–61. doi: 10.1038/1717. [DOI] [PubMed] [Google Scholar]
- 46.van Tuyl M, Post M. From fruitflies to mammals: Mechanisms of signalling via the Sonic hedgehog pathway in lung development. Respir Res. 2000;1:30–35. doi: 10.1186/rr9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bellusci S, Furuta Y, Rush MG, Henderson R, Winnier G, Hogan BL. Involvement of Sonic hedgehog (Shh) in mouse embryonic lung growth and morphogenesis. Development (Cambridge, England) 1997;124:53–63. doi: 10.1242/dev.124.1.53. [DOI] [PubMed] [Google Scholar]
- 48.Watkins DN, Berman DM, Burkholder SG, Wang B, Beachy PA, Baylin SB. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature. 2003;422:313–317. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- 49.Watkins DN, Berman DM, Baylin SB. Hedgehog signaling: Progenitor phenotype in smallcell lung cancer. Cell Cycle. 2003;2:196–198. [PubMed] [Google Scholar]
- 50.Mucenski ML, Wert SE, Nation JM, Loudy DE, Huelsken J, Birchmeier W, et al. beta-Catenin is required for specification of proximal/distal cell fate during lung morphogenesis. J Biol Chem. 2003;278:40231–40238. doi: 10.1074/jbc.M305892200. [DOI] [PubMed] [Google Scholar]
- 51.Mucenski ML, Nation JM, Thitoff AR, Besnard V, Xu Y, Wert SE, et al. Beta-catenin regulates differentiation of respiratory epithelial cells in vivo. Am J Physiol. 2005;289:971–979. doi: 10.1152/ajplung.00172.2005. [DOI] [PubMed] [Google Scholar]
- 52.You L, He B, Xu Z, Uematsu K, Mazieres J, Mikami I, et al. Inhibition of Wnt-2-mediated signaling induces programmed cell death in non-small-cell lung cancer cells. Oncogene. 2004;23:6170–6174. doi: 10.1038/sj.onc.1207844. [DOI] [PubMed] [Google Scholar]
- 53.Uematsu K, He B, You L, Xu Z, McCormick F, Jablons DM. Activation of the Wnt pathway in non small cell lung cancer: Evidence of dishevelled overexpression. Oncogene. 2003;22:7218–7221. doi: 10.1038/sj.onc.1206817. [DOI] [PubMed] [Google Scholar]
- 54.Hommura F, Furuuchi K, Yamazaki K, Ogura S, Kinoshita I, Shimizu M, et al. Increased expression of beta-catenin predicts better prognosis in nonsmall cell lung carcinomas. Cancer. 2002;94:752–758. doi: 10.1002/cncr.10213. [DOI] [PubMed] [Google Scholar]
- 55.Retera JM, Leers MP, Sulzer MA, Theunissen PH. The expression of beta-catenin in non-small-cell lung cancer: A clinicopathological study. J Clin Pathol. 1998;51:891–894. doi: 10.1136/jcp.51.12.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Awaya H, Takeshima Y, Amatya VJ, Ishida H, Yamasaki M, Kohno N, Inai K. Loss of expression of E-cadherin and beta-catenin is associated with progression of pulmonary adenocarcinoma. Pathol Int. 2005;55:14–18. doi: 10.1111/j.1440-1827.2005.01784.x. [DOI] [PubMed] [Google Scholar]
- 57.Moens CB, Auerbach AB, Conlon RA, Joyner AL, Rossant J. A targeted mutation reveals a role for N-myc in branching morphogenesis in the embryonic mouse lung. Gen Dev. 1992;6:691–704. doi: 10.1101/gad.6.5.691. [DOI] [PubMed] [Google Scholar]
- 58.Moens CB, Stanton BR, Parada LF, Rossant J. Defects in heart and lung development in compound heterozygotes for two different targeted mutations at the N-myc locus. Development (Cambridge, England) 1993;119:485–499. doi: 10.1242/dev.119.2.485. [DOI] [PubMed] [Google Scholar]
- 59.Okubo T, Knoepfler PS, Eisenman RN, Hogan BL. Nmyc plays an essential role during lung development as a dosage-sensitive regulator of progenitor cell proliferation and differentiation. Development (Cambridge, England) 2005;132:1363–1374. doi: 10.1242/dev.01678. [DOI] [PubMed] [Google Scholar]
- 60.Brown KR, Jurisica I. Unequal evolutionary conservation of human protein interactions in interologous networks. Genome Biol. 2007;8:95. doi: 10.1186/gb-2007-8-5-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kanehisa M, Goto S, Kawashima S, Nakaya A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002;30:42–46. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lau SK, Boutros PC, Pintilie M, Blackhall FH, Zhu CQ, Strumpf D, et al. Three-gene prognostic classifier for early-stage non small-cell lung cancer. J Clin Oncol. 2007;25:5562–5569. doi: 10.1200/JCO.2007.12.0352. [DOI] [PubMed] [Google Scholar]
- 64.Raponi M, Zhang Y, Yu J, Chen G, Lee G, Taylor JM, et al. Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer Res. 2006;66:7466–7472. doi: 10.1158/0008-5472.CAN-06-1191. [DOI] [PubMed] [Google Scholar]
- 65.Blackhall FH, Wigle DA, Jurisica I, Pintilie M, Liu N, Darling G, et al. Validating the prognostic value of marker genes derived from a non-small cell lung cancer microarray study. Lung Cancer. 2004;46:197–204. doi: 10.1016/j.lungcan.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 66.Bianchi F, Nuciforo P, Vecchi M, Bernard L, Tizzoni L, Marchetti A, et al. Survival prediction of stage I lung adenocarcinomas by expression of 10 genes. J Clin Invest. 2007;117:3436–3444. doi: 10.1172/JCI32007. [DOI] [PMC free article] [PubMed] [Google Scholar]