Abstract
In gastric cancer, a new epigenetic mechanism of tumour suppressor loss has been suggested where the histone methyltransferase enhancer of zeste homolog 2 (EZH2) is responsible for loss of expression of RUNX3. This is consistent with EZH2 upregulation in multiple cancer types being associated with poor prognosis. We investigated whether EZH2 influences the expression of RUNX3 in colorectal cancer (CRC) and whether this is independent of methylation. We determined protein and messenger RNA (mRNA) levels of EZH2 and RUNX3 and assessed RUNX3 methylation with methylation-specific polymerase chain reaction using 72 human CRCs and 8 CRC cell lines. We assessed the effect of efficient RNA interference-mediated knockdown of EZH2 on RUNX3 levels, cell viability and H3K27 trimethylation of the RUNX3 promoter using chromatin immunoprecipitation. Despite higher levels of EZH2 and lower levels of RUNX3 in CRC specimens in general, no inverse correlation between EZH2 and RUNX3 in paired samples was found arguing against a major role for histone methylation in silencing RUNX3 in CRC. Conversely, downregulation of RUNX3 mRNA in the same tumours was associated with RUNX3 DNA methylation (P < 0.05). In cell lines, knockdown of EZH2 removed the repressive chromatin marks from RUNX3 but did not result in RUNX3 re-expression. However, it prevented the re-silencing of RUNX3 after the removal of demethylating agents. In conclusion, DNA methylation is primarily responsible for the transcriptional silencing of RUNX3 in CRC, but EZH2 and histone methylation are necessary for its methylation-dependent re-silencing after the removal of demethylating agents. These results would predict that inhibitors of EZH2 and histone methylation would enhance the effects of demethylating agents in cancer therapy.
Introduction
Chromatin changes have long been associated with cancer. The best characterized alteration is CpG DNA hypermethylation, which often accumulates in promoter regions of tumour suppressor genes, thereby contributing to tumour suppressor loss through epigenetic silencing. In addition to DNA methylation, epigenetic modification states of histones are also implicated in oncogenesis. Particular global patterns of acetylation and methylation of histones H3 and H4 are associated with multiple cancer types. These and other findings promote an emerging view that epigenetic changes in the cancer cell genome may contribute just as significantly to disease progression as do genetic alterations to DNA sequence. However, epigenetic changes can potentially be reversed with inhibitors that block the relevant chromatin-modifying enzymes. Thus, it is important to better understand the role of these epigenetic enzymes in cancer cells with an eventual goal of developing new cancer treatments.
Enhancer of zeste homolog 2 (EZH2) is the catalytic subunit of polycomb-repressive complex 2 (PRC2), which is a highly conserved histone methyltransferase that targets lysine-27 of histone H3. This methylated H3K27 chromatin mark is commonly associated with silencing of differentiation genes in organisms ranging from plants to flies to humans. Studies in human tumours show that EZH2 is frequently overexpressed in a wide variety of cancerous tissue types, including prostate and breast and is associated with poor prognosis (1,2). Functional links between EZH2-mediated histone methylation and DNA methylation suggest that the two mechanisms may act in partnership (3) but the mechanistic contributions of EZH2 to cancer progression have not yet been determined.
The same genes that are silenced by methylation of both alleles in cancer are marked with H3K27 methylation in normal cells also suggesting that the two phenomena are linked. However, H3K27 methylation is not sufficient in itself to recruit DNA methyltransferases in normal cells and other unknown changes occurring during carcinogenesis must control whether promoter methylation ultimately occurs (4). Although some authors have shown that DNA methylation and complete transcriptional silencing of cancer genes persist after depletion of EZH2 (5), others have found that depletion of EZH2 is sufficient to lead to the upregulation of gene expression independently of changes in the promoter methylation status. Fujii et al. demonstrated an inverse correlation between EZH2 and RUNX3 gene expression in gastric cancer cell lines and an inverse relationship of these proteins at the individual cell level in human gastric cancer specimens in the absence of DNA methylation in the RUNX3 promoter region. RNA interference-mediated knockdown of EZH2 resulted in an increase in expression of the RUNX3 gene and was not associated with any change in DNA methylation status (6), suggesting that EZH2 can be primarily responsible for the silencing of tumour suppressor genes independently of other factors in gastric cancer. If confirmed, this is highly significant finding.
RUNX3 belongs to the RUNX family of genes, which play important roles in mammalian development and neoplasia (7–10). RUNX proteins form complexes with Smad2 and Smad3 that transmit transforming growth factor β/activin signals (11). RUNX3 gene is localized at the 1p36 locus and has been linked to gastric epithelial homeostasis and gastric carcinogenesis. The 1p36 region is thought to harbour one or several tumour suppressor genes since this region exhibits frequent loss of heterozygosity events in colon, gastric, breast and ovarian cancers (12), and the introduction of a normal human 1p36 chromosome fragment into colon cancer cells suppresses their tumourigenicity (13). A considerable proportion of gastric cancers do not express RUNX3 due to hemizygous deletion and hypermethylation of the RUNX3 promoter region (14). The hypermethylation of the RUNX3 promoter has also been found in 21% of colon cancer specimens and 65% of colon cancer cell lines, suggesting that RUNX3 also has a tumour suppressive function in colorectal cancers (CRCs) (15). Moreover, it has been shown that RUNX3 attenuates Wnt signalling, which is overactivated and plays a major role in CRC, through interaction with the β-catenin/transcription factor 4 complex and reduction of its transactivating potential (16).
In the present study, we aimed to investigate the role of EZH2 and RUNX3 in sporadic CRCs and CRC cell lines and specifically whether EZH2-mediated chromatin changes are responsible for RUNX3 gene silencing and therefore represent a new mechanism of tumour suppressor loss.
Materials and methods
Cell culture
CACO2, DLD1, SW480, LOVO, SW48, HT29, RKO and HCT116 colon cancer cell lines were obtained from the American Type Culture Collection and cultured in Dulbecco's modified Eagle's medium (Gibco, Paisley, Scotland) with 4.5 g/l glucose and l-glutamine, penicillin (50 U/ml), streptomycin (50 μg/ml) and 10% fetal calf serum (Gibco).
Immunoblotting
Cells were scraped into sample buffer (125 mM Tris/HCl, pH 6.8, 4% sodium dodecyl sulfate, 2% β-mercaptoethanol, 20% glycerol, 1 mg bromophenol blue). Protein concentration was measured using the RC DC protein assay kit (Bio-Rad, Hercules, CA). The lysates were sonicated and then heated at 95° for 5 min. Fifty micrograms of protein was loaded onto sodium dodecyl sulfate–polyacrylamide gel electrophoresis and blotted onto polyvinylidene difluoride membrane (Millipore, Bedford, MA). The blots were blocked in block buffer (2% low fat milk powder in Tris-buffered saline with 1% Triton) and incubated overnight at 4°C with primary antibody in Tris-buffered saline with 1% Triton with 0.2% low fat milk powder. Primary antibodies to EZH2 (mouse monoclonal, 1:1000) were from BD Transduction Laboratories (Breda, The Netherlands), to active form of RUNX3 (mouse monoclonal R3-5G4, 1:200) were from Abcam (Cambridge, UK) and actin (rabbit polyclonal) were from Santa Cruz Biotechnology (Santa Cruz, CA). Blots were then incubated for 1 h at room temperature in 1:2000 horseradish peroxidase-conjugated corresponding secondary antibody (Dako, Glostrup, Denmark) in block buffer. Finally, blots were developed using Lumilite Plus (Roche, Woerden, The Netherlands) and a Lumi-Imager (Bio-Rad).
Selection of patient material
Tissue from 72 CRC cases from the archives of the Pathology Department at the Academic Medical Centre, Amsterdam, was used for the compilation of the tissue microarray (TMA). The clinicopathological characteristics of patients used in the TMAs used in this study have been described previously (17). For the RNA isolation, frozen tissue from 47 CRC patients from archive of Gastroenterology and Hepatology Department of Leiden University Medical Center was used. The study was performed according to the instructions and guidelines of the Academic Medical Center (Amsterdam) and Leiden University Medical Center Medical Ethics Committees.
Construction of the tissue microarray
A Manual Tissue Arrayer MTA-1 (Beecher Instruments, Sun Prairie, WI) was used for the construction of the TMA. Three cores of tissue from each cancer specimen were used and for each cancer case one core from the corresponding normal colon.
Immunohistochemistry
TMA blocks were sectioned (4 μm), deparaffinized, immersed in 0.3% H2O2 in methanol for 20 min and heat treated at 100°C (pH 9) for 10 min. Sections were blocked with TENG-T [10 mmol/l Tris, 5 mmol/l ethylenediaminetetraacetic acid, 0.15 mol/l NaCl, 0.25% gelatin, 0.05% (vol/vol) Tween 20, pH 8.0] for 30 min. Slides were incubated with primary antibodies to EZH2 (1:12 000) and to RUNX3 (1:1000) overnight at 4°C in phosphate-buffered saline with 0.1% Triton and 1% bovine serum albumin. The Powervision+poly-HRP detection system (ImmunoVision Technologies, Daly City, CA) was used to visualize the antibody-binding sites. Sections were counterstained with haematoxylin. Negative control sections for all antibodies were processed in an identical manner after omitting the primary antibody and showed no staining.
TMA analysis
The cellular localization and pattern of immunoreactivity were examined by two investigators independently in a blinded fashion. Expression was graded from 0 to 2 for EZH2 (0 = positive nuclear staining in <10% of the epithelial cells, 1 = positive nuclear staining in <70% of the epithelial cells, 2 = positive nuclear staining in >70% of the cells), where score 2 was assumed as overexpression of the EZH2. A dichotomized scale was used to measure the intensity of the RUNX3 expression. Samples with <10% positive nuclear or cytoplamic staining in tumour cells were classified as negative and as positive if >10% of cells had staining intensity greater than that of negative control slides.
RNA isolation and real-time reverse transcription–polymerase chain reaction
Total RNA was isolated using Trizol (Invitrogen, Breda, The Netherlands) according to the manufacturer’s instructions. Complementary DNA was synthesized from 1 μg of total RNA using random primers (Promega, Leiden, The Netherlands) and MMLV-reverse transcriptase (Invitrogen). Polymerase chain reaction (PCR) for EZH2 and RUNX3 was performed with primers as in (ref. 6) using iCycler Thermal Cycler and iQ5 Multicolor Real-Time PCR Detection System (Bio-Rad). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression was used to normalize for variance.
Methylation analysis
DNA was extracted using the DNeasy Tissue Kit (Qiagen, Venlo, The Netherlands). Bisulphite treatment was performed using the EZ DNA Methylation kit (Zymo Research, Orange, CA) according to the manufacturer's instructions. The primer sequences for methylation-specific PCR for RUNX3 were as used in (15) and are depicted in supplementary Figure 1 (available at Carcinogenesis Online). PCR was performed with 40 cycles of 94°C, 67°C and 72°C of 1 min each, preceded by a 5 min denaturing step at 94°C and followed by a 10 min extension step at 72°C. The products were electrophoresed on 5% agarose gel. Human genomic DNA from peripheral blood lymphocytes was used as an unmethylated control. Human genomic DNA treated in vitro with Sss I methyltransferase (New England Biolabs, Beverly, MA) was used as a positive control for the methylated reaction.
RNA interference
Cells were transfected with either negative control small interfering RNA (siRNA) or EZH2-targeting siRNA (Ambion, ID 107417; Applied Biosystems, Nieuwerkerk a/d IJssel, The Netherlands) using DharmaFECT transfection reagent (Dharmacon, Etten-Leur, The Netherlands) according to the manufacturer's instructions. At various time points after transfection, cells were harvested and subjected to real-time reverse transcription–polymerase chain reaction (RT–PCR) and immunoblotting.
Chromatin immunoprecipitation assay
The chromatin immunoprecipitation assay was performed using Chromatin Immunoprecipitation Assay Kit, (Upstate, Lake Placid, NY). Five micrograms of HsK27me3 antibody (Upstate) was used. The two pairs of primers for RUNX3 promoter region were used as in (ref. 6) and are depicted in supplementary Figure 1 (available at Carcinogenesis Online). Preliminary PCRs were performed to determine the optimal PCR conditions to assure linear amplification of DNA. PCR products were electrophoresed on a 6% polyacrylamide gel. To measure the level of HsK27me3 in each immunoprecipitate, the ratios between the intensity of the PCR product in immunoprecipitated DNA versus input DNA (total chromatin) amplified by PCR in the linear range were calculated.
Treatment of cells with 5-aza-2′-deoxycytidine (5-aza-dC) and combined treatment of 5-aza-dC and trichostatin A
Cells were split 24 h before treatment and were then treated with either 5-aza-dC (5 μM) or 5-aza-dC with trichostatin A (150 nM) for 72 h.
MTT assay
Either 72 or 12 h after transfection of cells with control siRNA or EZH2 siRNA, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution was added (final concentration 0.5 mg/ml, stock solution 5 mg/ml MTT in phosphate-buffered saline), for 3 h. Cells were lysed in acidified 2-propanol and absorbance measured at 550–560 nm.
Statistical analysis
Data are shown as mean ± SD if not differently indicated. Statistical analysis was performed using the Statistical Package of Social Science (SPSS) version 16.0 for Windows. The χ2 test and Fisher’s exact test were used as appropriate. Spearman's rank correlation coefficient was calculated to analyse the association between EZH2 and RUNX3 messenger RNA (mRNA) expression. For comparison between normally distributed variables of interest, the one-way analysis of variance was used with Tukey's multiple comparison test when appropriate. P < 0.05 was considered statistically significant. Univariate analyses of time to death as a result of cancer were performed using the Kaplan–Meier method.
Results
RUNX3 expression is frequently lost and corresponds with methylation and not EZH2 expression in CRC cell lines
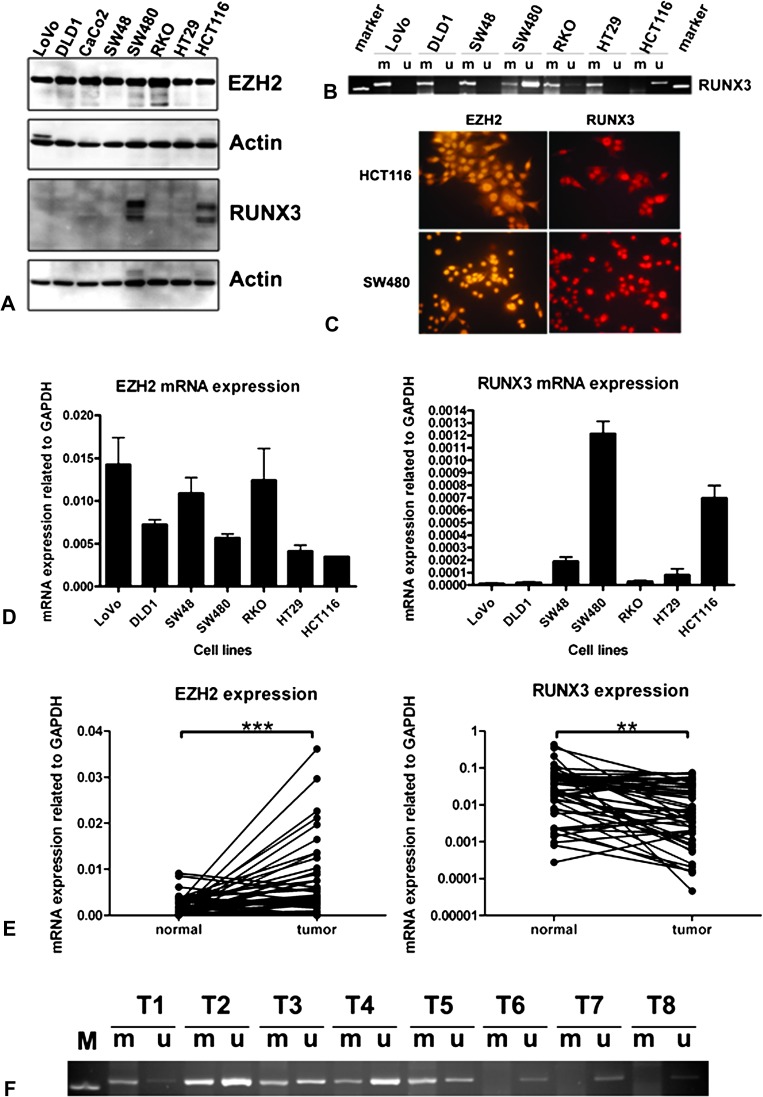
Seven CRC cell lines were investigated for the expression of EZH2 and RUNX3 at protein level by immunoblotting (Figure 1A). EZH2 is expressed at the protein level in all CRC cell lines tested. RUNX3 protein, however, is expressed only in two cell lines—in microsatellite unstable (MSI) HCT116 cells known as a cell line with the hypermethylator phenotype and in the microsatellite stable (MSS) SW480, which has no widespread aberrant methylation. This expression pattern concurs exactly with the pattern of methylation of the RUNX3 promoter found previously in these cell lines (15). We also repeated methylation-specific PCR for these seven cell lines and confirmed these results (Figure 1B). Expression of EZH2 and RUNX3 was also investigated by immunofluorescence showing positive staining for both in HCT116 and in SW480 cells (Figure 1C) and positive for EZH2 and negative for RUNX3 in RKO and DLD1 cells (Figure 3F).
Fig. 1.
Expression of EZH2 and RUNX3 in CRC cell lines and CRC patient specimens. (A) Immunoblots of colon cancer cell lines for EZH2 and RUNX3 with actin as a loading control. (B) Methylation-specific PCR (MSP) analysis of the CpG island methylation status of the RUNX3 promoter region in CRC cell lines. PCR products specific for unmethylated (U) and methylated (M) CpG sites were analysed in 2.5% agarose gels. (C) Positive nuclear expression of EZH2 and RUNX3 in HCT116 and SW480 CRC cell lines by immunofluorescence. (D) Quantitative RT–PCR analysis of EZH2 and RUNX3 expression in CRC cell lines. (E) Quantitative RT–PCR analysis of EZH2 (linear scale) and RUNX3 expression (log10 scale) in CRC patients. (F) MSP for RUNX3 promoter region MSP in a subgroup of CRC patients. T1–T5 shows positive methylated and unmethylated signals, whereas T6–T8 shows only an unmethylated band.
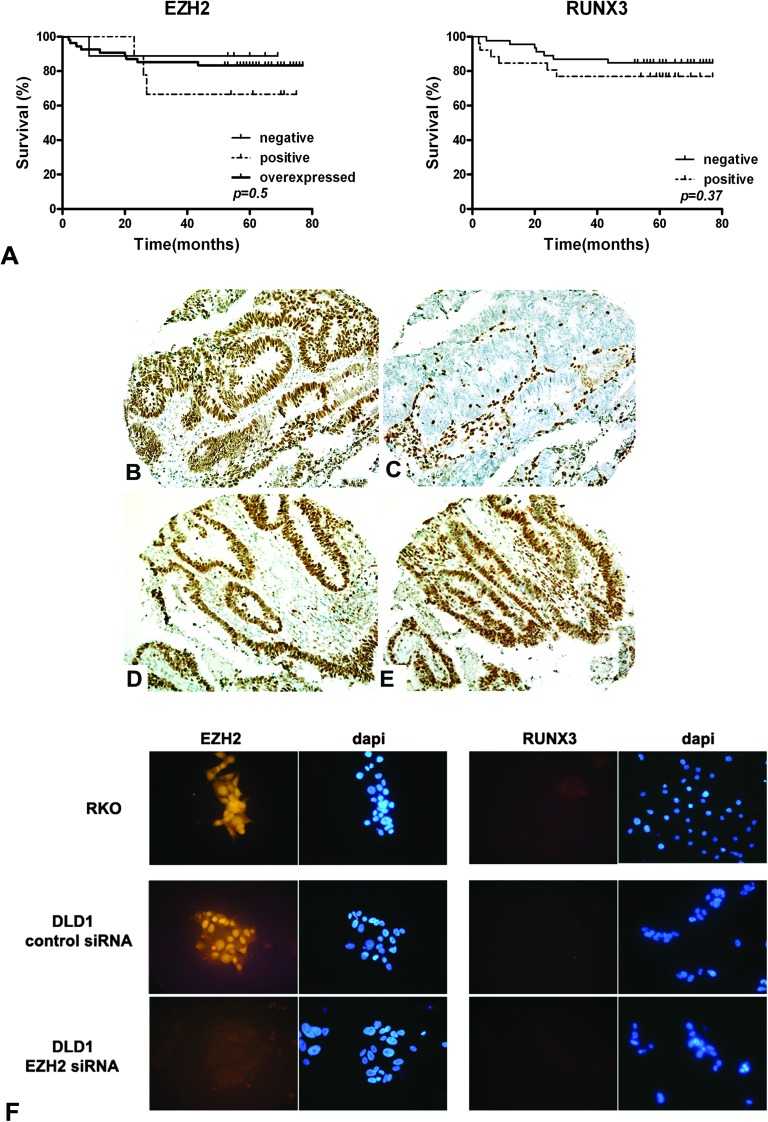
Fig. 3.
Co-localized expression patterns of EZH2 and RUNX3 in CRC specimens. (A) Kaplan–Meier survival analysis of CRC patients according to the expression of EZH2 and RUNX3. No significant association was found. (B) Overexpression of EZH2 coincides with (C) negative expression of RUNX3. (D) Overexpression of EZH2 co-localizes with (E) strong nuclear positivity of RUNX3. (F) Immunofluorescence images of CRC cells stained with EZH2 and RUNX antibodies. Nuclei were stained with 4′,6-diamidino-2-phenylindole. RKO and DLD1 cells show negative expression of RUNX3. The RUNX3 expression does not change after efficient EZH2 knockdown in DLD1 cells.
mRNA levels of EZH2 and RUNX3 as judged by real-time RT–PCR performed in the same cell lines correlate well with the protein levels (Figure 1D). All cell lines express EZH2 with highest relative values for LoVo, SW48 and RKO. SW480 and HCT116 exhibit much higher RUNX3 mRNA level than other cell lines, supporting the data from western blotting. These results suggest a primary role for methylation rather than EZH2 in controlling RUNX3 expression.
Downregulation of RUNX3 mRNA in human cancer tissue correlates significantly with RUNX3 promoter methylation but not with EZH2 overexpression
We next performed quantitative RT–PCR on complementary DNA from 47 CRCs and corresponding normal tissues. We found that expression of EZH2 mRNA is significantly increased in tumours (mean ± SEM 0.00673 ± 0.0003, relative values to GAPDH expression) compared with normal tissue (mean ± SEM 0.00195 ± 0.0003, relative values to GAPDH expression) (P < 0.001), whereas RUNX3 mRNA expression was significantly lower in CRCs (mean ± SEM 0.01749 ± 0.003238, relative values to GAPDH expression) compared with normal specimens (mean ± SEM 0.05301 ± 0.012, relative values to GAPDH expression) (P < 0.01) (Figure 1E).
When the expression of EZH2 and RUNX3 in cancer specimens was analysed for the degree of association on a per patient basis, we did not find a significant correlation (P = 0.2, Fisher’s exact test), supplementary Table 1 (available at Carcinogenesis Online). We also calculated Spearman's rank correlation coefficient and found no significant association (r =−0.302, P > 0.05, ns), although we see a trend.
We then performed methylation-specific PCR for the RUNX3 promoter region and found a good correlation between CpG island DNA promoter methylation (27/47 cancers) and downregulated RUNX3 mRNA levels (23/27 cancers) (P < 0.05). Eleven cancers had reduced RUNX3 mRNA levels without corresponding methylation but only six of these had upregulation of EZH2. Interestingly, 16 patients with downregulated RUNX3 expression had both methylation of the RUNX3 promoter region and upregulation of EZH2 mRNA (Table I, Figure 1F), suggesting that EZH2 may play a role together with methylation in the downregulation of RUNX3 expression but is not absolutely required.
Table I.
Association between RUNX3 methylation and downregulation of RUNX3 mRNA
| EZH2 |
P | |||
| No methylation (%) | Methylation (%) | |||
| RUNX3 | No change versus N | 9 (19.1) | 4 (8.5) | 0.045 |
| Downregulated versus N | 11 (23.4) | 23 (48.9) | ||
Note. N, normal colon tissue.
EZH2 protein overexpression does not correlate inversely with RUNX3 expression
We used a TMA and performed immunohistochemical staining for EZH2 and RUNX3 on tissue from 72 CRC specimens and corresponding normal tissue. The expression of EZH2 and RUNX3 was further investigated in normal colon using immunohistochemistry. In normal tissue, the expression of EZH2 is predominantly localized in the nuclei of the epithelial cells with a gradient decreasing from crypt to epithelial surface with low or no expression in the mature colonocytes of the surface epithelium (Figure 2A). RUNX3 is expressed in both epithelial and stromal cells in all corresponding normal tissues. The cellular staining pattern for RUNX3 in normal tissue is mainly cytoplasmatic. Weak nuclear positivity in normal epithelial cells contrasts with strong nuclear positivity in lymphocytes (which can be used as an internal positive control) (Figure 2B, supplementary Figure 2 is available at Carcinogenesis Online).
Fig. 2.
Expression of EZH2 and RUNX3 in normal human colon and in CRC specimens using immunohistochemistry. (A) Expression pattern of EZH2 in normal human colon. (B) Expression pattern of RUNX3 in normal human colon. (C) Positive nuclear EZH2 staining in CRC. (D) Positive RUNX3 expression in CRC. (E) Overexpression of EZH2 in cancer tissue. (F) Negative expression of RUNX3 in cancer tissue. (G) EZH2 shows negative staining in 9 (12.5%), positive staining in 9 (12.5%) and is overexpressed in 54 (75%) of CRC patients. RUNX3 is not expressed in 46 (64%) and exhibits positive nuclear or cytoplasmatic staining in 23 (36%) of CRC patients.
We found that EZH2 is not expressed in cancers from 9 patients (12.5%), exhibits positive staining in 9 patients (12.5%) and is overexpressed in 54 (75%) sporadic CRC specimens (Figure 2C, E and G). On the contrary, the majority (46 patients, 64%) of cancers show loss of expression of RUNX3 and only 26 patients (36%) demonstrate positive RUNX3 staining (Figure 2D, F and G and supplementary Figure 2 is available at Carcinogenesis Online), showing both cytoplasmatic and nuclear staining. We analysed our data in order to look for associations between EZH2 or RUNX3 expression and several clinicopathological and biological variables (sex, age, location, Dukes’ stage and grade of tumours) and no significant associations were observed (data not shown). Interestingly, we also do not see differences in expression of EZH2 and RUNX3 between MSS and MSI tumours, in contrast to a previous study (15). We found that EZH2 is overexpressed in 45 (75%) versus 9 (70%), positive in 6 (10%) versus 3 (23%) and not expressed in 8 (14%) versus 1 (8%) of MSS versus MSI tumours (P = 0.58, ns). RUNX3 staining is negative in 37 (63%) of MSS versus 9 (69%) of MSI tumours (P = 0.76, ns), supplementary Table 2 (available at Carcinogenesis Online). When we analysed the survival data of patients, we see no difference in 5 year survival when related to EZH2 or RUNX3 expression (Figure 3A).
We also analysed whether EZH2 protein overexpression inversely correlates with RUNX3 expression and in concordance with our findings at mRNA level found no such association (Figure 3B, C, D and E, supplementary Table 3 is available at Carcinogenesis Online). These data suggest that EZH2 can only play a role in RUNX3 downregulation in combination with other factors.
Knockdown of EZH2 does not lead to re-expression of RUNX3 in CRC cell lines
To determine whether EZH2 can independently downregulate RUNX3 expression, we performed RNA interference-mediated knockdown of EZH2 in seven CRC cell lines. The immunofluorescence analysis in DLD-1 cells showed complete knockdown of EZH2 96 h after transfection with siRNA but no re-expression of RUNX3 (Figure 3F). The level of EZH2 protein expression was undetectable by western blot in all cell lines, confirming an efficient knockdown (Figure 4A). Knockdown of EZH2 also resulted in a decrease of EZH2-specific chromatin repressive marks (HsK27me3) in the RUNX3 promoter detected by chromatin immunoprecipitation (Figure 4B) and leads to the re-expression of globin A (supplementary Figure 3 is available at Carcinogenesis Online), which is known to be controlled by EZH2-mediated silencing (18). The results of quantitative RT–PCR showed significantly decreased levels of EZH2 mRNA in all cell lines within 48–96 h after transfection with siRNA (Figure 4C). No changes in RUNX3 mRNA or protein levels were observed in any of the cell lines, regardless of their baseline expression of RUNX3 (Figure 4A and C).
Fig. 4.
EZH2 knockdown is not sufficient to restore RUNX3 expression in CRC cell lines. (A) Immunoblots of colon cancer cell lines for EZH2 and RUNX3 with actin as a loading control. For every cell line, the first lane is loaded with negative control siRNA-transfected cell lysate and the second lane with EZH2 siRNA-transfected cell lysate. Despite undetectable levels of EZH2 protein after siRNA-mediated knockdown, RUNX3 protein levels remain unchanged. (B) Chromatin immunoprecipitation analysis of the RUNX3 promoter in DLD1 cells treated with control or EZH2 siRNA. Two different pairs of primers for RUNX3 promoter region were used (chromatin immunoprecipitations 1 and 2). Levels of tri-methylated histone 3 K27 are reduced by siRNA-mediated knockdown of EZH2. (C) Left panel. Quantification of the efficiency of the siRNA-mediated knockdown of EZH2 on mRNA levels. The EZH2 mRNA expression in negative control siRNA-transfected cells is set at 100%. (C) Right panel. Quantitative RT–PCR shows no differences in RUNX3 mRNA expression after EZH2 knockdown. (D) Quantitative RT–PCR analysis of RUNX3 mRNA levels in cells treated with 5-aza-2’-deoxycytidine (5-aza-dC) and combined treatment of 5-aza-dC with trichostatin A (TSA) shows a considerable increase in RUNX3 mRNA level.
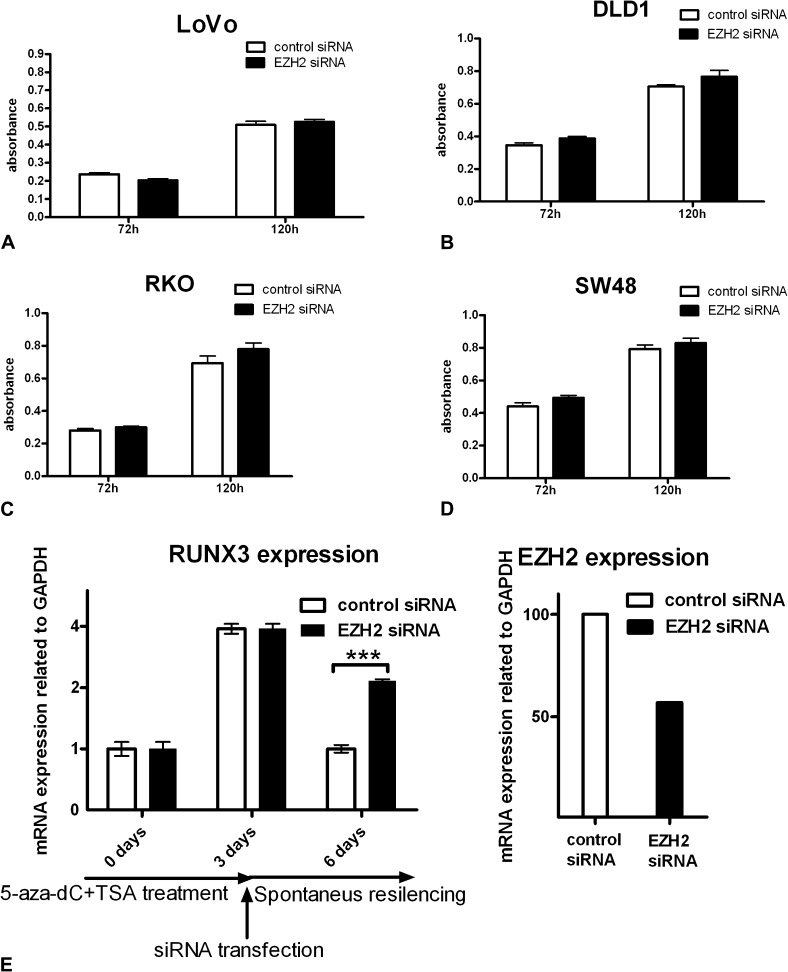
In contrast, treatment of DLD1 and LoVo cells with the demethylating agent, 5-aza-2’-deoxycytidine (5-aza-dC) or combined treatment with 5-aza-dC and the histone deacetylase inhibitor trichostatin A, did lead to a considerable increase in RUNX3 mRNA level (Figure 4D). These data suggest that EZH2 knockdown is not sufficient to restore RUNX3 expression in CRC cell lines. To investigate the possibility that EZH2 knockdown influences the expression of other unidentified tumour suppressor genes as suggested in prostate cancer, we assessed the influence of EZH2 knockdown on cell growth and viability. As seen in Figure 5A, B, C and D, efficient EZH2 knockdown does not affect cell growth or viability as assessed by the MTT assay.
Fig. 5.
(A, B, C and D) Efficient EZH2 knockdown does not affect cell growth or viability as assessed by the MTT assay in different CRC cell lines. (E) DLD1 cells were demethylated by treatment with 5-aza-dC and trichostatin A (TSA) for 3 days (72 h) leading to re-expression of RUNX3 mRNA. The same cells were subsequently transfected either with control siRNA or with EZH2 siRNA and the level of RUNX3 expression was evaluated 72 h after transfection (6 days after the beginning of the experiment). Knockdown of EZH2 significantly inhibits the re-silencing of RUNX3 after the removal of demethylating agents.
Knockdown of EZH2 prevents the re-silencing of RUNX3 after the removal of demethylating agents
Contrary to findings in gastric cancer, our results provide no evidence for an independent role for EZH2 in RUNX3 silencing in CRC, but nevertheless, a considerable proportion of patients with RUNX3 downregulation show both EZH2 overexpression and methylation of the RUNX3 promoter suggesting that the two processes may still be linked. It has been shown that genes that are silenced in cancer are often the same genes marked by EZH2 in normal cells, an observation suggesting that EZH2 acts mainly upstream of methylation or in its initiation. We hypothesized that EZH2 might be responsible for the phenomenon of re-methylation and re-silencing of tumour suppressors seen when demethylating agents are removed. To test this, we treated DLD1 cells with 5-aza-dC and trichostatin A for 72 h resulting in re-expression of RUNX3. In these same cells, we then knocked down EZH2 and determined RUNX3 expression 72 h later. We found that EZH2 knockdown prevented the re-silencing of RUNX3 (Figure 5E). This suggests that even though EZH2 does not play an independent role in silencing of tumour suppressors in CRC, it is required for the phenomenon of re-methylation and re-silencing of tumour suppressor genes after the removal of demethylating agents.
Discussion
EZH2 is the catalytic subunit of the PRC2, which is involved in gene silencing and histone H3 lysine 27 methylation. In embryonic stem cells, PRC2 controls the expression of a special set of developmental genes that must be repressed to maintain pluripotency and that are poised for activation during embryonic stem cell differentiation. RUNX3 belongs to this subset of genes (19). EZH2 and DNA methyltransferases co-immunoprecipate in vivo and EZH2 directly controls methylation in a small subgroup of genes in CRC cells (3). Genes that undergo silencing by de novo methylation in cancer are the same subset of genes that are marked with the repressive chromatin mark HsK27me3 in normal colonic cells. However, how or if one leads to the other is unclear as HsK27me3 in normal cells does not lead to recruitment of DNA methyltransferases and methylation (4). It is probably that additional changes arising during the process of carcinogenesis such as those found in leukaemia (20) are required. In addition, genes, where methylation occurs in only one allele in combination with mutation or deletion of the other, show no HsK27me3 in normal cells suggesting that although HsK27me3 targets a subset of genes for methylation in cancer, methylation can also be driven by clonal natural selection in genes without pre-existent HsK27me3.
In a recent publication, it has been shown that the putative tumour suppressor RUNX3, which is known to be hypermethylated in its promoter region in 20% of CRCs, is a target for repression by EZH2, and its expression is inversely correlated with EZH2 expression in gastric cancer cell lines and gastric cancer specimens. Moreover, the expression of RUNX3 was restored by EZH2 knockdown while no changes in DNA methylation status of the RUNX3 promoter were observed, suggesting a crucial role for EZH2 in maintaining RUNX3 gene silencing independently of promoter methylation and suggesting a novel mechanism of tumour suppressor inactivation (6). Other studies confirm that disruption of the PRC2 is sufficient to lead to the re-expression of a subset of genes in cancer (21).
In this study, we used seven CRC cell lines, extracted RNA from 47 human CRC patient specimens and used a TMA including 72 cancers to investigate the relationship between EZH2 and RUNX3 expression in CRC and compare expression with corresponding normal colon tissue. Here, we show that EZH2 is overexpressed in the majority of CRCs and significantly upregulated at mRNA level in CRCs compared with corresponding normal tissue. This is in agreement with a study published while this work was in progress (22). However, in this study, overexpression of EZH2 was associated with poor patient survival in a subgroup of Dukes B cancers. In contrast, in 72 CRC specimens unselected for stage, we do not see a significant difference in survival associated with EZH2 overexpression. This would be supported by our findings of unaltered cell growth and viability on EZH2 knockdown. The relatively low number of cases as well as better than average 5 year survival of the patients in our population (∼80%) limits the statistical power of our survival data. However, we do see a significant association with loss of SMAD4 in the same tumours (R.J.Jacobs, L.L.Kodach, J.C.H.Hardwick, unpublished results). In contrast to a previous study, we see no association between EZH2 and RUNX3 expression and MSI status. Although this could also be attributed to our small sample size, we have shown a highly significant association between BMPR2 expression and MSI in the same tumours (17).
Although overall EZH2 mRNA is upregulated in CRC versus corresponding normal tissue, RUNX3 expression is reduced, raising the possibility of a causative relationship. However, we do not find a significant inverse correlation between expression of EZH2 and RUNX3 on an individual specimen basis, but we do find a significant association between RUNX3 downregulation and RUNX3 methylation militating against a primary role for EZH2 in RUNX3 silencing. Our results are supported by data of Subramaniam et al. (23) showing that inactivation of RUNX3 was significantly associated with RUNX3 promoter methylation in majority of the colorectal polyps (75%, P = 0.022). In the remaining few cases of RUNX3 downregulation without promoter, hypermethylation was associated with mislocalization of the inactive protein. Interestingly, in our study, 16 of the 27 patients with downregulated RUNX3 expression have both methylation of the RUNX3 promoter region and upregulation of EZH2 mRNA, which might suggest interplay between EZH2 and methylation in downregulation of RUNX3 expression.
Similarly, at protein level, we do not see a significant correlation between EZH2 and RUNX3 expression on an individual patient basis. In our TMA, EZH2 overexpression is frequently (46%) associated with loss of RUNX3 staining, but almost as frequently overexpression of EZH2 coincides with positive RUNX3 expression (30%), suggesting that EZH2 overexpression does not necessarily cause RUNX3 gene silencing and implying that other mechanisms are involved. Interestingly, we see almost no nuclear staining for RUNX3 in normal tissue, whereas in some cancer specimens, we notice very strong nuclear positivity. For a transcription factor, where activity should correspond with nuclear localization, and a putative tumour suppressor, these results are the opposite of what would be expected. On the other hand, previously published work has shown strong nuclear positivity in normal tissue using an antibody they generated themselves directed against amino acids 191–300 of RUNX3 (23). This could be explained by the use of different antibodies, although we have used a commercially available antibody directed against the same epitope. Results similar to ours have been obtained in early-onset gastric carcinomas, where RUNX3 protein expression was not detected in normal tissue, but nuclear RUNX3 staining was observed in some tumour cells (24). Although others have reported an association between poorer survival and loss of nuclear RUNX3 expression, this was only true for a small subgroup (9%) where RUNX3 expression is retained but restricted to the cytoplasm. This sort of change is probably to be due to mutations in RUNX3 rather than transcriptional silencing due to methylation. No survival difference was seen between cancers with and without general cellular RUNX3 expression, a distinction that correlates well with methylation (25).
In contrast to gastric cancer (6), we do not see re-expression of the RUNX3 gene after knockdown of EZH2 in a panel of CRC cell lines, whereas we can restore RUNX3 expression using demethylating agents. Our data are more consistent with other reports, which show that EZH2 knockdown causes loss of HsK27me3 at the MLH1 promoter but not gene re-expression or DNA methylation changes (5). Our data suggest that depletion of EZH2 is insufficient to result in RUNX3 re-expression in the face of the extensive promoter hypermethylation of the RUNX3 promoter found in CRC cell lines and that CpG island methylation is the principal factor involved in maintaining the epigenetic silencing of genes in cancer cells.
Although knockdown of EZH2 does not lead to the re-expression of densely methylated and silenced genes such as RUNX3, we hypothesized that it may nevertheless play a role in marking chromatin for re-methylation. This is a clinically relevant issue as it is known that re-silencing of tumour suppressor genes occurs after the removal of demethylating agents (26) such as those now in various stages of clinical testing (27). This is thought to be a major obstacle in successful chemotherapy using these agents. We investigated this by first demethylating cells and then knocking down EZH2 while removing the demethylating agents. We show that knocking down EZH2 interferes with the re-silencing of RUNX3 after the removal of demethylating agents, in line with reports suggesting that EZH2 is important in the regulation of the de novo DNA methylation process.
In conclusion, we find no evidence from expression patterns in patient material for a role for EZH2 in primarily controlling RUNX3 expression or influencing prognosis. EZH2 knockdown is not sufficient to restore RUNX3 expression, suggesting that even if EZH2 is a regulator of DNA methylation and gene silencing in cancer, it plays a limited role after the establishment of dense promoter methylation. However, EZH2 is involved in promoter re-methylation and the re-silencing of RUNX3 after the removal of demethylating agents. Strategies aimed at inhibition of EZH2 may have additive effects when combined with demethylating agents.
Supplementary material
Supplemental Figures 1–3 Tables 1–3 can be found at http://carcin.oxfordjournals.org/.
Funding
NW0 (ZonMw VENI 916.76.087) to J.C.H.H; KWF (KWF 2007–3725) to L.L.K and R.J.J) Dutch Cancer Society to L.L.K, R.J.J.
Supplementary Material
Acknowledgments
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- CRC
colorectal cancer
- EZH2
enhancer of zeste homolog 2
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- mRNA
messenger RNA
- MSI
microsatellite unstable
- MSS
microsatellite stable
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PCR
polymerase chain reaction
- PRC2
polycomb-repressive complex 2
- RT–PCR
reverse transcription–polymerase chain reaction
- siRNA
small interfering RNA
- TMA
tissue microarray
References
- 1.Varambally S, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 2.Kleer CG, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl Acad. Sci. USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vire E, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 4.Schlesinger Y, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 5.McGarvey KM, et al. DNA methylation and complete transcriptional silencing of cancer genes persist after depletion of EZH2. Cancer Res. 2007;67:5097–5102. doi: 10.1158/0008-5472.CAN-06-2029. [DOI] [PubMed] [Google Scholar]
- 6.Fujii S, et al. Enhancer of zeste homologue 2 (EZH2) down-regulates RUNX3 by increasing histone H3 methylation. J. Biol. Chem. 2008;283:17324–17332. doi: 10.1074/jbc.M800224200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okuda T, et al. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 8.Levanon D, et al. Spatial and temporal expression pattern of Runx3 (Aml2) and Runx1 (Aml1) indicates non-redundant functions during mouse embryogenesis. Mech. Dev. 2001;109:413–417. doi: 10.1016/s0925-4773(01)00537-8. [DOI] [PubMed] [Google Scholar]
- 9.Woolf E, et al. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc. Natl Acad. Sci. USA. 2003;100:7731–7736. doi: 10.1073/pnas.1232420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li QL, et al. Transcriptional silencing of the RUNX3 gene by CpG hypermethylation is associated with lung cancer. Biochem. Biophys. Res. Commun. 2004;314:223–228. doi: 10.1016/j.bbrc.2003.12.079. [DOI] [PubMed] [Google Scholar]
- 11.Hanai J, et al. Interaction and functional cooperation of PEBP2/CBF with Smads. Synergistic induction of the immunoglobulin germline Calpha promoter. J. Biol. Chem. 1999;274:31577–31582. doi: 10.1074/jbc.274.44.31577. [DOI] [PubMed] [Google Scholar]
- 12.Ragnarsson G, et al. Loss of heterozygosity at chromosome 1p in different solid human tumours: association with survival. Br. J. Cancer. 1999;79:1468–1474. doi: 10.1038/sj.bjc.6690234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka K, et al. Suppression of tumourigenicity in human colon carcinoma cells by introduction of normal chromosome 1p36 region. Oncogene. 1993;8:2253–2258. [PubMed] [Google Scholar]
- 14.Li QL, et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109:113–124. doi: 10.1016/s0092-8674(02)00690-6. [DOI] [PubMed] [Google Scholar]
- 15.Goel A, et al. Epigenetic inactivation of RUNX3 in microsatellite unstable sporadic colon cancers. Int. J. Cancer. 2004;112:754–759. doi: 10.1002/ijc.20472. [DOI] [PubMed] [Google Scholar]
- 16.Ito K, et al. RUNX3 attenuates beta-catenin/T cell factors in intestinal tumorigenesis. Cancer Cell. 2008;14:226–237. doi: 10.1016/j.ccr.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 17.Kodach LL, et al. The bone morphogenetic protein pathway is inactivated in the majority of sporadic colorectal cancers. Gastroenterology. 2008;134:1332–1341. doi: 10.1053/j.gastro.2008.02.059. [DOI] [PubMed] [Google Scholar]
- 18.Garrick D, et al. The role of the polycomb complex in silencing {alpha}-globin gene expression in nonerythroid cells. Blood. 2008;112:3889–3899. doi: 10.1182/blood-2008-06-161901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee TI, et al. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di CL, et al. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science. 2002;295:1079–1082. doi: 10.1126/science.1065173. [DOI] [PubMed] [Google Scholar]
- 21.Kirmizis A, et al. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 2004;18:1592–1605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fluge O, et al. Expression of EZH2 and Ki-67 in colorectal cancer and associations with treatment response and prognosis. Br. J. Cancer. 2009;101:1282–1289. doi: 10.1038/sj.bjc.6605333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramaniam MM, et al. RUNX3 Inactivation in colorectal polyps arising through different pathways of colonic carcinogenesis. Am. J. Gastroenterol. 2009;104:426–436. doi: 10.1038/ajg.2008.141. [DOI] [PubMed] [Google Scholar]
- 24.Carvalho R, et al. Exclusion of RUNX3 as a tumour-suppressor gene in early-onset gastric carcinomas. Oncogene. 2005;24:8252–8258. doi: 10.1038/sj.onc.1208963. [DOI] [PubMed] [Google Scholar]
- 25.Soong R, et al. The expression of RUNX3 in colorectal cancer is associated with disease stage and patient outcome. Br. J. Cancer. 2009;100:676–679. doi: 10.1038/sj.bjc.6604899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Egger G, et al. Inhibition of histone deacetylation does not block resilencing of p16 after 5-Aza-2'-deoxycytidine treatment. Cancer Res. 2007;67:346–353. doi: 10.1158/0008-5472.CAN-06-2845. [DOI] [PubMed] [Google Scholar]
- 27.Egger G, et al. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.